Abstract
The membrane-bound tetrachloroethene reductive dehalogenase (PCE-RDase) (PceA; EC 1.97.1.8), the terminal component of the respiratory chain of Dehalobacter restrictus, was purified 25-fold to apparent electrophoretic homogeneity. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a single band with an apparent molecular mass of 60 ± 1 kDa, whereas the native molecular mass was 71 ± 8 kDa according to size exclusion chromatography in the presence of the detergent octyl-β-d-glucopyranoside. The monomeric enzyme contained (per mol of the 60-kDa subunit) 1.0 ± 0.1 mol of cobalamin, 0.6 ± 0.02 mol of cobalt, 7.1 ± 0.6 mol of iron, and 5.8 ± 0.5 mol of acid-labile sulfur. Purified PceA catalyzed the reductive dechlorination of tetrachloroethene and trichloroethene to cis-1,2-dichloroethene with a specific activity of 250 ± 12 nkat/mg of protein. In addition, several chloroethanes and tetrachloromethane caused methyl viologen oxidation in the presence of PceA. The Km values for tetrachloroethene, trichloroethene, and methyl viologen were 20.4 ± 3.2, 23.7 ± 5.2, and 47 ± 10 μM, respectively. The PceA exhibited the highest activity at pH 8.1 and was oxygen sensitive, with a half-life of activity of 280 min upon exposure to air. Based on the almost identical N-terminal amino acid sequences of PceA of Dehalobacter restrictus, Desulfitobacterium hafniense strain TCE1 (formerly Desulfitobacterium frappieri strain TCE1), and Desulfitobacterium hafniense strain PCE-S (formerly Desulfitobacterium frappieri strain PCE-S), the pceA genes of the first two organisms were cloned and sequenced. Together with the pceA genes of Desulfitobacterium hafniense strains PCE-S and Y51, the pceA genes of Desulfitobacterium hafniense strain TCE1 and Dehalobacter restrictus form a coherent group of reductive dehalogenases with almost 100% sequence identity. Also, the pceB genes, which may code for a membrane anchor protein of PceA, and the intergenic regions of Dehalobacter restrictus and the three desulfitobacteria had identical sequences. Whereas the cprB (chlorophenol reductive dehalogenase) genes of chlorophenol-dehalorespiring bacteria are always located upstream of cprA, all pceB genes known so far are located downstream of pceA. The possible consequences of this feature for the annotation of putative reductive dehalogenase genes are discussed, as are the sequence around the iron-sulfur cluster binding motifs and the type of iron-sulfur clusters of the reductive dehalogenases of Dehalobacter restrictus and Desulfitobacterium dehalogenans identified by electron paramagnetic resonance spectroscopy.
Tetrachloroethene (PCE), a frequently detected groundwater contaminant, is used by different bacteria as a terminal electron acceptor in a process called dehalorespiration (10, 14, 16, 20, 21, 32, 35, 40, 46). Six such isolates belong phylogenetically to the subphylum Firmicutes of the gram-positive bacteria, whereas other isolates are affiliated with phylogenetic groups such as the δ and ɛ subclasses of the Proteobacteria and the green nonsulfur bacteria. Hence, the physiological ability to use PCE as an electron acceptor apparently evolved in parallel in several phyla of eubacteria. Eight isolates use dihydrogen and/or formate as an electron donor, which implies that there is energy conservation via a chemiosmotic mechanism. The formation of a proton motive force upon dihydrogen oxidation coupled to PCE reduction was unequivocally shown by oxidant pulse experiments performed with Dehalobacter restrictus (33). Similar to the fumarate respiration system of Wolinella succinogenes (15), the proton electrochemical potential in Dehalobacter restrictus was shown to be generated by the proton-producing hydrogenase facing the periplasm and transferring the electrons to the cytoplasm-oriented PCE reductive dehalogenase (PCE-RDase) via menaquinone. Desulfitobacterium hafniense strain PCE-S (formerly Desulfitobacterium frappieri strain PCE-S) has a similar arrangement of the enzymes involved in the respiration chain (21), whereas the PCE-RDase of Dehalospirillum multivorans is located in the cytoplasm (23).
PCE dechlorination activity can be measured in in vitro systems with the artificial low-potential electron donor methyl viologen. A ferredoxin isolated from Dehalospirillum multivorans also serves as in vitro electron donor but only at a rather low rate (1%) compared to the rates obtained with reduced methyl viologen (23). Hence, the natural electron donor is not yet known. Photoreversible inactivation of PCE reduction in cell extracts by 1-iodopropane indicated that a cob(I)amid is involved in the catalysis of this reductive dechlorination reaction (21, 25, 29, 34, 41, 42). Cob(I)alamin in its free form has previously been shown to reductively dechlorinate PCE and other chlorinated ethenes in homogeneous aqueous solutions (7, 11). It has been suggested that the first step of PCE reduction by cob(I)alamin is a dissociative one-electron transfer that yields a vinyl radical and cob(II)alamin (11). Several chloroethene RDases have been purified and characterized (18, 22, 27, 29, 34, 41, 42). The majority of these enzymes have been isolated as monomeric enzymes with molecular masses around 50 or 60 kDa. The only exception is the PCE-RDase of Clostridium bifermentans DPH-1, which seems to be a homodimeric enzyme with a subunit molecular mass of 35 kDa (29). All chloroethene RDases characterized so far contain one cobamide, and for some of them the presence of approximately eight atoms of iron and eight acid-labile sulfur atoms per mole of enzyme has been determined, indicating that two iron-sulfur clusters are cofactors. An electron paramagnetic resonance (EPR) spectroscopic study of the PCE-RDase of Dehalobacter restrictus demonstrated that this enzyme contains two [4Fe-4S] clusters with low but different redox potentials (Em, <−480 mV) and one cobalamin with an Em(Co1+/2+) of −350 mV as cofactors (34).
The chloroethene RDase genes of monomeric enzymes with similar molecular masses characterized so far (two pceA genes and one tceA gene) have rather low levels of sequence identity (27 to 32%) (18, 27, 41). The common features are a twin-arginine signal sequence that may be involved in the translocation of the enzyme into or across the cytoplasmic membrane (4, 45), iron-sulfur cluster binding motifs, and no known binding motif for the cobamide. In addition, the pceA and tceA genes have been found to be linked with open reading frames (ORFs) designated pceB and tceB coding for a small hydrophobic protein with two (27) or three (18, 22, 41) transmembrane helices. The cotranscription of both genes, as shown for Dehalospirillum multivorans by reverse transcription-PCR (27), suggests that there is functional linkage of the two gene products. However, despite the indications on a genetic level, the roles of PceB and TceB remain to be confirmed on the protein level.
Here, we describe complete purification and biochemical characterization of the PCE-RDase of Dehalobacter restrictus and cloning and sequencing of the pceA and pceB genes of Dehalobacter restrictus and Desulfitobacterium hafniense strain TCE1 (formerly Desulfitobacterium frappieri strain TCE1). This allowed for the first time the comparison of the EPR spectroscopic data and the gene sequence of a chloroethene RDase. In addition, the sequence data for all RDases that have been biochemically characterized are compared, and the consequences of this information for annotation of putative RDase genes are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Dehalobacter restrictus DSMZ 9455T was cultivated anaerobically with PCE as the electron acceptor and H2 as the electron donor in a two-liquid-phase system in 1-liter flasks as previously described (33). Desulfitobacterium hafniense strain TCE1 (= DSMZ 12704) was cultivated under anaerobic conditions in a medium described previously (9). The culture was incubated at 30°C and 100 rpm. Escherichia coli DH5α [deoR endA1 gyrA96 hsdR17(rk− mk+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80lacZΔM15 F− (12)] was used as the host for cloning vectors. This strain was cultivated in Luria-Bertani liquid medium and on plates containing 100 μg of ampicillin per ml at 37°C.
Protein purification.
