Abstract
The gene encoding a putative nitrilase was identified in the genome sequence of the photosynthetic cyanobacterium Synechocystis sp. strain PCC6803. The gene was amplified by PCR and cloned into an expression vector. The encoded protein was heterologously expressed in the native form and as a His-tagged protein in Escherichia coli, and the recombinant strains were shown to convert benzonitrile to benzoate. The active enzyme was purified to homogeneity and shown by gel filtration to consist probably of 10 subunits. The purified nitrilase converted various aromatic and aliphatic nitriles. The highest enzyme activity was observed with fumarodinitrile, but also some rather hydrophobic aromatic (e.g., naphthalenecarbonitrile), heterocyclic (e.g., indole-3-acetonitrile), or long-chain aliphatic (di-)nitriles (e.g., octanoic acid dinitrile) were converted with higher specific activities than benzonitrile. From aliphatic dinitriles with less than six carbon atoms only 1 mol of ammonia was released per mol of dinitrile, and thus presumably the corresponding cyanocarboxylic acids formed. The purified enzyme was active in the presence of a wide range of organic solvents and the turnover rates of dodecanoic acid nitrile and naphthalenecarbonitrile were increased in the presence of water-soluble and water-immiscible organic solvents.
Nitrilases that hydrolyze organic nitriles to carboxylic acids and ammonia are a commercially very interesting group of enzymes because nitriles are important intermediates in the chemical synthesis of various products. Several biotransformation processes have been described which utilize the chemo-, regio-, or enantioselectivity of nitrilases (3, 9, 17, 25, 26, 34). All nitrilases studied in some detail contain a cysteine residue in their catalytical center and catalyze the hydrolysis of their substrates via the intermediate formation of a covalently bound acyl residue (Fig. 1) (15, 30). Nitrilases from a variety of prokaryotic and eukaryotic organisms have been described (10, 11, 33, 39). The main source of nitrilases are bacterial species, which are usually obtained by enrichments from environmental samples with the nitriles as sole source of nitrogen. These enrichments result in most cases in the isolation of bacteria that either belong to the Proteobacteria (such as Acinetobacter, Alcaligenes, or Pseudomonas strains) (21, 22, 40, 41) or to the high-GC gram-positive bacteria (mostly rhodococci) (10, 16, 18, 21, 36). Many of the known nitrilases possess considerable disadvantages (e.g., insufficient stability or selectivity or low specific activities) that prevent their industrial application, and there is therefore a constant demand for new nitrilases. Because the traditional enrichment strategies usually result in the isolation of a rather restricted group of microorganisms, we decided to perform an in silico screening for new nitrilases from DNA sequences obtained in the course of genome projects and deposited in the databases. Therefore, these databases were screened for the presence of putative nitrilase genes in mesophilic microorganisms that are only distantly related to the typical nitrilase-containing organisms previously described. These sequence comparisons suggested that an interesting nitrilase might be present in the cyanobacterium Synechocystis sp. strain PCC6803, which was therefore investigated in the present study.
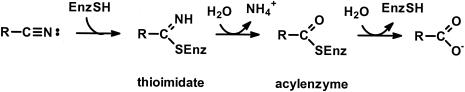
FIG. 1.
Proposed mechanism for the nitrilase reaction (30).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli JM109 was used for all cloning experiments. The strain was routinely cultured in Luria-Bertani (LB) medium (plus 100 μg of ampicillin/ml if appropriate). The plasmids pJOE2702 and pJOE2775 have been described before (37, 38).
Amplification of the nitrilase gene by PCR and subsequent cloning of the gene.
The gene was amplified from the total DNA of Synechocystis sp. strain PCC6803 (kindly provided by Anke Engels, University of Tübingen, Tübingen, Germany) by using the two oligonucleotide primers 5′-TTG GAT CCT TAA TGG CTA AAA ATC CG-3′ (SycnitBI) and 5′-AAA AAT TCA TAT GCT GAA TTA TAC AAA AAA T-3′ (SycNitNu), which incorporated BamHI and NdeI restriction sites, respectively. The amplification was performed in a final volume of 40 μl, and the reaction mixtures contained template DNA, 10 pmol of each primer (MWG Biotech, Ebersberg, Germany), a 220 μM concentration of each deoxynucleotide triphosphate, 1× PCR buffer, 2 U of Taq DNA polymerase (Boehringer Roche), and 5 mM MgCl2.
The following PCR program was used for the amplification of an 1,041-bp DNA fragment: 3 min of denaturing at 93°C; 30 cycles of denaturing (1 min at 93°C), annealing (1 min at 40°C), and polymerization (25 s at 72°C) followed. The program was completed by an additional polymerization step (5 min at 72°C).
The amplified 1,041-bp DNA fragment was cut with NdeI and BamHI, and the resulting fragment was ligated into vector pJOE2702, which had been cut with the same restriction enzymes. The resulting plasmid was designated pDHE21.
In order to obtain a nitrilase derivative with a His6 tag, the nitrilase gene was amplified with the primer pair 5′-TTG GAT CCA TGG CTA AAA ATC GGA TT-3′ (SycnitBIHis) and SycNitNu (see above) by using pDHE21 as a template under the conditions described above. The amplified fragment was also cut with NdeI and BamHI, and the resulting fragment was ligated into the NdeI/BamHI-cut vector pJOE2775, giving pDHE22.
Expression of the nitrilase in E. coli.
For the induction of the nitrilase in the recombinant E. coli strains, precultures (2 ml each) were grown in LB-ampicillin medium. From these precultures, Erlenmeyer flasks (usually with a volume of 300 ml) containing 0.2% (wt/vol) l-rhamnose in LB-ampicillin medium were inoculated to an initial optical density at 546 nm of 0.02. The bacterial cultures were incubated at 30°C on an orbital shaker at 120 rpm. The cells were harvested ca. 13 h later.
For the preparation of resting cells, exponentially growing bacterial cultures expressing the nitrilase activity were harvested by centrifugation (15 min, 4°C, 9,500 × g) and washed and resuspended in 50 mM Na-K phosphate buffer (pH 7.0).
Analytical methods.
