Abstract
The nucleotide sequence of the biphenyl catabolic transposon Tn4371 has been completed and analyzed. It confirmed that the element has a mosaic structure made of several building blocks. In addition to previously identified genes coding for a tyrosine recombinase related to phage integrases and for biphenyl degradation enzymes very similar to those of Achromobacter georgiopolitanum KKS102, Tn4371 carries many plasmid-related genes involved in replication, partition, and other, as-yet-unknown, plasmid functions. One gene cluster contains most of the genes required to express a type IV secretion-mating pair formation apparatus coupled with a TraG ATPase, all of which are related to those found on IncP and Ti plasmids. Orthologues of all Tn4371 plasmid-related genes and of the tyrosine recombinase gene were found, with a very similar organization, in the chromosome of Ralstonia solanacearum and on the yet-to-be-determined genomic sequences of Erwinia chrysanthemi and Azotobacter vinelandii. In each of these chromosomal segments, conserved segments were separated by different groups of genes, which also differed from the Tn4371 bph genes. The conserved blocks of genes were also identified, in at least two copies, in the chromosome of Ralstonia metallidurans CH34. Tn4371 thus appears to represent a new family of potentially mobile genomic islands with a broad host range since they reside in a wide range of soil proteobacteria, including plant pathogens.
Tn4371 is a 55-kb transposable element, which allows its host to degrade biphenyl and 4-chlorobiphenyl. It was isolated after a mating between Ralstonia sp. strain A5 (a strain resembling Ralstonia oxalatica [P. Vandamme, unpublished data]) carrying the broad-host-range conjugative plasmid RP4 and Ralstonia metallidurans CH34. Selection was applied for transconjugants that expressed the heavy metal resistances from CH34 and grew with biphenyl as a sole source of carbon and energy (31). This provided transconjugants, which carried an RP4 plasmid with a 55-kb insert near its tet resistance operon. The insert was shown to transpose to other locations and hence was called Tn4371 (20, 30, 31). Previous partial sequencing revealed a modular structure formed of groups of genes, which have orthologues in widely divergent bacteria and mobile elements. These include (i) an int gene encoding a tyrosine recombinase of the same family as many bacteriophage, conjugative transposons, and pathogenicity island integrases; (ii) biphenyl catabolic genes (bph) very similar in organization and nucleotide sequence to the bph gene cluster characterized in Achromobacter georgiopolitanum KKS102 (formerly named Pseudomonas sp. strain KKS102 [16]); and (iii) at least one RP4- and Ti-plasmid-like gene, trbI (20), involved in the formation of conjugating mating pairs.
Tn4371 transposition most likely involves a site-specific excision/integration process since the ends of the element can be detected covalently bound (20). In the CH34 chromosome and on the pMOL30 plasmid of that strain, transposition is targeted to a low number of sites, as it is the case on the RP4 plasmid where 2 sites were identified so far. The main target site in RP4 consists of a 5′-TTTTTCAT-3′ sequence, which is also present between the covalently joined ends of the transposon (20).
We now report the complete nucleotide sequence of Tn4371, which was found to contain several plasmid-related genes, in addition to a complete type IV secretion gene cluster and an orthologue of the TraG motor protein responsible for DNA transfer during conjugation (see reference 9 for a short review). Comparison of the whole Tn4371 sequence with both complete bacterial genomic sequences and bacterial genomic sequences still being determined revealed the presence in Ralstonia solanacearum (27), R. metallidurans (http://www.jgi.doe.gov/JGI_microbial/html/ index.html), Azotobacter vinelandii (NZ_AAAD01RO000088), and Erwinia chrysanthemi (http://tigrblast.tigr.org/ufmg/index.cgi?database =e_chrysanthemi|seq) of chromosomal regions closely related in their sequence and organization to several segments of Tn4371, including the left and right regions flanking the bph genes, but in which the bph gene cluster was in all cases replaced by a different set of open reading frames (ORFs).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and chemicals.