Cells (approximately 10 liters of culture per harvest event) were harvested with a continuous centrifuge (50,000 × g; 25 liters h−1; 4°C; Carl Padberg, Lahr, Germany) that was flushed with N2 during centrifugation, and they were stored in liquid nitrogen as concentrated cell suspensions (125 g [wet weight] of cells liter−1 in 25 mM Tris buffer [pH 7.8]). A yield of 300 to 400 mg (wet weight) of cells liter of culture−1 was obtained. All subsequent steps were carried out at 4°C with exclusion of oxygen inside a glove box filled with N2-H2 (96:4, vol/vol). Crude extract was produced by the procedure described previously (33) by ultrasonication of a thawed concentrated cell suspension. The resulting crude extract was stirred (15 min, 20°C) in the presence of 0.5 M KCl and 0.1% (wt/vol) octyl-β-d-glucopyranoside (OGP) and subsequently fractionated by centrifugation (200,000 × g, 1 h, 4°C). The supernatant was quantitatively decanted, and the pellet was resuspended in 25 mM Tris buffer (pH 8) and stirred (30 min, 20°C) after addition of Triton X-100 to a final concentration of 1.2% (wt/vol) to obtain a protein/detergent ratio of 1:1 (wt/wt). After centrifugation (200,000 × g, 1 h, 4°C) the membrane extract (supernatant) was loaded on a Mono Q column (0.5 by 5 cm) connected to a Jasco high-performance liquid chromatography (HPLC) system (OmniLab AG, Mettmenstetten, Switzerland). The PCE-RDase was eluted (flow rate, 1 ml min−1) with 30 ml of a linear NaCl gradient (0 to 0.35 M) in 25 mM Tris buffer (pH 8)-0.1% Triton X-100, and the elution maximum was at 0.11 M NaCl. The fractions containing PCE-RDase activity were pooled and concentrated by ultrafiltration (PM-30 membrane; Amicon, Witten, Germany). The PCE-RDase activity was determined in microtiter plates containing reaction buffer with reduced methyl viologen and PCE in each well. The reaction was started by adding a droplet of a fraction and was considered positive when comparably fast decolorization of the reduced methyl viologen occurred. In the final step, aliquots of the concentrated PCE-RDase pool were loaded on a Superose 6 10/30 size exclusion chromatography column (Pharmacia-LKB, Freiburg, Germany) equilibrated with 25 mM Tris buffer (pH 8)-0.1% Triton X-100-150 mM NaCl. The protein was eluted at a flow rate of 0.4 ml min−1, and 0.5-ml fractions were collected. PCE-RDase fractions with identical elution volumes from several runs were pooled, concentrated by ultrafiltration, and stored in liquid nitrogen.
Analytical methods.
The purity of PCE-RDase was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in 12% polyacrylamide separation gels, followed by subsequent silver staining (30). Six proteins from the high-range Combithek of Boehringer-Mannheim were used as molecular mass markers (Boehringer-Mannheim, Rotkreuz, Switzerland). The molecular mass of the native protein was determined in the presence of Triton X-100 (0.1%, wt/vol) or OGP (0.1%, wt/vol) and 150 mM NaCl by size exclusion chromatography as described above for the purification procedure. The standards used for the latter procedure included amylase (206 kDa), alcohol dehydrogenase (146.8 kDa), albumin (66.2 kDa), carboanhydrase (29 kDa), and cytochrome c (12.4 kDa).
The PCE-reducing activity was measured by a photometric assay with dithionite-reduced methyl viologen as the artificial electron donor as previously described (33). Oxidation of concentrations of reduced methyl viologen of >0.2 mM were monitored at 700 nm (ɛ = 2.31 mM−1 cm−1) (26). Kinetic data obtained with different substrate concentrations were modeled by using Michaelis-Menten kinetics without inhibition and with uncompetitive inhibition when the enzyme substrate complex was catalytically inactivated.
Inactivation of PCE reduction by 1-iodopropane (5 μM) was tested with a photometric assay in the dark. Reversion of 1-iodopropane inactivation was assessed by short exposure of the cuvette to a 20-W halogen lamp. The protein content was determined with bicinchoninic acid by using 4 to 20 μg of bovine serum albumin as the standard (38).
Cobamide extraction from the purified enzyme and reversed-phase HPLC analysis were done by an equivalent procedure as previously described (39). Identically treated cyanocobalamin (vitamin B12) was the standard. UV/visible spectra were recorded with a Hitachi U-2000 spectrophotometer (Hitachi, Tokyo, Japan) by using 1-cm quartz cuvettes.
The total iron content was determined by atomic absorption spectroscopy (model 2100; Perkin-Elmer, Überlingen, Germany) with Fe(NO3)3 · 9H2O in 0.5 M HNO3; 1.000 ± 0.002 g of Fe (Merck, Darmstadt, Germany) liter−1 was used as the standard. The cobalt in PCE-RDase samples was determined by inductively coupled plasma-mass spectrometry (ELAN 500; Perkin-Elmer). Acid-labile sulfide was determined as described by Rabinowitz (31), with the modifications of Beinert (3). Commercially available [2Fe-2S] ferredoxin from the red marine alga Porphyra umbilicalis (2) was used as the reference protein. The N-terminal sequence of PCE-RDase purified in the presence of OGP was determined by using Edman degradation.
Dechlorination products were analyzed by gas chromatography with a GC Varian Star 3400CX equipped with a GS-GasPro column (30 m by 0.32 mm; J&W Scientific, MSP Friedly & Co, Koeniz, Switzerland) coupled to a flame ionization detector. The carrier gas utilized was nitrogen at a flow rate of 1.3 ml/min. The initial temperature was 45°C; the column was kept at 45°C for 3 min, and then the temperature was raised to 75°C at a rate of 15°C/min and then to 200°C at a rate of 25°C/min and finally kept at 200°C for 5 min. Medium containing the chloride ions released during dechlorination was analyzed by titration with a Chlor-o-Counter (FLOHR Instrumenten, Nieuwegein, The Netherlands).
DNA isolation.
Cells were harvested by centrifugation at 5,000 rpm for 10 min, resuspended in a lysis buffer (pH 8) containing 20 mM Tris (pH 8), 10 mM NaCl, 1 mM EDTA, 100 μg of proteinase K per ml, and 0.5% SDS, and incubated for 6 h at 50°C. One volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added. After incubation for 10 min at room temperature and centrifugation at 6,000 × g and 10°C for 20 min, the aqueous phase was transferred into a fresh tube and again extracted with 1 volume of phenol-chloroform-isoamyl alcohol. The sample was mixed and incubated on ice for 5 min and finally centrifuged at 18,000 × g and 4°C for 15 min. The aqueous phase was again transferred, and the remaining phenol was extracted with 1 volume of diethyl ether. Finally, the DNA was precipitated with ethanol, washed, and dried in a vacuum centrifuge. Plasmid DNA isolation was performed by using a QIAprep spin miniprep kit (Qiagen AG, Basel, Switzerland).
DNA amplification.
The oligonucleotides (Microsynth GmbH, Balgach, Switzerland) used in this study were DR3f (5′-GA[C/T] ATI GTI GCI CCI ATI AC-3′), DR4r (5′-CC[A/G] AA[A/G] TCI ATI GG[C/T] TT[A/G] TCI GG-3′), PCE1f (5′-ATG CAA TTA TTA TTA AGG AGG AAG-3′), PCE2r (5′-CTA AGC AGA AAT AGT ATC CGA ACT-3′), T7 promoter primer (5′-TAA TAC GAC TCA CTA TAG GG-3′), and SP6 promoter primer (5′-ATT TAG GTG ACA CTA TAG-3′). In order to avoid too much degeneration of primers DR3f and DR4r, inosine (I) was inserted for three- to fourfold-degenerated bases.
Degenerate PCR was performed under the following conditions. A 50-μl PCR mixture contained 5 μl of 10× Taq DNA polymerase buffer, 2.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 5 μM degenerate primer DR3f, 5 μM degenerate DR4r, and 2.5 U of Taq DNA polymerase (Promega, Catalys AG, Wallisellen, Switzerland). Fifty nanograms of Dehalobacter restrictus genomic DNA was used as the template. The DNA was amplified with a T3 thermocycler (Biometra, Biolabo Scientific Instruments, ChÂtel-St-Denis, Switzerland) with the following program: 3 min of preheating at 94°C, 36 cycles of 30 s of denaturation at 94°C, 1 min of primer annealing at 50°C, and 2 min of elongation at 72°C. A final extension step of 10 min at 72°C was included.