Benzonitrile and benzoate were analyzed by reversed-phase high-pressure liquid chromatography (HPLC) by using Millenium Chromatography Manager 2.0 equipped with a photodiode array detector model 996 and HPLC pump model 510 (Waters Associates, Milford, Mass.). A reversed-phase column (125 by 4.0 mm [internal diameter]; Grom, Herrenberg, Germany) packed with 5-μm particles of Grom-Sil 100 Octyl was used. The separated compounds were detected photometrically at 210 nm with a photodiode array detector. The eluting solvent system was composed of 79.7% (vol/vol) water, 20% (vol/vol) acetonitrile, and 0.3% H3PO4. The usual flow rate was 0.7 ml/min.
Enzyme assays.
The standard assays contained 50 mM Na-K phosphate buffer (pH 7.0), benzonitrile (10 mM), and preparations of the purified nitrilase or resting cells of the recombinant E. coli strains. For the preparation of resting cells, exponentially growing bacterial cultures expressing the nitrilase activity were harvested by centrifugation (15 min, 4°C, 9,000 × g) and then washed and resuspended in 50 mM Na-K phosphate buffer (pH 7.0). The reaction mixtures were incubated in a Thermomixer (CLF-Schutron, Emersacker, Germany) at 30°C and 300 rpm. Aliquots (usually 50 to 100 μl) were taken after different time intervals, the reactions were stopped by the addition of 10% (vol/vol) 1 M HCl, and the cells or precipitated proteins were removed by centrifugation (16,000 × g, 2 min). The turnover of benzonitrile was usually analyzed by HPLC (see above). The conversion of other (especially aliphatic nitriles) was usually determined by the quantitation of the amount of ammonia (in 1 ml of reaction volume), which was enzymatically released from the substrates by using a commercially available ammonia test system relying on the indophenol method (Spectroquant 14752; Merck, Darmstadt, Germany). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of product (benzoate or ammonia) per min.
Preparation of cell extracts.
The cultures of E. coli JM109(pDHE21) or E. coli JM109(pDHE22) were harvested by centrifugation (9,500 × g, 15 min, 4°C), washed, and resuspended in Na-K phosphate buffer (50 mM, pH 7.5) or Tris-HCl (50 mM, pH 7.5), respectively. The cells were disrupted by using a French press (Aminco, Silver Springs, Md.) at 80 MPa. Cell debris was removed by centrifugation at 110,000 × g for 60 min at 4°C. Protein was determined by the method of Bradford (2) with bovine serum albumin as a standard.
Purification of the native enzyme.
Protein was purified at 4°C by use of a fast-performance liquid chromatography system consisting of a LCC 501 plus controller, two P-500 pumps, a UV-1 monitor, a REC-112 recorder, and a FRAC-100 autosampler from Pharmacia (Uppsala, Sweden).
Crude extract was applied to a Q Sepharose column (XK 16/10; Pharmacia). The adsorbed proteins were eluted with a linear gradient of 0 to 400 mM NaCl in Tris-HCl (50 mM, pH 7.5) at a flow rate of 2 ml/min. Fractions (1.5 ml) were collected, and enzyme activity was determined with benzonitrile as the substrate by using the Spectroquant system. The nitrilase was eluted at a concentration of ca. 0.2 M NaCl. The active fractions were pooled, transferred to a hydroxyapatite column (XK 16/10, ceramic hydroxyapatite type I, 20-μm particle size; Bio-Rad), and eluted with a linear gradient (10 to 400 mM) of K phosphate buffer (pH 7.5) at a flow rate of 1 ml/min, and fractions of 0.9 ml were then collected. The enzyme activity eluted at a concentration of 135 mM phosphate. The pooled fractions (1.8 ml) were concentrated by ultrafiltration (Vivaspin 2; Vivascience, Hannover, Germany) to a volume of 0.6 ml, and proteins were finally chromatographed on a Superdex 200 Prep-Grade HiLoad column (HR 16/60; Amersham Biosciences) by using Na-K phosphate buffer (50 mM, pH 7.3) plus 150 mM NaCl as the elution buffer.
Purification of the His-tagged enzyme.
Cell extracts of E. coli JM109(pDHE22) were prepared in Tris-HCl buffer (50 mM, pH 7.5) as described above. The nickel-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany) was suspended in Tris-HCl (20 mM, pH 7.5) and transferred (4 ml) to an empty 10 ml-polypropylene column. The filled column was equilibrated with a buffer system (pH 7.5) consisting of Tris-HCl (50 mM), NaCl (300 mM), and imidazole (40 mM). The cell extracts (50 to 70 mg of protein) were applied to the column, and the protein was eluted with subsequent steps of the Tris-HCl-NaCl buffer (2 to 4 ml each) with increasing imidazole concentrations (100, 150, 300, and 500 mM). The fractions with nitrilase activity eluted at an imidazole concentration of 500 mM. This fraction was finally concentrated by ultracentrifugation (Vivaspin2, 10,000 MWCO PES; Vivascience) and resuspended in Na-K phosphate buffer (50 mM, pH 7.4) plus NaCl (300 mM) in order to remove the imidazole which disturbed the ammonia test.
PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (20). Gels were silver stained by the method of Shevchenko et al. (35).
Determination of molecular weight.
The relative molecular mass of the native enzyme was determined by gel filtration by using a Superdex 200 Prep-Grade HiLoad 16/60 column (Pharmacia). For the determination of the molecular mass of the nitrilase, the column was calibrated with thyroglobulin (Mr 670,000), ferretin (Mr 440,000), catalase (Mr 232,000), aldolase (Mr 158,000), and albumin (Mr 67,000) as references (from the high- and low-molecular-weight gelfiltration kits from Amersham Pharmacia).
Substrate specificity of the purified nitrilase.
The purified enzyme (20 μg) was incubated for 30 to 240 min in 0.2 ml of Na-K phosphate buffer (pH 7.0, 50 mM) plus 5% (vol/vol) methanol and 5 mM concentrations of the respective nitriles. The reaction was stopped by the addition of 20 μl of 1 M HCl, and precipitated protein was removed by centrifugation in an Eppendorf centrifuge (16,000 × g, 2 min). The supernatant was diluted with H2O to 1 ml, and the ammonia content was determined as suggested by the manufacturer of the test kit (Spectroquant 14752; Merck). A control experiment without added enzyme was performed for each substrate, and the amount of ammonia found in the reaction mixture was corrected for the value found in the control experiment.