The plasmids used in the present study derive from pLAFR3 (32) (pECG212, pECG236, and pECG293), pBluescript II SK(+) (Stratagene) (pECG319, pECG316, pECG328, pECG327, pECG332, pECG344, pECG346, pECG317, and pECG345), or pDrive (Qiagen) (pDrive2kbTn4371 and pDrive0.3kbTn4371). The first plasmids were isolated from a cosmid library built by cloning fragments resulting from a partial SauIIIA digestion of RP4::Tn4371 in pLAFR3. The pECG300 series contain DNA fragments resulting from PstI digestion of appropriate cosmid clones (19). The origin of the pDrive series is described below. L broth (17) was used as a rich medium to grow Escherichia coli containing the plasmids, at 37°C. Where needed, tetracycline, ampicillin, and chloramphenicol were added to the broth at 20, 50, and 30 mg liter−1, respectively.
Sequencing strategy.
DNA for sequencing was isolated by Qiagen plasmid Midi kit (Qiagen). PstI fragments from characterized members of a Tn4371 library in pLAFR3 (19) were sequenced by primer walking from both ends after a cloning step in vector pBluescript II SK(+). Junctions between the fragments present on pECG319 and pECG316 and between fragments present on pECG316 and pECG328 were sequenced by primer walking on cosmid clones pECG212, pECG293, and pECG236, starting from primer sequences chosen on the basis of the sequences obtained for the pBluescript clones. Two remaining gaps between fragments present on pECG317 and pECG345 and between fragments present on pECG345 and pECG322 were sequenced after the corresponding region had been amplified by PCR by using the following primers: pECG317fw (5′-GCAATCAGATGTACCTCGATGC-3′) and pECG345rv (5′-TGGTCAGCTTGAACTCGATCAG-3′) for the pECG317-pECG345 gap and pEG345fw (5′-CTTGTCATCACCACAAGCCG-3′) and pEG322rv (5′-AACTGGATGTAGACCTTCTGGCC-3′) for the pECG345-pECG322 gap. Amplification was performed after a short denaturation cycle of 3 min at 95°C by using 35 cycles as follows: 95°C for 20 s, 58°C for 20 s, and 72°C for 30 s for the 0.3-kb fragment and 2 min for the 2-kb fragment, with a final elongation cycle at 72°C for 10 min. The PCR products were then cloned into the pDrive cloning vector from the Qiagen PCR cloning kit. The generated pDrive2kbTn4371 and pDrive0.3kbTn4371 were used for sequencing by primer walking. Sequencing reactions were performed with the BigDye terminator sequencing master mix (Applied Biosystems). Sequences available under accession numbers AJ012075, Y10831, X97984, X98271, and Y10832 were included where appropriate. Nucleotide sequencing was performed either on a Pharmacia ALF or an ABI 310 genetic analyzer from Applied Biosystems or by GenomExpress (Grenoble, France). The complete Tn4371 sequence was annotated by using the iAnt environment (27) and is available under accession no. AJ536756 and at http://graton.ulb.ac.be/Tn4371/. Preliminary sequence data for E. chrysanthemi was obtained from The Institute for Genomic Research (http://www.tigr.org) and from a collaborative annotation effort (8). Sequence data for R. metallidurans CH34 are available online (http://www.jgi.doe.gov/JGI_microbial/html/ralstonia /ralston_homepage.html/). Comparisons between Tn4371 ORF products and these unfinished sequences were performed by using local TBLASTN access (2).
RESULTS
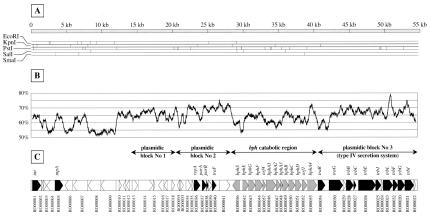
The nucleotide sequence of Tn4371 was completed, analyzed, and annotated as described in Materials and Methods. It consists of 54,657 bp. As previously reported, it is flanked by an 8-bp direct repeat (5′-TTTTTCAT-3′), which might be part of its site-specific recombination site (20). A restriction digest simulated from the sequence (Fig. 1A) confirmed the previous experimental observation that restriction sites tend to cluster in two different regions (0 to 12 kb and 20 to 40 kb on the map), suggesting some heterogeneity in Tn4371 DNA composition (19). The average Tn4371 percent G+C (%GC) content was 63.5% (molar ratio) close to that found in the genus Ralstonia (63.8 to 68.3%) but was not uniform (10). Figure 1B shows the %GC calculated in a 500-bp window moved along the sequence by 10-bp steps. The %GC fluctuated between 51.2 and 78.6%, appearing as a succession of peaks sitting on a basal platform. The platform stood at 54% from 0 to 12 kb, 63% from 12 to 24 kb, 67% from 24 to 28 kb, 60% from 28 to 41 kb, and 65% from 41 to 54 kb, likely reflecting the trace of building blocks of different origins that came together to constitute Tn4371.