Specific PCR with primers PCE1f and PCE2r was performed under the following conditions. A 50-μl PCR mixture contained 5 μl of 10× Pfu DNA polymerase buffer, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 1 μM, and 5 U of proofreading Pfu DNA polymerase (Promega, Catalys AG). The PCR was performed by using 30 cycles of 30 s of denaturation at 94°C, 1 min of primer annealing at 56°C, and 2 min of elongation at 72°C. A final 10-min extension step at 72°C was included.
PCR product purification, cloning, and selection of clones.
PCR products were analyzed by agarose gel electrophoresis and were purified by using a Minelute PCR purification kit (Qiagen AG), and they were eluted in 10 μl (final volume). Before ligation 7 μl of purified PCR products amplified with Pfu DNA polymerase was incubated for 30 min at 70°C with 1 μl of a solution containing 10× Taq DNA polymerase buffer, 0.2 mM dATP, and 5 U of Taq DNA polymerase.
For cloning, PCR products were ligated into the pGEM-T Easy vector (Promega, Catalys AG) according to the manufacturer's instructions. Ligated products were transformed into CaCl2-competent E. coli DH5α cells by using the standard heat shock protocol. Transformed cells were incubated for 1 h at 37°C on a rotary shaker at 200 rpm before they were plated onto Luria-Bertani plates containing 100 μg of ampicillin per ml, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml (blue/white selection).
White E. coli colonies were resuspended in 10 μl of distilled H2O, lysed for 10 min at 95°C, and subsequently briefly centrifuged. One microliter of supernatant was used as a template in a 10-μl PCR mixture by using 1 μM T7 and 1 μM SP6 as the promoter primers. The PCR products were analyzed by agarose gel electrophoresis. Clones containing DNA fragments of the expected length were selected.
DNA sequencing and sequence analysis.
Cycle sequencing reactions were performed by using a BigDye Terminator v3.0 Ready Reaction kit (Applied Biosystems, Rotkreuz, Switzerland) according to the manufacturer's instructions. The following primers were used: T7 and SP6 promoter primers and pceAB-specific internal primers. Samples were analyzed with an ABI Prism 3100 genetic analyzer (Applied Biosystems).
Sequence alignment was performed by using a local version of T-coffee Mocca (28). Comparison with databank sequences was done with Blast (1).
Nucleotide sequence accession numbers.
The nucleotide sequences of the pceAB genes of Dehalobacter restrictus and Desulfitobacterium hafniense strain TCE1 have been deposited in the EMBL database under accession numbers AJ439607 and AJ439608, respectively. The nucleotide sequences of the pceAB genes of Desulfitobacterium hafniense strain PCE-S have been deposited in the GenBank database under accession number AY216592.
RESULTS AND DISCUSSION
Purification of PCE-RDase.
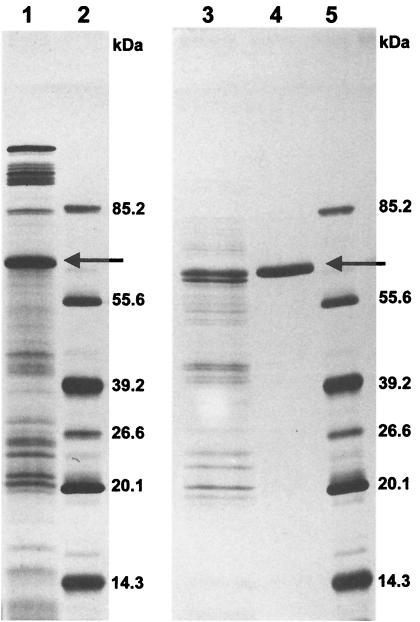
More than 95% of the PCE-RDase activity of Dehalobacter restrictus was recovered in the membrane fraction (33). The activity could not be solubilized by 0.5 M KCl or a low concentration of the detergent OGP (0.1%, wt/vol), showing that the PCE-RDase is tightly bound to the membrane fraction. Crude extract was therefore pretreated with 0.5 M KCl and 0.1% OGP before ultracentrifugation to eliminate loosely associated proteins from the membranes. The PCE-RDase activity was subsequently extracted from the membrane fraction with either 1% Triton X-100 or 1% OGP. Both detergents extracted approximately 30% of the membrane proteins that contained 91 to 95% of the PCE-RDase activity. Because precipitates formed during concentration by ultrafiltration in the presence of OGP, the detergent Triton X-100 was used for routine purification. However, all purification steps could also be carried out in the presence of OGP if special care was taken during the concentration step (keeping the ultrafiltration cell at 4°C, low gas pressure). After two subsequent chromatography steps, the PCE-RDase was purified to electrophoretic homogeneity (Fig. 1). The purification factor, 22.3-fold, indicated that more than 4% of the total cellular protein consisted of PCE-RDase (Table 1). Trichloroethylene (TCE) RDase activity always copurified with PCE-RDase activity, and no additional TCE-RDase activity was detected in other fractions after chromatography.
FIG. 1.
SDS-PAGE of the PCE-RDase of Dehalobacter restrictus. The SDS-12% polyacrylamide gel was silver stained. Lane 1, membrane fraction after extraction (10 μg of protein); lane 3, fraction after anion-exchange chromatography column (3.5 μg); lane 4, purified PCE-RDase which eluted from the size exclusion chromatography column (2 μg); lanes 2 and 5, molecular mass markers, including fructose-6-phosphate kinase (85.2 kDa), glutamate dehydrogenase (55.6 kDa), aldolase (39.2 kDa), triose phosphate isomerase (26.6 kDa), trypsin inhibitor (20.1 kDa), and lysozyme (14.3 kDa). The arrow indicates the PCE-RDase band.
TABLE 1.
Purification of PCE-RDase of Dehalobacter restrictusa
| Purification step | Sp act (U/mg of protein)b | Purification (fold) | Total activity (U) | Yield (%) |
|---|---|---|---|---|
| Crude extractc | 0.61 | 1.0 | 1,818 | 100 |
| Membrane fraction | 0.82 | 1.3 | 1,443 | 79 |
| Membrane extract | 2.64 | 4.3 | 1,322 | 73 |
| Mono Q | 9.15 | 15.0 | 1,067 | 59 |
| Superose 6 | 13.60 | 22.3 | 964 | 53 |
TCE-RDase activity always copurified with PCE-RDase activity.
One unit of PCE-RDase activity was defined as oxidation of 2 μmol of methyl viologen/μmol of Cl− formed/min at 30°C, as determined from initial rates of methyl viologen oxidation.
18.6 g (wet weight) of cells was used for purification.
The PCE-RDase of Dehalospirillum multivorans is a cytoplasmic enzyme, accounts for a little less than 1% of the cellular protein, and does not require the presence of a detergent for purification (26). The same is true for the PCE-RDases of C. bifermentans (29) and Desulfitobacterium sp. strain Y51 (41). However, there have been indications that the PCE-RDase of the latter bacterium is localized in the periplasmic fraction. The PCE-RDases of Desulfitobacterium sp. strains PCE-S (22), PCE1 (42), and TCE1 (42) are membrane associated (30 to 74% of the activity is in the membrane fraction), account for 0.4 to 1.0% of the cellular protein, and have been purified in the presence of 0.1% Triton X-100. The membrane-bound PCE-RDase of Dehalococcoides ethenogenes 195 was extracted with 0.1% Triton X-100 and accounted for 1.3% of the membrane proteins (19). A remarkable difference compared to the PCE-RDases of other organisms whose TCE dechlorination activity copurified with the PCE dechlorination activity is the fact that the PCE-RDase activity of Dehalococcoides ethenogenes 195 did not include TCE reduction activity (Table 2). The majority of the PCE-RDases dechlorinate PCE and TCE at similar rates. Only the PCE-RDase of Desulfitobacterium sp. strains PCE1 and Y51 showed significantly different rates. Whereas the PCE-RDase of the former bacterium dechlorinated PCE 10 times faster than it dechlorinated TCE (42), the opposite was observed for the PCE-RDase of strain Y51 (41). In Dehalococcoides ethenogenes, workers have identified a TCE-RDase that completely dechlorinates TCE via cis- and trans-1,2-dichloroethenes (DCEs) and vinyl chloride to ethene (19). This RDase was also found exclusively in the membrane fraction and accounted for up to 4% of the total membrane protein.