Determination of the temperature and pH optima.
For the determination of the temperature maximum, the purified nitrilase (80 μg of protein) was incubated in 0.34 ml of Na-K phosphate buffer (50 mM, pH 7.0) with benzonitrile (10 mM) in Eppendorf reaction tubes in a thermoshaker at 35, 40, 45, 50, or 55°C. Aliquots (50 μl) were taken after 10, 20, 40, 60, and 120 min, and the formation of benzoate was quantified by HPLC.
The pH optimum of the nitrilase was determined by using a universal buffer according to the method of Theorell and Stenhagen (32). The buffer consisted of a mixture of citric acid, phosphoric acid, boric acid, and NaOH, which was titrated with HCl to obtain the desired pH value (32). The purified nitrilase (55 μg of protein) was incubated in 0.1 ml of the respective buffer with benzonitrile (10 mM) in Eppendorf reaction tubes in a thermoshaker at 30°C. The reactions were stopped after 60 min by the addition of 10 μl of 1 M HCl, and the formation of benzoate was quantified by HPLC.
Chemicals.
The nitriles were obtained from Fluka (Neu-Ulm, Germany), Lancaster (Mühlheim-Main, Germany), Merck (Darmstadt, Germany), Serva (Heidelberg, Germany), Bayer (Leverkusen, Germany), Sigma-Aldrich (Munich, Germany), and the Sigma-Aldrich Library of Rare Chemicals (Milwaukee, Wis.).
RESULTS
Cloning of the nitrilase gene from Synechocystis sp. strain PCC6803.
The published genome sequence of the cyanobacterium Synechocystis sp. strain PCC6803 (NCBI accession number D64005) contained an open reading frame encoding a putative nitrilase. The deduced protein sequence (NCBI accession no. NP_442646) demonstrated ca. 36% sequence identity to the nitrilase from Rhodococcus rhodochrous J1 (NCBI accession no. A45070). The putative nitrilase gene from Synechocystis sp. strain PCC6803 contained at the 5′ end a set of two ATG triplets separated by 15 nucleotides. It appeared more probable that the second ATG triplet functioned in vivo as a start codon because a putative Shine-Dalgarno sequence was found upstream and also the sequence alignment of the corresponding gene product was in better agreement with published nitrilase sequences. The corresponding gene was therefore amplified from the complete DNA of strain PCC6803 by PCR as described in Materials and Methods and expressed under the control of a rhamnose-inducible promoter (in plasmid pDHE21) or under the same promoter as a (carboxy-terminally) His-tagged protein (in plasmid pDHE22).
Demonstration of nitrilase activity.
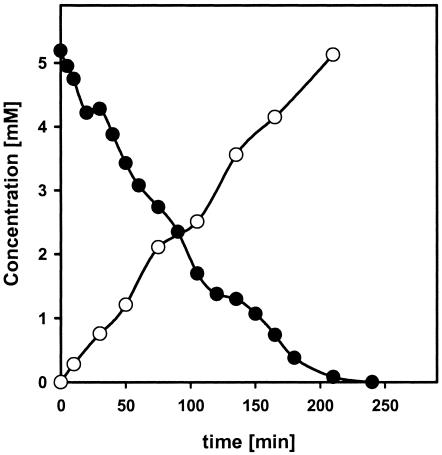
The putative nitrilase was induced in E. coli JM109(pDHE21) by the addition of rhamnose (see Materials and Methods), and resting cells were then incubated with benzonitrile. The recombinant cells converted benzonitrile almost stoichiometrically to benzoate with a specific activity corresponding to 0.02 U/mg of protein (Fig. 2). In a control experiment with the plasmid-free strain E. coli JM109, no turnover of benzonitrile was detected.
FIG. 2.
Demonstration of a nitrilase activity in E. coli(pDHE22) which harbors the recombinant nitrilase from Synechocystis sp. strain PCC6803. The reaction mixture contained, in a total volume of 5 ml, 50 mM Na-K phosphate buffer, 5 mM benzonitrile, and resting cells of E. coli(pDHE22) corresponding to an optical density at 546 nm of 4. Aliquots (50 μl each) were removed after different time intervals, the reaction was terminated by the addition of 5 μl of 1 M HCl, and cells and precipitated protein were removed by centrifugation (2 min, 16,600 × g). The conversion of benzonitrile to benzoate was determined by HPLC.
Purification of the nitrilase.
The nitrilase was purified from cell extracts of the recombinant E. coli strains carrying plasmids pDHE21 (wild-type nitrilase) or pDHE22 (carrying the carboxy-terminal His tag) as described in Materials and Methods. The purification of the wild-type enzyme by anion-exchange chromatography, hydroxyapatite chromatography, and gel filtration resulted in a pure enzyme preparation (as shown by SDS-PAGE and silver staining) with a rather low yield and low specific activity (0.21 U/mg of protein) for the conversion of benzonitrile (Table 1). The purified nitrilase (7 μg of protein) was incubated in Na-K phosphate buffer (50 mM, pH 7.0) with benzonitrile (10 mM) at 30°C. The turnover of the substrate was analyzed for several hours, and it was observed that the increase in the formation of benzoate was almost linear for at least 5 h before the reaction slightly slowed down.
TABLE 1.
Purification of nitrilase from Synechocystis sp. strain PCC6803a
| Purification step | Total protein (mg) | Sp act (U/mg) | Total activity (U) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 227 | 0.03 | 6.8 | 100 | 1.00 |
| Ion exchange | 40 | 0.11 | 4.4 | 65 | 3.7 |
| Hydroxyapatite | 10.7 | 0.13 | 1.4 | 21 | 4.3 |
| Gel filtration | 1.7 | 0.21 | 0.36 | 5.3 | 7.0 |
Experimental details are given in Materials and Methods. The enzymatic activity was determined with benzonitrile as a substrate, and the amount of ammonia released was spectrophotometrically determined.