FIG. 1.
Tn4371 Characteristics. (A) Conceptual restriction map based on the complete nucleotide sequence of Tn4371. (B) %GC calculated from a 500-bp window moved along the sequence by 10-nucleotide steps. (C) Predicted genetic organization of Tn4371. Gray arrows represent the bph gene cluster, black arrows indicate ORFs sharing homology with known genes, and white arrows are used for the other genes.
Fifty-three ORFs were identified (using the FrameD software included in the iANT annotation package, Fig. 1C). Functional assignment was possible for 20 of them, based on their similarity with known proteins in the National Center for Biotechnology Information (NCBI) database. All of the others but four had an orthologue of unknown function in the database. Details are available online at http://graton.ulb.ac.be/Tn4371/.
Comparison with the sequences in the databases further demonstrated, to the right of the previously sequenced left end of Tn4371 (defined as 5′-TTTTTCAT-3′ and the integrase-tyrosine recombinase gene int) the presence of plasmid related genes (orfRO00013 and -14; repA, parAB, and traF; and the traG-trb gene cluster) organized in three conserved blocks (orfRO00013 to -18, orfRO00055 and -33 to -41, orfRO00054, and all of the ORFs to its right). The first set included two conceptual proteins similar to proteins of unknown functions encoded by genes located near the transfer origin of E. coli plasmid F (Q9WTE4 and Q9S4W2). The genes located between int and orfRO00013 could not be assigned with any function, except for an insertion sequence (IS) element transposase (orfRO0005) closely related to R. metallidurans IS1090 (GenBank accession no. AJ010060) and R. solanacearum ISRso7 of the IS256 family (see the IS database at http://www-is.biotoul.fr/). Alignments with the related IS suggested that this one was truncated. The second set of Tn4371 plasmid-related genes contained ORFs whose translated products were, respectively, related to (i) the RepA protein of Erwinia stewartii plasmid pSW500 (GenBank accession no. S65577), Pseudomonas aeruginosa plasmid pVS1 (GenBank accession no. BAA96327), and plasmid pEMT8 (GenBank accession no. CAC94910) isolated from a polluted environment; (ii) a ParA partition protein of the type Ib family (7) and its associated ParB protein, whose AUG start codon overlapped the ParA UGA stop codon by one base; and (iii) the conjugation protein TraF (one of the pilus assembly proteins) of IncP plasmids. The third and largest cluster of plasmid related genes mapped to the right of the bph gene cluster. Their translated products were very similar to the so called T4CP, i.e., type IV coupling proteins TrwB/TraG/VirD4 (4a) and to proteins of the mating-pair formation (mpf) apparatus, related to the type IV secretion system, from plasmids RP4, R388, and Ti. The mpf genes were named trbB-I according to their orthologues in RP4 despite their different organization [BC(D)EFG(H)IJ(K)L in RP4 and BCEJLFGI in Tn4371].
As shown in Fig. 1B and C, the successive platforms in GC content aligned with the different functional blocks, supporting the hypothesis that these originated from various sources and were brought together through successive rounds of recombination events, most likely via horizontal transfer.
Related gene clusters in other bacterial chromosomes.