TABLE 2.
Biochemical characteristics of purified chloroethene RDasesa
| Enzyme | Organism | Molecular
mass
(kDa)
|
Localizationb | Apparent
Km
(μM)
|
Sp act
(nkat/mg)
|
Cofactor(s) | |||
|---|---|---|---|---|---|---|---|---|---|
| SDS-PAGE | Gel filtration | PCE | TCE | PCE | TCE | ||||
| PCE-RDase | Dehalococcoides ethenogenes | 51 | NDc | m | ND | 342 | Corrinoid, Fe/Sd | ||
| Desulfitobacterium sp. strain PCE-1 | 48 | ND | m | ND | 92 | Corrinoidd | |||
| PCE/TCE-RDase | Dehalospirillum multivorans | 57 | 58 | s | 200 | 240 | 2,640 | ∼2,200 | 1 Corrinoid, 8Fe/8Sf |
| Dehalobacter restrictus | 60 | 164/71g | m | 20 | 24 | 250 | 338 | 1 Corrinoid, 2[4Fe/4S]h | |
| Desulfitobacterium hafniense strain PCE-S | 65 | 198 | m | 10 | 4 | 650 | 690 | 1 Corrinoid, 8Fe/8Sf | |
| Desulfitobacterium hafniense strain TCE1 | 59 | ND | m | ND | ND | 190 | 157 | Corrinoidd | |
| Desulfitobacterium sp. strain Y51 | 58 | 67 | s | 106 | 535 | 2.7 | 13.5 | Corrinoidd | |
| Clostridium bifermentans strain DPH-1 | 35 | 70 | s | 12 | 1.2 | Corrinoidd | |||
| TCE-RDase | Dehalococcoides ethenogenes | 61 | ND | m | ND | 200 | Corrinoid, Fe/Sd | ||
| pH opti- mum | Temp optimum (°C) | Oxygen sensitivity half-life (min) | Inhibitors | Other substrates | Compounds not dechlorinated | N-terminal amino acid sequence | Reference(s) | ||
| NRe | NR | NR | 1-Iodoethane, sulfite, dithionite, cyanide, azide | NR | TCE, 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE, VC | NR | 19 | ||
| NR | NR | NR | 1-Iodopropane, N2O, sulfite | TCE | cis-1,2-DCE | GQESESAIVXFAVQXV | 42 | ||
| 8 | 42 | 120 | 1-Iodopropane, sulfite, cyanide, nitrite, N2O, EDTA, chlorinated methanes | trans-1,3-DCP, 2,3-DCP, 1,1,3-TCP | 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE, HCA, 1,1,1-TCA, 3-CB, 3,4-DCB, 4-CP | GVPGANAAEKEKNAA EIRQQFAMTAGS | 25-27 | ||
| 8.1 | NR | 280 | 1-Iodopropane | CT, HCA, 1,1,2,2-TeCA, 1,1,1,2-TeCA, 1,1,2-TCA, (1,1,1-TCA) | 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE | ADIVAPITETSEFPYKV DAK | 33, 34; this study | ||
| 7.2 | 50 | 50 | 1-Iodopropane, cyanide, azide, sulfite, EDTA | NR | CT, 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE | ADIVAPITETSEFPYKV DAK | 21, 22 | ||
| NR | NR | NR | NR | NR | cis-1,2-DCE, chlorophenols | ADIVAPITEXTEFPYPV | 42 | ||
| 7.2 | 37 | 330 | 1-Iodopropane, sulfite | HCA, PCA, 1,1,2,2-TeCA, 1,1,1,2-TeCA | 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE, 1,1,1-TCA, 1,1,2-TCA | ADIVAPITETSEFPYKV DAK | 41 | ||
| 7.5 | 35 | 1,200 | 1-Iodopropane | 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE, 1,1,1-TCA, 1,1,2-TCA | VC | AEVYNKDGNKLDLYGK VDGLHYFSNDT | 29 | ||
| NR | NR | NR | 1-Iodopropane, sulfite, dithionite, cyanide, azide, Cu2+, Zn2+ | 1,1-DCE, cis-1,2-DCE, trans-1,2-DCE, (VC), (VB), 1,2-DCA, 1,2-DBA | PCE, 1,1,2,2-TeCA | KDVDDLLSAGKALEGD HANKVNNHPWW | 18, 19 | ||
Abbreviations for compounds: VC, vinyl chloride; DCP, dichloropropene; TCP, trichloropropene; CT, carbon tetrachloride; HCA, hexachloroethane; PCA, pentachloroethane; TeCA, tetrachloroethane; TCA, trichloroethane; DCA, dichloroethane; VB, vinyl bromide; DBA, dibromoethane; CB, chlorobenzoate; DCB, dichlorobenzoate; CP, chlorophenol.
m, membrane fraction; s, soluble fraction.
ND, not determined.
Determined by photo-reversible inhibition of the reduced enzyme by iodo-alkanes.
NR, not reported.
The corrinoid was determined by photo-reversible inhibition of the reduced enzyme by iodo-alkanes and was quantified by analysis of cobalt content and by extraction of the corrinoid from purified enzyme, followed by spectroscopic analysis. The 8Fe/8S was quantified by atomic absorption spectroscopy and by standard procedures for acid-labile sulfide.
The molecular mass depended on the detergent used; it was 164 kDa with Triton X-100 (micelle mass, 90 kDa), and it was 71 kDa with OGP (micelle mass, 8 kDa).
The corrinoid was determined by photo-reversible inhibition of the reduced enzyme by iodo-alkanes and by EPR spectroscopy and was quantified by analysis of cobalt content and by extraction of corrinoid from purified enzyme, followed by spectroscopic analysis. The presence of two [4Fe/4S] clusters was determined by EPR spectroscopy, and the iron and sulfur contents were quantified by atomic absorption spectroscopy and by standard procedures for acid-labile sulfide.
Molecular properties of PCE-RDase.
SDS-PAGE revealed a single protein band at an apparent molecular mass of 60 ± 1 kDa (Fig. 1). Size exclusion chromatography in the presence of Triton X-100 indicated that the apparent molecular mass of the native enzyme was 164 ± 17 kDa, whereas in the presence of OGP the apparent molecular mass of the native enzyme was 71 ± 8 kDa (data not shown). The difference in the native molecular masses with two different detergents indicated that the PCE-RDase in its native form is a monomeric enzyme and that the high apparent molecular mass in the presence of Triton X-100 is the result of interactions with the detergent micelles. Triton X-100 has a critical micelle concentration of 0.02% and an estimated micelle mass of up to 90 kDa, whereas OGP forms micelles at a critical micelle concentration of 0.5% with a micelle mass of only 8 kDa (13). The addition of the molecular mass of the detergent micelles to the molecular mass of the protein band determined after SDS-PAGE explains the apparent molecular mass of the native PCE-RDase determined by size exclusion chromatography. A similar effect was found for the PCE-RDase of Desulfitobacterium hafniense strain PCE-S (22). In this case, the high molecular mass of the native enzyme (200 kDa) determined by size exclusion chromatography in the presence of Triton X-100 was explained by trimer formation that occurred as an artifact during protein purification.
The iron content of the PCE-RDase was 7.1 ± 0.6 mol of iron/mol of PCE-RDase, and the acid-labile sulfur content was 5.8 ± 0.5 mol of sulfur/mol of enzyme. This is in good agreement with the values for the two [4Fe-4S] clusters that were shown by EPR spectroscopy to be present in this enzyme (34). Similar amounts of iron and acid-labile sulfur were determined for the PCE-RDases of Dehalospirillum multivorans and Desulfitobacterium hafniense strain PCE-S, indicating that these enzymes also contain two iron-sulfur clusters (Table 2) (22, 26). No data are available for other chloroethene RDases. The redox potentials of the two [4Fe-4S] clusters of the PCE-RDase of Dehalobacter restrictus have been determined by EPR spectroelectrochemical titrations to be as low as approximately −480 mV, indicating that these clusters function as electron transfer devices rather than for storage of reducing equivalents (34). This in contrast to the EPR analysis of the chlorophenol RDase of Desulfitobacterium dehalogenans, in which a [4Fe-4S] cluster with a low redox potential (−440 mV) and a [3Fe-4S] cluster with a high redox potential (70 mV) have been identified (43).