The purification of the His-tagged protein variant from E. coli JM109(pDHE22) by nickel-agarose chromatography resulted in a nitrilase preparation with an ∼2 times-higher specific activity. This might suggest that the nitrilase was partially inactivated during the time-consuming multistep purification of the native enzyme.
Enzyme stability.
The His-tagged enzyme was moderately stable during storage under different conditions. After 10 days of storage at 5°C in Na-K phosphate buffer (50 mM, pH 7.5) or in the same buffer plus 1 mg of bovine serum albumin/ml, 0.3 M NaCl, or 1% (vol/vol) PEG 4000, ca. 20, 40, 15, or 10%, respectively, of the initial activity could be detected. After storage at −20°C under the same conditions, between 58 and 68% of the initial activity was recovered.
Molecular weight and subunit structure.
The purified wild-type enzyme gave a single band by SDS-PAGE (2 to 6 μg of enzyme) with a molecular weight of ca. 40,000. A protein with the same molecular weight was already very prominent in the cell extract. The molecular weight of the holoenzyme was estimated by gel filtration to be 390,000. Therefore, it can be assumed that the enzyme consists of ca. 10 subunits of identical size.
Temperature and pH optimum.
The temperature optimum of the nitrilase was at 50°C if the reaction was monitored only for 10 min, but the enzyme rapidly lost its activity under these conditions. If the reaction was monitored for 2 h, a temperature optimum of 40 to 45°C was determined, but also under these conditions only 60% of the initital activity could be recovered at the end of the incubation period. The enzyme showed maximal activity at pH 7.0. At pH 5.0 and 8.0 ca. 50 to 65% of the maximal activity was found. No enzymatic activity was detected at pH 3 or 12.
Substrate specifity.
The purified His-tagged enzyme was incubated with more than 50 different nitriles (5 mM each), and the relative activities determined by the quantitation of the amount of ammonia released. The enzyme showed a clear preference for fumarodinitrile (trans-1,2-dicyanoethene), which was converted with a >100-fold-higher relative activity than benzonitrile (Table 2). Also, some larger benzene derivatives, heterocyclic compounds, and long-chain aliphatic (di)nitriles were converted with higher relative activities than benzonitrile. In order to obtain a better estimation of the catalytic properties of the Synechocystis nitrilase, the basic catalytic constants were determined for some selected substrates. A comparison of the specificity constants (kcat/Km) (Table 3) confirmed the assumption that aliphatic dinitriles were the preferred substrates of the nitrilase.
TABLE 2.
Conversion of various nitriles by purified His-tagged nitrilase from Synechocystis sp. strain PCC6803
| General compound group | Substrate | Relative activity (%)a |
|---|---|---|
| Benzene derivatives | Benzonitrile | 100 |
| 4-Aminobenzonitrile | 28 | |
| Benzylcyanide | 52 | |
| 2-Chlorobenzylcyanide | 6 | |
| 3-Chlorobenzylcyanide | 316 | |
| 4-Chlorobenzylcyanide | 11 | |
| 2-Methylbenzylcyanide | 207 | |
| 4-Hydroxybenzylcyanide | 10 | |
| 2-3 Ring systems | 2-Naphthalenecarbonitrile | 757 |
| 2-Naphthaleneacetonitrile | 8 | |
| 9-Anthracenecarbonitrile | 10 | |
| 3-Phenoxyphenylacetonitrile | 35 | |
| Heterocyclic compounds | 2-Thienylacetonitrile | 311 |
| 3-Thienylacetonitrile | 102 | |
| 3-Cyanopyridine | 245 | |
| Indole-3-acetonitrile | 354 | |
| Saturated aliphatic mononitriles | Propionitrile | 33 |
| 2-Chloropropionitrile | 270 | |
| 3-Dimetylaminopropionitrile | 8 | |
| Butyronitrile | 109 | |
| Valeronitrile | 222 | |
| Isovaleronitrile | 9 | |
| 2-Methylglutaronitrile | 394 | |
| Octanoic acid nitrile | 303 | |
| Dodecanoic acid nitrile | 36 | |
| Saturated aliphatic dinitriles | Adipinic acid dinitrile | 528 |
| Octanoic acid dinitrile | 588 | |
| Decanoic acid dinitrile | 476 | |
| Unsaturated aliphatic mononitriles | Acrylonitrile | 68 |
| 2-Butenenitrile | 49 | |
| 3-Butenenitrile | 409 | |
| Unsaturated aliphatic dinitriles | Fumarodinitrile | 12,000 |
| 3-Hexenoic acid dinitrile | 14 |
The activity with benzonitrile was 100%. Less than 5% relative activity was found with the following substrates. 2-Ethyl-2-phenylbutyronitrile, 2-phenylvaleronitrile, 2,6-dichlorobenzylcyanide, naproxen nitrile, ketoprofen nitrile, acetonitrile, malononitrile, 2-methyl-3-butenenitrile, diethyldicyanofumarate, isobutyronitrile, 2-methylbutyronitrile, pentadecanenitrile, and nonadecanenitrile.
TABLE 3.
Kinetic constants of nitrilase with different substrates
| Substrate | Vmax (U mg−1) | Km (mM) | Kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| Benzonitrile | 0.33 | 0.97 | 0.22 | 227 |
| Indole-3-acetonitrile | 1.5 | 2.4 | 1.0 | 417 |
| 2-Chloropropionitrile | 1.4 | 5.2 | 0.92 | 177 |
| Valeronitrile | 0.7 | 4.4 | 0.47 | 107 |
| Octanoic acid dinitrile | 0.7 | 0.2 | 0.47 | 2,350 |
| Decanoic acid dinitrile | 0.4 | 0.3 | 0.27 | 900 |
| Fumarodinitrile | 109 | 28 | 72.7 | 2,600 |
Conversion of aliphatic dinitriles.