Sequence comparison with the genome sequence of the phytopathogenic bacterium R. solanacearum revealed, on its chromosome, four blocks of genes with a significant level of similarity and the same organization as in Tn4371 (27) (see also Fig. 2). Comparison with available bacterial genome sequences, including some currently being determined, uncovered further related chromosomal segments. Some were found in R. metallidurans CH34, split over two or more as-yet-undetermined contigs (contigs 696, 668, 600, 620, 573, 185, and 373 [http://www.jgi.doe.gov/JGI_microbial/html/ralstonia/ralston _homepage.html]). One single segment in the E. chrysanthemi 3937 and one in the A. vinelandii chromosome (GenBank accession no. NZ_AAAD01000088, ORFs avin3078 to avin3126), contained an int-related gene at one end and the same three sets of conserved plasmid-related genes with the same relative organization (Fig. 2). The int gene was in all cases followed by nonconserved ORFs. The E. chrysanthemi segment missed the parAB related ORFs. It carried orthologues (Ech00008 and -9) of plasmid F genes coding for the poison antidote proteins CcdA and CcdB (18) at about the same position as those of orfRO00002 and orfRO00003 in Tn4371. In view of their size and organization, these orthologues could encode the products of another poison antidote system. Possible orthologues of RadC lay in more or less the same position in the R. solanacearum, E. chrysanthemi, A. vinelandii, and R. metallidurans contig C668, although in the latter case it was split by several int-related ORFs. In all of the DNA segments that carried a trb gene cluster, it was flanked by two additional conserved ORFs. The one on the left was an orthologue of bphR in Tn4371, despite the absence of the other bph genes in the other sequences.
FIG. 2.
Genetic organization and alignment of Tn4371 and related chromosomal islands. The GenBank accession numbers and references for the sequences used to build the figure are in the text. For R. metallidurans only unassembled contigs were available for comparison. The presence (or absence) of a type IV secretion gene cluster in the vicinity of contig 668 and 696 remains to be tested experimentally. Contigs 636, 620, 573, and 373 are represented in continuity, separated by spacers because PCR experiments have confirmed their linkage, although the gaps remain to be sequenced (Monchy and Mergeay, unpublished). Orthologues are represented in the same color. White arrows represent ORFs, which have no orthologue in the other islands. Space between genes does not reflect any discontinuity unless stated otherwise. In a few cases, one orthologue appeared split in two: traR in E. chrysanthemi (Ech0023 and Ech00024), radC in R. metallidurans contig 668, and trbI in A. vinelandii (Avin3123 and Avin3124). This might result from sequencing errors since these sequencing projects are in a preliminary phase. Sequence comparison between orthologous regions was performed by using NCBI BLAST (http://www.ncbi.nlm.nih.gov:80/BLAST/) or the sequence comparison tools at the Microbial Genome Database website (http: //mbgd.genome.ad.jp/).
Orthologues of many of the above-mentioned genes were also found in Mesorhizobium loti (GenBank accession no. NC_002682; see also Fig. 2) in the region of the pMLb plasmid that carries nodulation genes. However, their relative positions were not the same as those in the chromosomal regions considered so far.
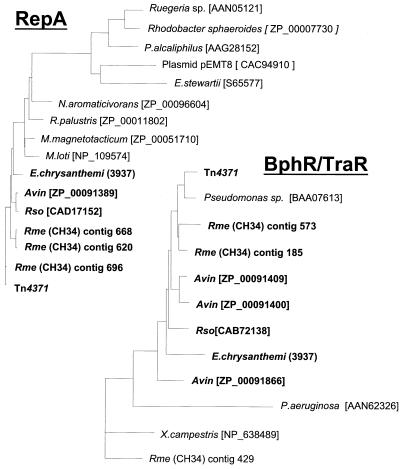
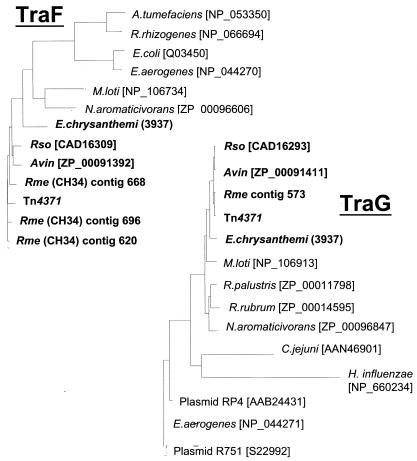
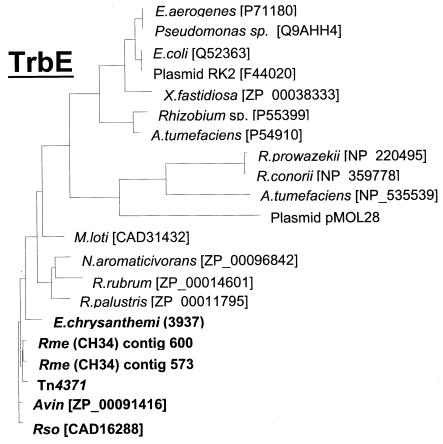
Pairwise comparisons between Tn4371 gene products and their orthologues in all of the bacterial and plasmid sequences mentioned thus far showed that conservation was always stronger between Tn4371, R. metallidurans, R. solanacearum, A. vinelandii, and E. chrysanthemi than with the next-closest relative in the databases. Clearly, the levels of sequence similarity do not match the taxonomic proximities. In phylogenetic trees built from multiple alignments of these amino acid sequences, each family of orthologues showed very similar clustering of the set of genes from Tn4371, R. metallidurans CH34, R. solanacearum, E. chrysanthemi, and A. vinelandii. The M. loti pMLb-encoded proteins again appeared to be less related based on these criteria (see Fig. 4 for a few examples). This did not hold true for the E. chrysanthemi integrases, which were, respectively, ca. 12 and 15% identical to their orthologues.