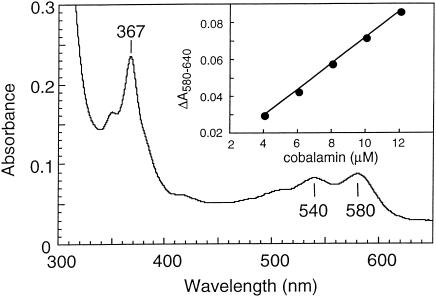
The cobalt content determined by ICP-MS was 0.59 ± 0.02 mol of cobalt/mol of enzyme. The corrinoid extracted from the enzyme by cyanolysis had the same spectroscopical features as cobalamin (Fig. 2) and the same retention time in a reversed-phase HPLC. Cultivation of Dehalobacter restrictus depends on the presence of cobalamin in the medium (14), and growth on cobinamide-containing medium ceased after a few transfers (data not shown). All these results indicated that the corrinoid present in PCE-RDase is a cobalamin, which is in contrast to the recent findings obtained with Dehalospirillum multivorans; for the latter organism it has been shown that the corrinoid isolated from the PCE-RDase had different catalytic dechlorination properties than commercially available cobalamin (24). The cobalamin content determined spectrophotometrically by measuring the difference between A580 and A640 was 0.97 ± 0.07 mol of cobalamin/mol of enzyme (Fig. 2). The same corrinoid content has been found for PCE-RDases of Dehalospirillum multivorans and Desulfitobacterium hafniense strain PCE-S (22, 26). An unusually high redox midpoint potential for the Co(I/II) (−350 mV) has been reported previously for the PCE-RDase of Dehalobacter restrictus, and the cob(II)alamin was present in the base-off form in the isolated enzyme (34). The significance of these features for the reaction mechanism remains to be elucidated.
FIG. 2.
UV/visible electronic absorption spectrum of the dicyanocobamide extracted by cyanolysis from purified PCE-RDase of Dehalobacter restrictus and purified by HPLC. (Inset) Standard curve obtained with purchased and analogically treated cobalamin, which allowed estimation of the amount of corrinoid per mole of PCE-RDase.
Catalytic properties.
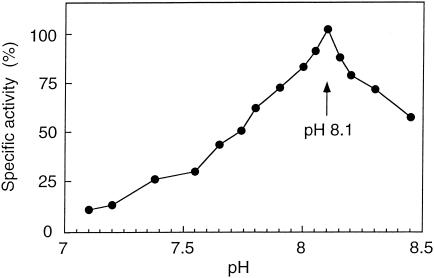
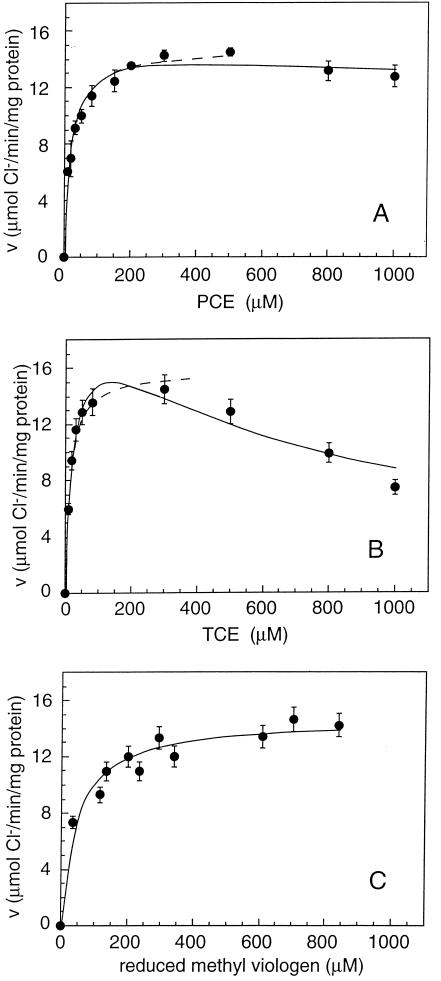
The purified PCE-RDase exhibited a pH optimum for activity of pH 8.1 (Fig. 3). PCE and TCE were reduced at maximal specific activities of 250 ± 12 and 338 ± 30 nkat/mg of protein, respectively, if inhibition constants for noncompetitive inhibition by the substrates themselves of 8.3 and 0.76 mM, respectively, were used in the calculations (Fig. 4). Fitting of the data by considering competitive or noncompetitive inhibition did not result in a reasonable description of the data. Inhibition patterns similar to that described above were also observed for Desulfitobacterium hafniense strain PCE-S and Dehalospirillum multivorans; the latter, however, had much higher inhibition constants for PCE and TCE (18 and 39 mM, respectively) (22, 26). Since the inhibition constants of all three organisms are quite high and sometimes even greater than the solubility in aqueous solution, it is possible that the inhibition patterns observed do not have any physiological significance. For the PCE-RDase of Dehalobacter restrictus, the apparent half-velocity constants (Km) for PCE, TCE, and reduced methyl viologen were 20.4 ± 3.2, 23.7 ± 5.2, and 47 ± 10 μM, respectively. The PCE-RDase of Dehalobacter restrictus reductively dechlorinated PCE and TCE with rates that were similar to those of the enzymes of Desulfitobacterium hafniense strain TCE1 (42) and Dehalococcoides ethenogenes (19) but were 3 and 10 times lower than those of the PCE-RDases of Desulfitobacterium hafniense strain PCE-S and Dehalospirillum multivorans, respectively (22, 26) and 100 to 200 times higher than those of the PCE-RDases of Desulfitobacterium sp. strain Y51 (41) and C. bifermentans DPH-1 (29) (Table 2). The Km values were on the same order of magnitude as those of Desulfitobacterium hafniense strain PCE-S and were about 10 times lower than those of Dehalospirillum multivorans and Desulfitobacterium sp. strain Y51 (22, 26, 41).
FIG. 3.
pH dependence of PCE-RDase activity. The pH meter was calibrated at 30°C, and the data points are means of three independent measurements; 100% activity was defined as 250 ± 12 nkat/mg of protein.
FIG. 4.
PCE-RDase activity (v) as a function of the concentration of PCE (A), TCE (B), and reduced methyl viologen (C). In each case the solid line is the best fit obtained by using the model with inhibition constants for noncompetitive inhibition by PCE and TCE of 8.3 and 0.76 mM, respectively. The dashed lines are fits according to Michaelis-Menten kinetics without inhibition.
The PCE-RDase of Dehalobacter restrictus had quite a broad substrate spectrum (Table 3). Besides PCE and TCE, trichlorofluoroethene, tetrachloromethane, hexachloroethane, tetrachloroethane, trichloroethane, and 1,1,1-trichloro-2,2,2-trifluoroethane caused PCE-RDase-dependent oxidation of reduced methyl viologen. DCEs were not reduced at all. Whether the chlorinated compounds that were dechlorinated by the PCE-RDase could also be utilized as terminal electron acceptors by Dehalobacter restrictus is not known. With the exception of the PCE-RDase of C. bifermentans DPH-1 (29), the PCE-RDases did not reduce DCEs (Table 2) (22, 26, 41, 42). The TCE-RDase of Dehalococcoides ethenogenes, an enzyme that does not dechlorinate PCE, reduces all DCEs (19). Interestingly, tetrachloromethane caused methyl viologen oxidation in the presence of PCE-RDase of Dehalobacter restrictus but was not reduced by the same enzyme of Dehalospirillum multivorans (26). Tetrachloromethane even inhibited PCE reduction by the enzyme of the latter organism.
TABLE 3.