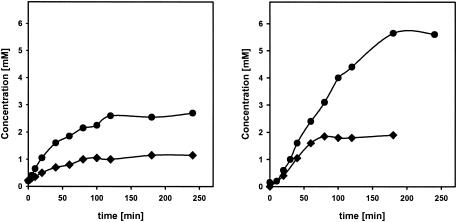
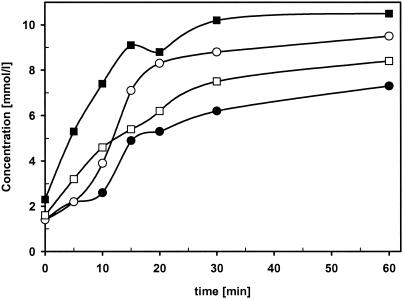
It has been repeatedly shown that many nitrilases convert dinitriles preferentially to mononitrile-monocarboxylic acids (1, 6, 12, 19). We therefore tested whether this was also valid for the nitrilase from Synechocystis sp. strain PCC6803. The purified enzyme was incubated with fumarodinitrile, succinodinitrile, adipodinitrile, 3-hexenoic acid dinitrile, octanoic acid dinitrile, or decanoic acid dinitrile (1 or 3 mM, each), and the time-dependent formation of ammonia was analyzed. The results suggested that the reactions stopped with all dinitriles with fewer than six carbon atoms after the release of 1 mol of ammonia per mol of the respective substrates. In contrast, 2 mol of ammonia were formed from 1 mol of octanoic acid dinitrile or decanoic acid dinitrile (Fig. 3).
FIG. 3.
Hydrolysis of fumarodinitrile and decanoic acid dinitrile by the purified nitrilase from Synechocystis sp. strain PCC6803. The reactions were performed in 1 ml of Na-K phosphate buffer (pH 7.0) containing 1 mM (♦) or 3 mM (•) fumarodinitrile (left panel) or decanoic acid dinitrile (right panel) and 80 μg of purified enzyme. Aliquots (100 μl each) were removed after different time intervals, the reactions were terminated by the addition of 10 μl of HCl (1 M), and precipitated protein was removed by centrifugation (2 min, 16,600 × g). The concentration of ammonia was subsequently determined by using the Spectroquant test.
Stability of the nitrilase in the presence of organic solvents.
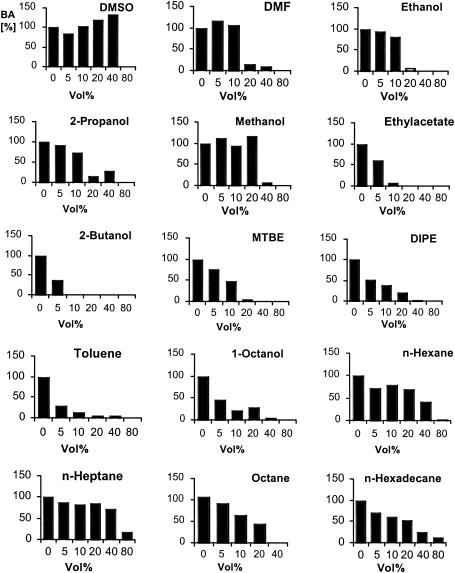
The analysis of the substrate specifity of the nitrilase from Synechocystis sp. strain PCC6803 demonstrated that long-chain aliphatic compounds and rather hydrophobic aromatic compounds such as 2-naphthalenecarbonitrile were preferentially hydrolyzed. The enzymatic conversion of hydrophobic compounds in aqueous solutions can be limited by the low bioavailability of the hydrophobic substrates. It has been demonstrated that these limitations might be overcome by the inclusion of organic solvents into the medium (for recent reviews about the effects of organic solvents on enzymatic reactions, see reference 4 and 14). To the best of our knowledge, there is as yet only one study available in the literature which demonstrated that a bacterial nitrilase (from a pseudomonad) acted in the presence of organic solvents (23). Because the nitrilase from Synechocystis seemed to possess a preference for hydrophobic substrates, it appeared possible that the conversion of this type of compounds could be further increased by the addition of organic solvents. Therefore, the tolerance of the enzyme against organic solvents was tested, and it was found that the enzyme was active in the presence of a wide range of organic solvents (Fig. 4).
FIG. 4.
Hydrolysis of benzonitrile by the purified nitrilase from Synechocystis sp. strain PCC6803 in the presence of different concentrations of various organic solvents. The reaction mixtures contained 19 μg of the purified (His-tagged) nitrilase in 250 μl of 50 mM Na-K phosphate buffer (pH 7.0) plus 0 to 200 μl of the solvents indicated. These enzyme preparations were incubated for 10 min at room temperature. Subsequently, benzonitrile (10 mM) was added, and the reaction mixtures were shaken at 30°C. The reactions were stopped after 30 min by the addition of 10% (vol/vol) 1 M HCl. Precipitated protein was removed by centrifugation (2 min, 16,000 × g), and the concentration of benzoate formed was determined in the aqueous phase by HPLC. The enzyme preparation formed under these conditions in a purely aqueous phase 0.7 mM concentration of benzoate (100%). The relative amounts of benzoate (BA) formed in correlation to the control experiment in a purely aqueous phase are presented. DMSO, dimethyl sulfoxide; DMF, dimethyl formamide, MTBE, methyl tert-butyl ether; DIPE, diisopropyl ether.
Conversion of long-chain aliphatic nitriles in the presence of organic solvents.
The conversion of (liquid) aliphatic nitriles with different chain lengths (4 to 19 carbon atoms) was analyzed in the presence of different concentrations of methanol, which were tolerated by the enzyme (0 to 20% [vol/vol]). These experiments demonstrated that the addition of methanol indeed resulted in increased conversion rates for aliphatic nitriles with chain lengths of more than 12 carbon atoms and that this effect most pronounced with dodecanoic acid nitrile. Thus, within 30 min in the presence of 10% (vol/vol) methanol, approximately twice the amount of ammonia was released compared to the control without added methanol. In contrast, no enhancement of the reaction rates was observed with aliphatic nitriles with shorter chain lengths or if the concentration of the organic solvent was increased to 40% (vol/vol) methanol.
The turnover of the aliphatic nitriles with different chain lengths was also analyzed in two-phase systems with hexane or heptane as organic phases (50% [vol/vol] each). No increase in the conversion rates of dodecanoic acid nitrile could be observed under these conditions.