FIG. 4.
Phylogenetic trees of Tn4371 RepA, TraF, TraG, TrbE, TraR, and their orthologues. The phylogenetic trees were built by using the PHYLIP package. Proteins similar to individual Tn4371 ORF products were identified by searching the NCBI protein database by using BLASTP. Multiple alignments in the PHYLIP format were performed by using CLUSTALW software (http://www.infobiogen.fr/services/analyseq/cgi-bin /clustalw_in.pl). The alignments were analyzed with Protdist (PHYLIP package), which uses protein sequences to compute a distance matrix. The distance for each pair of species estimates the total branch length between the two species and was used in the distance matrix programs NEIGHBOR with the Jones et al. (13) model of amino acid change. The Protdist output was then analyzed with the NEIGHBOR program (PHYLIP package), which implements the neighbor-joining method of Saitou and Nei (26) (26) and the UPGMA (unweighted pair-group method with arithmetic averages) method of clustering. NEIGHBOR constructs a tree by successive clustering of lineages, setting branch lengths as the lineages join. The resulting tree was displayed by using Treeview in a phylogram format. Avin, Rme, and Rso stand for A. vinelandii, R. metallidurans, and R. solanacearum, respectively. pMOL28 is one of the two endogenous plasmids of R. metallidurans CH34. Identity levels between Tn4371-encoded proteins and their orthologues from A. vinelandii, R. metallidurans, and R. solanacearum ranged from 39 to 49% identity for the less-conserved TrbJ up to 97% for the most-conserved protein, ParA. It was ca. 80% for most of the proteins. E. chrysanthemi proteins were less conserved. Identity levels were about 60% for the majority of the proteins and ranged from 35 to >80% for the less- and most-conserved ones, TrbJ and RepA, respectively. BphR (BAA07613) belongs to the Achromobacter georgiopolitanum KKS102 bph gene cluster, which is very closely related to that of Tn4371 (19, 22).
In our attempts to define the ends of the potential genomic islands, we found that the Tn4371, R. solanacearum, R. metallidurans CH34 C696 and C668, and A. vinelandii sequences upstream of the int gene could be aligned at the nucleotide level (see Fig. 3). These alignments all included the 5′-TTTTTCAT-3′ sequence, which in Tn4371 is duplicated upon insertion of the transposon in its preferred target site on RP4 (20) and which could constitute one end of the islands. However, the other end of Tn4371 (400 last bp), including the 5′-TTTTTCAT-3′ repeat, had no significant similarity to any of the other genome sequences, even farther away and on either side of the last conserved ORF after trbI. This finding was unexpected since both ends of the islands should bear Int-binding sites and thus present some similarity. Consistent with the low level of similarity between E. chrysanthemi encoded Int proteins and their orthologues on the other islands, no similarity could be detected between their upstream region and the Tn4371 left end. It is thus still difficult to be sure which of the E. chrysanthemi int-related ORFs belongs to that island and to further assign its left end.
FIG. 3.