Substrate spectrum of the PCE-RDase of Dehalobacter restrictus
| Compounda | Concn (mM) | Sp act (%)b |
|---|---|---|
| Trichloroethene | 0.5 | 100 |
| Trichlorofluoroethene | 0.4 | 62 ± 8 |
| trans-1,2-DCE | 0.4 | 0 |
| cis-1,2-DCE | 0.4 | 0 |
| 1,1-DCE | 0.4 | 0 |
| Tetrachloromethane | 0.6 | 15 ± 7c |
| Hexachloroethane | 0.4 | 60 ± 11c |
| 1,1,2,2-Tetrachloroethane | 0.4 | 21 ± 1 |
| 1,1,1,2-Tetrachloroethane | 0.4 | 17 ± 6 |
| 1,1,1-Trichloro-2,2,2-trifluoroethane | 0.5 | 14 ± 7 |
| 1,1,2-Trichloroethane | 0.5 | 43 ± 13 |
| 1,1,1-Trichloroethane | 0.5 | 1.4 ± 0.1 |
| N2O | 20 | 13 ± 5 |
No oxidation of reduced methyl viologen was observed upon addition of 2-, 3-, or 4-chlorobenzoate, 2-, 3-, or 4-chlorophenol, pentachlorophenol, nitrate, nitrite, thiosulfate, sulfite, acrylate, or fumarate at a concentration of 0.4 mM.
One unit of PCE-RDase activity corresponded to the amount of protein that catalyzed the oxidation of 2 μmol of methyl viologen radicals/μmol of Cl− formed/min at 30°C, as determined from initial rates of methyl viologen oxidation. A PCE-RDase specific activity of 250 ± 12 nkat/mg of protein was defined as 100%.
The abiotic oxidation that occurred in the presence of only reduced methyl viologen was subtracted from the significantly higher oxidation in the presence of the PCE-RDase.
Ammonium, which stimulated the PCE-RDase of Dehalospirillum multivorans (26), had an inhibitory effect on the PCE-RDase of Dehalobacter restrictus; there was a loss of about 25% of the activity in the presence of 4 mM NH4+ compared to the activity in control assays in which no ammonium was added (data not shown). No effect of ammonium ions was observed on the PCE-RDase of Desulfitobacterium hafniense strain PCE-S (22). The PCE-RDase of Dehalobacter restrictus was oxygen sensitive, as reported for other PCE-RDases (22, 26, 29, 41), and it had an activity half-life of 280 ± 10 min upon exposure to air.
Reaction mechanism.
Inhibition experiments with 1-iodopropane in cell suspensions indicated that a cobamide is involved in the reductive dechlorination of PCE (21, 25, 29, 34, 41, 42). The dechlorination reaction catalyzed by the pure enzyme was also inhibited by 1-iodopropane, and the enzyme could be reactivated by illumination (Fig. 5). Additional evidence for the involvement of cob(I)alamin was obtained by the slow inactivation of the enzyme by N2O (data not shown). Studies with free cobalamin indicated that PCE and cob(I)alamin react by a dissociative one-electron transfer, yielding cob(II)alamin and a trichlorovinyl radical (11, 36). Similar experiments with PCE-RDase of Dehalobacter restrictus and increasing amounts of the Ḋ donor d7-isopropyl alcohol providing evidence for a radical mechanism with free cobalamin did not indicate that a dissociative one-electron transfer is involved in the cobalamin enzyme-catalyzed reaction (data not shown). The dechlorination of trans-1,3-dichloropropene to a mixture of trans-1-chloropropene, cis-1-chloropropene, and 3-chloropropene by the PCE-RDases of Dehalospirillum multivorans and Desulfitobacterium hafniense strain PCE-S, on the other hand, is in accordance with a dechlorination mechanism involving a radical intermediate (24). Rapid freezing experiments combined with EPR analysis might provide additional indications of the reaction mechanism involved.
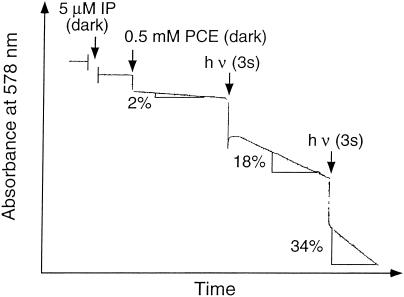
FIG. 5.
Photoreversible inactivation of PCE-RDase activity by 1-iodopropane (IP). The reaction was carried out in a cuvette with purified enzyme in the dark, and the oxidation of reduced methyl viologen was recorded spectrophotometrically at 578 nm. Reactivation occurred by illumination with a 20-W halogen lamp. The arrows indicate the times of addition of 1-iodopropane or illumination; the percentages indicate the amounts of activity present after illumination compared with the amount of activity in a cuvette assay to which 1-iodopropane was not added.
N-terminal sequence.
The N-terminal sequence of the first 20 amino acids (ADIVA PITET SEFPY KVDAK) was identical to the sequences of the PCE-RDases of Desulfitobacterium hafniense strain PCE-S (22) and Desulfitobacterium sp. strain Y51 (41) and very similar to the sequence of the PCE-RDase of Desulfitobacterium hafniense strain TCE1 (42) (Table 2). The high degree of similarity among the N-terminal sequences also suggested that the remaining gene sequence is very similar, a hypothesis that guided the cloning strategy described below.
PCR amplification, cloning, and sequencing of the pceAB genes of Dehalobacter restrictus and Desulfitobacterium hafniense strain TCE1.
The forward primer DR3f was designed based on the N-terminal sequence of the purified PCE-RDase (PceA) of Dehalobacter restrictus (DIVAPIT), which resulted in twofold degeneration, but the sequence contained five neutral bases (inosine). The reverse primer DR4r was designed based on a conserved 8-amino-acid stretch found in PceA of Dehalospirillum multivorans and in CprA of Desulfitobacterium dehalogenans (PDKPIDFG) (27, 43), which resulted in a 16-fold degenerate primer containing three inosines. When genomic DNA of Dehalobacter restrictus was used as the template, the degenerate PCR resulted in a product that was approximately 1,100 bp long, which was purified, cloned (resulting in plasmid pDR1), and sequenced. Sequence analysis revealed an 1,126-bp DNA fragment. A comparison with the sequences of other RDases showed that this sequence exhibited 97% identity at the protein level with the sequence of PceA of Desulfitobacterium hafniense strain PCE-S (G. Diekert, personal communication). Therefore, new primers specific for the beginning of the pceA gene and for the end of the pceB gene of Desulfitobacterium hafniense strain PCE-S were designed, which allowed isolation of the pceAB gene clusters from Dehalobacter restrictus (resulting in plasmid pDR2). Since the N-terminal sequence of PceA of Desulfitobacterium hafniense strain TCE1 was found to be very similar to the N-terminal sequences described previously (42), the same pair of specific primers was used to isolate the pceAB genes from this strain, resulting in plasmid pTCE. Plasmids pDR2 and pTCE were sequenced completely in both directions.
Sequence analysis.
Plasmids pDR2 and pTCE both had ORFs that were 1,656 and 318 bp long, which were designated pceA and pceB and were analogous to the pceA and pceB genes found in Dehalospirillum multivorans (27). As indicated by the very similar N-terminal sequences of the proteins, the sequences of the pceAB gene clusters of Dehalobacter restrictus and Desulfitobacterium hafniense strain TCE1 were found to be very similar to each other (13 of 551 amino acids were different) and almost identical to the pceAB sequences of Desulfitobacterium hafniense strain PCE-S and Desulfitobacterium sp. strain Y51 (41).
Figure 6 shows a physical map of the pceA and pceB genes of the four bacterial strains mentioned above. The pceA gene is 1,656 bp long and codes for a 551-amino-acid protein which has a theoretical molecular mass of 61,299 Da in its unprocessed form. As observed for all other chloroethene RDases, this pceA product also contains a leader sequence that is 39 amino acids long and contains a twin arginine motif (RRxFLK) that is usually found in proteins that are exported to the periplasm and contain redox cofactors (45). However, the residue in front of the two arginine residues is not a serine or threonine, as has been speculated to be typical for this motif (4), but it is an asparagine, which is found quite frequently in RDases. In fact, only TceA of Dehalococcoides ethenogenes and PceA of Dehalospirillum multivorans (18, 27) have the twin arginine motif proposed by Berks (4); all the other molecules have an asparagine instead. The signal peptide cleavage site (ADA↓ADIVA) respects the −1/−3 rule as defined by von Heijne (44). The theoretical molecular mass of the processed PceA is 57,372 Da. Taking into account the eight iron and sulfur atoms and the cobalamin cofactor, the molecular mass is 59,426 Da, which is close to the 60.1 kDa estimated from the purified PceA of Dehalobacter restrictus on an SDS gel (Fig. 1).