Effects of organic solvents on the conversion of naphthalenecarbonitrile.
Finally, the influence of organic solvents on the conversion of almost water-insoluble solid nitriles was tested with naphthalenecarbonitrile as the substrate. In this system, an increase in the reaction rates was observed in the presence of methanol and a slightly more pronounced effect in two-phase systems with water and hexadecane. The analysis of different ratios of hexadecane and the aqueous phase demonstrated that the maximal conversion of naphthalenecarbonitrile was achieved in the presence of 40% (vol/vol) hexadecane (Fig. 5).
FIG. 5.
Conversion of naphthalenecarbonitrile by the purified nitrilase from Synechocystis sp. strain PCC6803 in the presence of different volumes of hexadecane. The reaction mixtures contained 580 μg of the purified (His-tagged) nitrilase in 200 μl of 50 mM Na-K phosphate buffer (pH 7.0) plus 10 mM naphthalenecarbonitrile. Different volumes of hexane were added (0% [vol/vol] [•], 20% [vol/vol] [○], 40% [vol/vol] [▪], or 60% [vol/vol] [□]), and the reaction mixtures were shaken at 30°C in a Thermomixer at 1,000 rpm. The reactions were stopped after different time intervals by the addition of 10% (vol/vol) 1 M HCl. Precipitated protein was removed by centrifugation (2 min, 16,000 × g), and the concentration of ammonia formed was determined in the aqueous phase by using the Spectroquant ammonia test.
DISCUSSION
We attempted here for the first time to obtain a novel bacterial nitrilase from the sequence information generated by various sequencing projects currently performed on bacterial genomes. The search for DNA sequences with sequence homologies to known nitrilases suggested that genes homologous to known nitrilases might be present in different microorganisms but that they are much less widespread compared, for example, to amidase genes. The successful cloning of a functional nitrilase demonstrated that this approach is potentially useful but also demonstrated the pitfalls of this strategy, especially that no selection takes place in order to obtain enzymes with high specific activities and that even screening of a huge spectrum of putative substrates will not result in a conclusive identification of the natural substrate(s) of the enzymes. In a different approach in order to obtain novel nitrilases without the traditional enrichment strategies, it was recently shown that active nitrilases can also be obtained from genomic libraries constructed from the “metagenome” of (uncultured) microorganisms by expression cloning (5).
The present study clearly demonstrated that the cyanobacterium Synechocystis sp. strain PCC6803 harbors a gene that encodes a nitrilase activity, which could allow the organism to grow with various nitriles as nitrogen sources. Cyanobacteria are able to use some simple organic nitrogen compounds, such as amino acids, purines, urea, or acetamide, as nitrogen sources (7, 13, 28) but, to the best of our knowledge, there is no published information that describes the utilization of nitriles by cyanobacteria. Furthermore, the genetic organization of the nitrilase gene within the chromosome of Synechocystis. spp. PCC6803 also did not suggest a putative natural function of the nitrilase because the gene was surrounded by two open reading frames, which encoded hypothetical proteins of unknown function, and the gene was not included in any obvious operon structure. Thus, we are currently unable to suggest any physiological function of the nitrilase in this microorganism, but it may be worthwhile to test whether Synechocystis sp. strain PCC6803 can utilize some of the nitriles that are substrates of the nitrilase as nitrogen sources.
The fundamental enzyme characteristics that have been determined for the nitrilase from Synechocystis sp. strain PCC6803 clearly resemble the data obtained with nitrilases from mesophilic chemotrophic bacteria: thus, similar temperature optima (∼40 to 45°C) and pH optima (pH 7 to 7.5) have been described for the nitrilases from R. rhodochrous J1 or Alcaligenes faecalis JM3 (16, 27). The observed substrate specificity of the Synechocystis nitrilase suggested that the enzyme most closely resembled the previously described group of aliphatic nitrilases, although the ability to hydrolyze aromatic substrates puts the enzyme somehow outside the established classification system for nitrilases. Previously, three bacterial nitrilases with a preference for aliphatic substrates have been described from Acidovorax facilis 72W, Comamonas testosteroni, and R. rhodochrous K22 (8, 18, 19, 24). Unfortunately, there is only very limited information available regarding the substrate specifities of these reference enzymes, but it is striking that fumarodinitrile was identified as the preferred substrate for the nitrilase from Synechocystis sp. strain PCC6803 and also for the aliphatic nitrilase from Acidovorax facilis strain 72W (8), although the nitrilase from Synechocystis was clearly differentiated by its ability to convert substrates such as benzylcyanide, butyronitrile, crotononitrile, or valeronitrile from the Acidovorax nitrilase. Only the nitrilase from R. rhodochrous K22 has been purified and biochemically characterized in greater detail from the enzymes that have been previously described as aliphatic nitrilases (18). This enzyme demonstrated with almost all substrates rather low specific activities (<1 U/mg of protein) and reached the highest specific activities (3.3 U/mg of protein) with glutaronitrile (18). In contrast, for several aromatic nitrilases and arylacetonitrilases, specific activities of 15 to nearly 150 U/mg of protein have been described for the conversion of specifically favored substrates (10, 16, 27, 40). The observation that the purified Synechocystis nitrilase demonstrated a Vmax of >100 U/mg of protein with its preferred substrate fumarodinitrile demonstrated that nitrilases can also convert aliphatic substrates with rather high catalytic efficiencies.