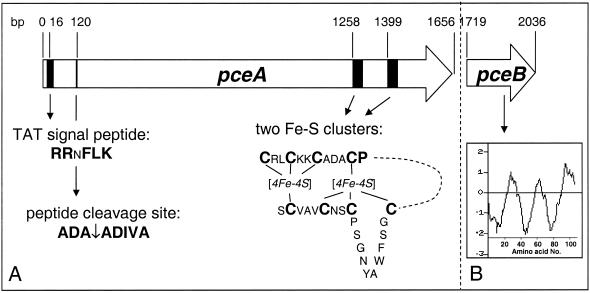
FIG. 6.
Physical representation of the PceA and putative PceB RDases of D. restrictus. (A) PceA and its features, including the TAT signal peptide with a conserved RRxFLK signature, the peptide cleavage site, and two [4Fe-4S] clusters towards the C-terminal end. (B) PceB putative protein and a hydrophobicity plot indicating the presence of three transmembrane α-helices. The Kyle-Doolittle hydrophobicity plot was obtained by using the software Protein Hydrophilicity/Hydrophobicity Search (Bioinformatics Unit, Weizmann Institute of Science, Rehovot, Israel).
A motif very similar to the binding motifs for two [4Fe-4S] iron-sulfur clusters (5) was present towards the C-terminal part of the PceA (Fig. 6A). The motif for the first four cysteines (CX2CX2CX3CP) is found in all RDases, and some quite conserved residues in between lead to the consensus sequence CRXCKKCADXCP, which seems to be very specific for RDases, as determined by the pattern-searching software with general protein databases (PATTERNp [6]). The most variations in this consensus sequence were found in TceA of Dehalococcoides ethenogenes and PceA of Dehalospirillum multivorans (18, 27). The second half of the two [4Fe-4S] iron-sulfur cluster binding motifs is less conserved in RDases. In the four PceA discussed here these motifs can be defined as CX10CX2CX3C, which are missing a proline at the end of the motif and have the first two cysteine residues separated by 10 instead of 2 amino acids. In other RDases, the proline is present, and the first two cysteine residues are separated by variable stretches consisting of 2 residues for TceA of Dehalococcoides ethenogenes (leading to a motif identical to that found for Fe8S8 ferredoxins) (18), 10 residues for PceA of Dehalospirillum multivorans (27), and 12 residues for all CprAs (43; GenBank accession numbers AY013365 and AF204275). It is therefore not possible to deduce a general consensus sequence for the second half of this iron-sulfur cluster motif for all RDases.
EPR measurements with the purified enzymes allowed clear definition of the kind of iron-sulfur clusters present in CprA of Desulfitobacterium dehalogenans (43) and PceA of Dehalobacter restrictus (34). The RDase of Desulfitobacterium dehalogenans contains one [4Fe-4S] cluster and one [3Fe-4S] cluster, and the RDase of Dehalobacter restrictus contains two [4Fe-4S] clusters. In the amino acid sequences of both enzymes the first cysteine of the second group seems to be missing and is replaced by a glycine. However, both RDases contain a cysteine further upstream (12 amino acids upstream for Desulfitobacterium dehalogenans and 10 amino acids upstream for Dehalobacter restrictus). The 10-amino-acid stretch of the latter enzyme starts with a glycine and ends with a proline, which are two structure-breaking residues, indicating that there is formation of a loop in the tertiary structure which allows the participation of the 10-amino-acid-upstream cysteine as a ligand in a [4Fe-4S] cluster. The 12-amino-acid stretch of CprA of Desulfitobacterium dehalogenans does not start or end with a structure-breaking amino acid, indicating that the cysteine is not involved in iron-sulfur cluster binding. The presence of a [3Fe-4S] cluster in this enzyme corroborates this hypothesis.
Since the cobalt in cob(II)alamin of the PceA of Dehalobacter restrictus was not coordinated by a fifth ligand (34), it was not surprising that the PceA does not contain a corrinoid binding motif (DXHXXGSXLGG) found in vitamin B12-dependent mutases and methionine synthases, where the histidine is responsible for the binding of the corrinoid (17). This suggests that in RDases another binding motif is responsible for insertion of the corrinoid cofactor.
A 62-bp spacer separates the pceA and pceB genes of Dehalobacter restrictus (Fig. 6). Despite the very low sequence similarity of putative RdhB proteins, a common feature is their conserved secondary structure. The molecule almost always appears to be a stretch of three hydrophobic α-helices; the only exception is the putative PceB of Dehalospirillum multivorans, in which only two hydrophobic α-helices are observed (27). Hence, these putative RdhB proteins seem to be functionally conserved. It has been speculated that the putative RdhB protein is active in anchoring RdhA in or to the membrane, but no biochemical evidence that supports this hypothesis has been presented so far. The only indication of a functional RdhB protein is the coexpression of the rdhA and rdhB genes shown for the CprA of Desulfitobacterium dehalogenans (37) and the PceA of Dehalospirillum multivorans (27). In contrast to the pceAB gene clusters, all the cprB genes are located in the region directly upstream from their cprA counterparts in Desulfitobacterium dehalogenans (43), Desulfitobacterium chlororespirans (GenBank accession number AF204275), Desulfitobacterium sp. strains PCE1 and Viet-1 (GenBank accession numbers AY013360 and AF259791), and Desulfitobacterium hafniense strain DCB-2 (GenBank accession numbers AY013365,and AF403180 to AF403185).
Only nine RDases of anaerobic bacteria have been characterized on both the biochemical level and the genetic level so far (Fig. 7). Two groups of rather well-conserved RDases can be distinguished. The first group contains the PceA proteins of Dehalobacter restrictus, Desulfitobacterium hafniense strains TCE1 and PCE-S, and Desulfitobacterium sp. strain Y51, which exhibit very strong identity (97 to 99%) on the DNA level as well as on the amino acid level. The second group is composed of the chlorophenol RDases (CprA) of Desulfitobacterium dehalogenans and Desulfitobacterium hafniense strain DCB-2, two very closely related proteins with 99% identity, and the chlorophenol RDase of Desulfitobacterium chlororespirans, which is slightly divergent (64% identity with the other two proteins). Two chloroethene RDases, PceA of Dehalospirillum multivorans and TceA of Dehalococcoides ethenogenes, exhibit low levels of similarity with each other and also with the other RDases (only approximately 25% sequence identity). Despite the existence of two distinct groups of RDases based on gene sequence data, it is difficult to compare them on a biochemical level since biochemical characterization has not been done rigorously enough in all cases. Nevertheless, in the studies in which C-2 chlorinated compounds as well as aromatic compounds have been tested, it has been shown that chloroethene RDases cannot dechlorinate chlorophenols and vice versa (Table 2).
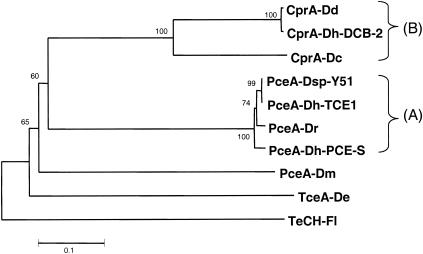
FIG. 7.
Tree-based representation of nine biochemically and genetically characterized RDases. The recently isolated PceA proteins from Dehalobacter restrictus (PceA-Dr) (EMBL accession number AJ439607) and Desulfitobacterium hafniense strain TCE1 (PceA-Dh-TCE1) (accession number AJ439608) are members of a very conserved group (group A) together with the PceA proteins of Desulfitobacterium hafniense strain PCE-S (PceA-Dh-PCE-S) (accession number AY216592) and Desulfitobacterium sp. strain Y51 (PceA-Dsp-Y51) (accession number AB070709). The group B RDases are chlorophenol RDases, including CprA of Desulfitobacterium chlororespirans (CprA-Dc) (accession number AF204275), CprA of Desulfitobacterium hafniense strain DCB-2 (CprA-Dh-DCB-2) (accession number AY013365), and CprA of Desulfitobacterium dehalogenans (CprA-Dd) (AF115542). TceA of Dehalococcoides ethenogenes (TceA-De) (accession number AF228507) and PceA of Dehalospirillum multivorans (PceA-Dm) (accession number AF022812) cannot be associated with any RDase group. The tree was constructed by using the neighbor-joining and bootstrapping tools of ClustalX and was rooted with the tetrachloro-p-hydroquinone RDase of Flavobacterium sp. (TeCH-Fl) (accession number PIR A40625).
In silico investigations of the genomes of Dehalococcoides ethenogenes (The Institute for Genomic Research, Bethesda, Md.) and Desulfitobacterium hafniense strain DCB-2 (DOE Joint Genome Institute, Walnut Creek, Calif.) showed that these genomes contain several putative RDase gene clusters (up to 17 in the case of Dehalococcoides ethenogenes). A comparison of the pceAB genes of the Dehalobacter-Desulfitobacterium group with the genome data for Desulfitobacterium hafniense strain DCB-2 revealed one contig with 65% identity (contig2389, submitted as clone 2977 by J. K. Davies and J. M. Tiedje [GenBank accession number AF403185]). Two overlapping ORFs (ORF1 and ORF2) match PceA, and a third ORF (ORF3) matches PceB. Suspecting a frameshift in the sequence data of the DOE Joint Genome Institute, we amplified a fragment covering this region by PCR from genomic DNA of Desulfitobacterium hafniense strain DCB-2 and cloned and sequenced it (data not shown). In this new sequence an additional cytosine was observed at position 1410 of the inverse complementary sequence of clone 2977 (position 309 of ORF1). Once the data were corrected, the stop codon of ORF1 (at position 457) did not exist anymore, resulting in a single 1,647-bp ORF instead of two overlapping ORFs. This ORF encodes a 548-amino-acid putative protein with 66% identity to the PCE-RDase of the Dehalobacter-Desulfitobacterium group. The similarity level is clearly greater than the typical level for comparisons of chlorophenol RDases and PCE-RDases (only ∼30%). In addition, ORF3, which has high levels of similarity with pceB genes, is located directly downstream of the ORF which is probably a putative pceA gene. Although the genes have been annotated as putative chlorophenol RDase genes and although Desulfitobacterium hafniense strain DCB-2 does not show PCE-RDase activity (8), the similarity level and the relative position of the pceA and pceB genes indicate that these ORFs should be considered putative chloroethene RDase genes.
Acknowledgments
This work was supported by Swiss National Science Foundation grants 3100-040855.94/1 and 3152-055413.
We are grateful to the group of G. Diekert for providing copies of accepted manuscripts and DNA sequence information prior to publication, to A. Ulrich for analysis of the content of iron (atomic absorption spectroscopy) and cobalt (ICP-MS), and to P. James and U. Kämpfer for determination of the N-terminal sequence.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrew, P. W., L. J. Rogers, D. Boulter, and B. G. Haslett. 1976. Ferredoxin from a red alga, Porphyra umbilicalis. Eur. J. Biochem. 69:243-248. [DOI] [PubMed] [Google Scholar]
- 3.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C. 1996. A common export pathway for protein binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 5.Bruschi, M., and F. Guerlesquin. 1988. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol. Rev. 54:155-176. [DOI] [PubMed] [Google Scholar]
- 6.Cockwell, K. Y., and I. G. Giles. 1989. Software tools for motif and pattern scanning: program descriptions including a universal sequence reading algorithm. Comput. Applic. Biosci. 5:227-232. [DOI] [PubMed] [Google Scholar]
- 7.Gantzer, C. J., and L. P. Wackett. 1991. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25:715. [Google Scholar]
- 8.Gerritse, J., O. Drzyzga, G. Kloetstra, M. Keijmel, L. P. Wiersum, R. Hutson, M. D. Collins, and J. C. Gottschal. 1999. Influence of different electron donors and accepters on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 65:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerritse, J., G. Kloetstra, A. Borger, G. Dalstra, A. Alphenaar, and J. C. Gottschal. 1997. Complete degradation of tetrachloroethene in coupled anoxic and oxic chemostats. Appl. Microbiol. Biotechnol. 48:553-562. [DOI] [PubMed] [Google Scholar]
- 10.Gerritse, J., V. Renard, T. M. P. Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 11.Glod, G., W. Angst, C. Holliger, and R. P. Schwarzenbach. 1997. Corrinoid-mediated reduction of tetrachloroethene, trichloroethene, and trichlorofluoroethene in homogeneous aqueous solution: reaction kinetics and reaction mechanisms. Environ. Sci. Technol. 31:253-260. [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hjelmeland, L. M., and A. Chrambach. 1984. Solubilization of functional membrane proteins. Methods Enzymol. 104:305-318. [DOI] [PubMed] [Google Scholar]
- 14.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and A. J. B. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 15.Kröger, A., V. Geisler, E. Lemma, F. Theis, and R. Lenger. 1992. Bacterial fumarate respiration. Arch. Microbiol. 158:311-314. [Google Scholar]
- 16.Krumholz, L. R. 1997. Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int. J. Syst. Bacteriol. 47:1262-1263. [Google Scholar]
- 17.Ludwig, M. L., and R. G. Matthews. 1997. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 66:269-313. [DOI] [PubMed] [Google Scholar]
- 18.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MaymoGatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 21.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168:513-519. [DOI] [PubMed] [Google Scholar]
- 22.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 23.Miller, E., G. Wohlfarth, and G. Diekert. 1996. Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch. Microbiol. 166:379-387. [DOI] [PubMed] [Google Scholar]
- 24.Neumann, A., A. Siebert, T. Trescher, S. Reinhardt, G. Wohlfarth, and G. Diekert. 2002. Tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans: substrate specificity of the native enzyme and its corrinoid cofactor. Arch. Microbiol. 177:420-426. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, A., G. Wohlfarth, and G. Diekert. 1995. Properties of tetrachloroethene and trichloroethene dehalogenase of Dehalospirillum multivorans. Arch. Microbiol. 163:276-281. [Google Scholar]
- 26.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 27.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 29.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 30.Rabilloud, T. 1990. Mechanisms of protein silver staining in polyacrylamide gels: a 10-year synthesis. Electrophoresis 11:785-794. [DOI] [PubMed] [Google Scholar]
- 31.Rabinowitz, J. C. 1978. Analysis of acid-labile sulfide and sulfhydryl groups. Methods Enzymol. 53:275-277. [DOI] [PubMed] [Google Scholar]
- 32.Scholz-Muramatsu, H., A. Neumann, M. Messmer, E. Moore, and G. Diekert. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48-56. [Google Scholar]
- 33.Schumacher, W., and C. Holliger. 1996. The proton/electron ratio of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethene in “Dehalobacter restrictus.” J. Bacteriol. 178:2328-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher, W., C. Holliger, A. J. B. Zehnder, and W. R. Hagen. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421-425. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, P., and P. McCarty. 1996. Isolation and characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl. Environ. Microbiol. 62:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shey, J., and W. A. van der Donk. 2000. Mechanistic studies on the vitamin B12-catalyzed dechlorination of chlorinated alkenes. J. Am. Chem. Soc. 122:12403-12404. [DOI] [PubMed] [Google Scholar]
- 37.Smidt, H., A. D. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria—molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 38.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 39.Stupperich, E., I. Steiner, and M. Ruhlemann. 1986. Isolation and analysis of bacterial cobamides by high-performance liquid chromatography. Anal. Biochem. 155:365-370. [DOI] [PubMed] [Google Scholar]
- 40.Suyama, A., R. Iwakiri, K. Kai, T. Tokunaga, N. Sera, and K. Furukawa. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474-1481. [DOI] [PubMed] [Google Scholar]
- 41.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Pas, B. A., J. Gerritse, W. M. de Vos, G. Schraa, and A. J. M. Stams. 2001. Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1. Arch. Microbiol. 176:165-169. [DOI] [PubMed] [Google Scholar]
- 43.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 44.von Heijne, G. 1984. How signal sequences maintain cleavage specificity. J. Mol. Biol. 173:243-251. [DOI] [PubMed] [Google Scholar]
- 45.Voordouw, G. 2000. A universal system for the transport of redox proteins: early roots and latest developments. Biophys. Chem. 86:131-140. [DOI] [PubMed] [Google Scholar]
- 46.Wild, A., R. Hermann, and T. Leisinger. 1997. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511. [DOI] [PubMed] [Google Scholar]