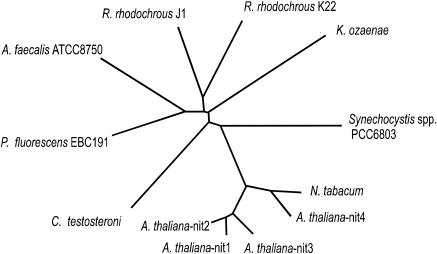
In order to predict a putative natural function for the nitrilase, sequence comparisons were performed with various known or predicted nitrilases from microbial or plant origin. These alignments demonstrated that the known bacterial aliphatic nitrilases did not form a distinct cluster within the nitrilase group. Obviously, the nitrilase from Synechocystis sp. strain PCC6803 did not cluster tightly with the known microbial nitrilases but was placed in the dendrograms somewhere intermediately between the bacterial and plant nitrilases (Fig. 6). Many plants (especially members of the Cruciferae) encode for several different nitrilases, and for Arabidopsis thaliana four different nitrilase genes have been identified. Three of these genes (Nit1 to Nit3) demonstrated high sequence similarities with each other, and the encoded nitrilases are able to convert indole-3-acetonitrile to the plant growth hormone indole-3-acetic acid. These three nitrilases also show a rather similar substrate specifity (39). In contrast, the fourth nitrilase (Nit4) differed according to its sequence and substrate specifity significantly from the three others, and it has been recently suggested that Nit4 and its orthologs in other plants encode for a β-cyano-l-alanine hydratase/nitrilase that is involved in cyanide detoxification in plants (31). Surprisingly, the Synechocystis nitrilase did not convert β-cyano-l-alanine (unpublished results), although it hydrolyzed the C-4 compounds fumarodinitrile with extremely high specific activities. Some interesting parallels could be found when the substrate specificity of the Synechocystis nitrilase was compared to the Nit1 nitrilase from Arabidopsis thaliana (29). Thus, both enzymes demonstrated a certain preference for long-chain aliphatic nitriles. Furthermore, both enzymes converted dinitriles with higher relative activities compared to the mononitriles of the same chain length and almost did not hydrolyze nitriles that carried in the α-position larger substituents (e.g., 2-phenylpropionitrile). Nevertheless, several differences between the two enzymes were also observed. For example, the nitrilase from Synechocystis converted benzonitrile and indole-3-acetonitrile with approximately the same or even higher relative activities compared to butyronitrile, whereas these substrates were only converted by the Arabidopsis nitrilase with <3% of the activity observed with butyronitrile (29).
FIG. 6.
Dendrogram resulting from pairwise alignments of amino acid sequences of different nitrilases. The following sequences with their relevant NCBI registration numbers were used: Alcaligenes faecalis JM3 (D13419); C. testosteroni (L32589); Klebsiella pneumoniae (ozaenae) (J03196); Pseudomonas fluorescens EBC 191 (C. Kiziak, unpublished data); R. rhodochrous K22 (D12583); R. rhodochrous J1 (D12583); Synechocystis sp. strain PCC 6803 (D64005); Nit4 from Arabidopsis thaliana (U09961) and Nicotina tabacum (D63331); and Nit1, Nit2, and Nit3 from Arabidopsis thaliana (Y07648). The sequences were aligned by using the program CLUSTAL X1.8, the dendrograms calculated by bootstrap neighbor joining, and the phylogenetic tree drawn by using the program TreeView 1.6.6, with the standard parameters.
In conclusion, it appears that the Synechocystis nitrilase hydrolyzes the broadest range of nitriles from all nitrilases currently known and that thus the approach used to obtain this nitrilase indeed resulted in the aspired islation of a novel type of nitrilase.
Acknowledgments
We thank the students of the practical course in microbiology at the University of Stuttgart—Bettina Buschhorn, Fabienne Fiesel, Ivica Granic, Ina Merz, Rosemarie Schäfer, and Arlette Wieland—for their assistance.
REFERENCES
- 1.Bengis-Gerber, C., and A. L. Gutman. 1989. Selective hydrolysis of dinitriles into cyano-carboxylic acids by Rhodococcus rhodochrous N.C.I.B. 11216. Appl. Microbiol. Biotechnol. 32:11-16. [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bunch, A. W. 1998. Nitriles, p. 277-324. In H. J. Rehm and G. Reed (ed.), Bio/technology, vol. 8a. Wiley-VCH, Weinheim, Germany.
- 4.Carrea, G., and S. Riva. 2000. Enzyme in organischen Lösungsmitteln: Eigenschaften und Einsatz in der Synthese. Angew. Chem. 112:2312-2341. [Google Scholar]
- 5.DeSantis, G., Z. Zhu, W. A. Greenberg, K. Wong, J. Chaplin, S. R. Hanson, B. Farwell, L. W. Nicholson, C. L. Rand, D. P. Weiner, D. E. Robertson, and M. J. Burk. 2002. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J. Am. Chem. Soc. 124:9024-9025. [DOI] [PubMed] [Google Scholar]
- 6.Effenberger, F., and S. Osswald. 2001. Selective hydrolysis of aliphatic dinitriles to monocarboxylic acids by a nitrilase from Arabidopsis thaliana. Synthesis 2001:1866-1872. [Google Scholar]
- 7.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487-517. In D. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 8.Gavagan, J. E., R. DiCosimo, A. Eisenberg, S. K. Fager, P. W. Folsom, E. C. Hann, K. J. Schneider, and R. D. Fallon. 1999. A gram-negative bacterium producing a heat-stable nitrilase highly active on aliphatic nitriles. Appl. Microbiol. Biotechnol. 52:654-659.
- 9.Gavagan, J. E., S. K. Fager, R. D. Fallon, P. W. Folsom, F. E. Herkes, A. Eisenberg, E. C. Hann, and R. DiCosimo. 1998. Chemoenzymatic production of lactams from aliphatic α,ω-dinitriles. J. Org. Chem. 63:4792-4801. [Google Scholar]
- 10.Harper, D. B. 1977. Microbial metabolism of aromatic nitriles: enzymology of the C-N cleavage by Nocardia sp. (rhodochrous group) N.C.I.B. 11216. Biochem. J. 165:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper, D. B. 1977. Fungal degradation of aromatic nitriles: enzymology of C-N cleavage by Fusarium solani. Biochem. J. 167:685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakeya, H., N. Sakai, A. Sano, M. Yokoyama, T. Sugai, and H. Ohta. 1991. Microbial hydrolysis of 3-substituted glutaronitriles. Chem. Lett. 1991:1823-1824.
- 13.Kapp, R., S. E. Stevens, Jr., and J. L. Fox. 1975. A survey of available nitrogen sources for the growth of the blue-green alga, Agmenellum quadruplicatum. Arch. Microbiol. 104:135-138. [DOI] [PubMed] [Google Scholar]
- 14.Klibanov, A. M. 2001. Improving enzymes by using them in organic solvents. Nature 409:241-246. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, M., H. Komeda, N. Yanaka, T. Nagasawa, and H. Yamada. 1992. Nitrilase from Rhodococcus rhodochrous J1: sequencing and overexpression of the gene and identification of an essential cysteine residue. J. Biol. Chem. 267:20746-20751. [PubMed] [Google Scholar]
- 16.Kobayashi, M., T. Nagasawa, and H. Yamada. 1989. Nitrilase of Rhodococcus rhodochrous J1: purification and characterization. Eur. J. Biochem. 182:349-356. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, M., and S. Shimizu. 1994. Versatile nitrilases: nitrile-hydrolysing enzymes. FEMS Microbiol. Lett. 120:217-224. [Google Scholar]
- 18.Kobayashi, M., N. Yanaka, T. Nagasawa, and H. Yamada. 1990. Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J. Bacteriol. 172:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, M., N. Yanaka, T. Nagasawa, and H. Yamada. 1990. Monohydrolysis of an aliphatic dinitrile compound by nitrilase from Rhodococcus rhodochrous K22. Tetrahedron 46:5587-5590. [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Layh, N., B. Hirrlinger, A. Stolz, and H.-J. Knackmuss. 1997. Enrichment strategies for nitrile-hydrolysing bacteria. Appl. Microbiol. Biotechnol. 47:668-674. [Google Scholar]
- 22.Layh, N., A. Stolz, S. Förster, F. Effenberger, and H.-J. Knackmuss. 1992. Enantioselective hydrolysis of O-acetylmandelonitrile to O-acetylmandelic acid by bacterial nitrilases. Arch. Microbiol. 158:405-411. [Google Scholar]
- 23.Layh, N., and A. Willetts. 1998. Enzymatic nitrile hydrolysis in low water systems. Biotechnol. Lett. 20:329-331. [Google Scholar]
- 24.Lévy-Schil, S., F. Soubrier, A.-M. Crutz-Le Coq, D. Faucher, J. Crouzet, and D. Pétré. 1995. Aliphatic nitrilase from a soil isolated Comamonas testosteroni sp.: gene cloning and overexpression, purification, and primary structure. Gene 161:15-20. [DOI] [PubMed] [Google Scholar]
- 25.Martinková, L., and V. Kren. 2002. Nitrile- and amide-converting microbial enzymes: stereo-, regio- and chemoselectivity. Biocatal. Biotrans. 20:79-93. [Google Scholar]
- 26.Mathew, C. D., T. Nagasawa, M. Kobayashi, and H. Yamada. 1988. Nitrilase-catalyzed production of nicotinic acid from 3-cyanopyridine in Rhodococcus rhodochrous J1. Appl. Environ. Microbiol. 54:1030-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagasawa, T., J. Mauger, and H. Yamada. 1990. A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Eur. J. Biochem. 194:765-772. [DOI] [PubMed] [Google Scholar]
- 28.Neilson, A. H., and T. Larsson. 1980. The utilization of organic nitrogen for growth of algae: physiological aspects. Physiol. Plant 48:542-553. [Google Scholar]
- 29.Osswald, S., H. Wajant, and F. Effenberger. 2002. Characterization and synthetic application of recombinant AtNIT1 from Arabidopsis thaliana. Eur. J. Biochem. 269:680-687. [DOI] [PubMed] [Google Scholar]
- 30.Pace, H. C., and C. Brenner. 2001. The nitrilase superfamily: classification, structure and function. Genome Biol. 2:0001.1-0001.9. [Online.] [DOI] [PMC free article] [PubMed]
- 31.Piotrowski, M., S. Schönfelder, and E. W. Weiler. 2001. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-alanine hydratase/nitrilase. J. Biol. Chem. 276:2616-2621. [DOI] [PubMed] [Google Scholar]
- 32.Rauscher, K., J. Voigt, I. Wilke, and K.-T. Wilke. 1982. Chemische Tabellen und Rechentafeln für die analytische Praxis 7. Überarbeitete Auflage, Verlag Harri Deutsch, Frankfurt am Main, Germany.
- 33.Rezende, R. P., J. C. T. Dias, V. Ferraz, and V. R. Linardi. 2000. Metabolism of benzonitrile by Cryptococcus sp. UFMG-Y28. J. Basic Microbiol. 40:389-392. [DOI] [PubMed] [Google Scholar]
- 34.Schulze, B. 2002. Hydrolysis and formation of C-N bonds, p. 699-715. In K. Drauz and H. Waldmann (ed.), Enzyme catalysis in organic synthesis, vol. II. Wiley-VCH, Weinheim, Germany.
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson, D. E., R. Feng, F. Dumas, D. Groleau, A. Mihoc, and A. C. Storer. 1992. Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol. Appl. Biochem. 15:283-302. [PubMed] [Google Scholar]
- 37.Stumpp, T., B. Wilms, and J. Altenbuchner. 2000. Ein neues, l-Rhamnose induzierbares Expressionssystem für Escherichia coli. Biospektrum 6:33-36. [Google Scholar]
- 38.Volff, J. N., C. Eichenseer, P. Viell, W. Piendl, and J. Altenbuchner. 1996. Nucleotide sequence and role in DNA amplification of the direct repeats composing the amplifiable element AUD1 of Streptomyces lividans 66. Mol. Microbiol. 21:1037-1047. [DOI] [PubMed] [Google Scholar]
- 39.Vorwerk, S., S. Biernacki, H. Hillebrand, I. Janzik, A. Müller, E. W. Weiler, and M. Piotrowski. 2001. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212:508-516. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, K., I. Fujimatsu, and K.-I. Komatsu. 1992. Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J. Ferment. Bioeng. 73:425-430. [Google Scholar]
- 41.Yamamoto, K., and K.-I. Komatsu. 1992. Purification and characterization of nitrilase responsible for the enantioselective hydrolysis from Acinetobacter sp. AK 226. Agric. Biol. Chem. 55:1459-1466. [PubMed] [Google Scholar]