Alignment of the left ends of Tn4371 and related chromosomal islands. A stretch of 200 bp from the Tn4371 left end was compared to individual genome sequences of E. chrysanthemi 3937 and R. metallidurans CH34 (contigs 668 and 696 [C668 and C696], resepctively), A. vinelandii OP (Avin), and R. solanacearum GMI1000 (TnRso) by using BLASTN. All of the aligned regions are located just to the left of the int orthologues in the islands shown in Fig. 2. E. chrysanthemi 3937 and CH34 contig 636 did not contain any sequence significantly similar to the other islands' left ends. The coordinates indicated are those of the Tn4371 sequence.
DISCUSSION
Completion of the nucleotide sequencing of the biphenyl transposon Tn4371 confirmed the presence of a complete mating-pair formation trb operon, of a traF gene responsible for the processing or circularization of the TrbC pilin protein (14), and of a motor protein TraG related to those of the RP4/Ti family of plasmids. A more surprising finding was the presence, in Tn4371, of additional plasmid-related genes, some of which appear to be involved in replication (repA) and partition (parAB), i.e., functions associated with the maintenance of an extrachromosomal genetic element. Indeed, the combined presence of a site-specific recombinase gene (int) and conjugative transfer machinery rather suggested that Tn4371 might be a conjugative transposon. The conjugative transfer of Tn4371 could, however, never be clearly demonstrated (20). Attempts to integrate the transposon in the chromosome of a plasmid-free derivative of R. metallidurans CH34, the only strain easily amenable to conjugation experiments and in which transposition of Tn4371 can be easily traced experimentally, led to the isolation of a new element, called Tnbph. It carries the Tn4371 right portion, including the complete bph and traG-trbI gene clusters. Tnbph transfers by conjugation between CH34 derivatives (20), between CH34 and Comamonas sp. strain BR60, and between BR60 derivatives (C. Wyndham, unpublished data). The discovery of at least one potentially complete traG-trb gene cluster in the CH34 chromosome (Fig. 2, contigs 573 and 373) now, however, introduces the possibility that conjugative transfer of either Tn4371 or/and Tnbph from CH34 could rely on host genes, for instance, to complement a transfer machinery that is not completely functional. Alternatively, Tnbph could be a recombinant between Tn4371 and homologous chromosomal genes. Experiments are in progress to test that hypothesis. The BR60 genome sequence is not yet available, and thus it is still impossible to draw any conclusions concerning the transfer of Tn4371 between BR60-derived strains. Further investigation is required to clarify this point.
Another puzzling observation is that no ORF encoding for a TraI relaxase and TraK orthologue could be identified on Tn4371. Thus far, these proteins, which organize and nick the DNA at oriT as the first step in conjugative transfer, were always found to be associated with the TraG-TypeIV secretion conjugation apparatus (for a review, see reference 36). Some plasmids use tyrosine recombinases to resolve dimers (1). This could also be the case for Tn4371, which, overall, would then rather appear as a plasmid. In CH34 at least, Tn4371 could not be detected as an autonomously replicating circular entity (20; unpublished results). All of the conclusions drawn thus far for Tn4371 also apply to the other DNA segments analyzed above. In R. solanacearum GMI1000 the island is on the chromosome (27). E. chrysanthemi 3937 and A. vinelandii strain OP (ATCC 13705), a derivative of which was used for the sequencing project, do not host any plasmid (15, 25), and the R. metallidurans CH34 contigs concerned do not belong to either the pMOL28 (J. Dunn, unpublished data) or the pMOL30 (M. Mergeay, unpublished data) plasmid endogenous to that strain. Several hypotheses appear to be reasonable to account for this apparent contradiction. All of the DNA islands discussed above could be plasmids that cannot assume their plasmid mode of life in their present host. However, the high conservation of such a large set of genes with the same organization would appear to be unlikely for a defective genetic element that did not recently undergo selective pressure for its maintenance functions. Alternatively, the plasmid maintenance functions could in these cases rather assume a function related to conjugative transfer, despite the absence of similarity with TraI and TraK. Their conceptual RepA proteins include several Y residues, but none of them is in a context that fits the TraI relaxase consensus sequence (36). In addition to their nicking activity, TraI relaxases have a helicase activity that is also essential for DNA transfer. This helicase activity could here depend on a second protein, for instance, the product encoded by orfRO00008 from Tn4371, which carries domains typical of DNA helicases (PRODOM domains PD460075, PD001658, and PD002094). RepA could conceivably here nick the DNA for transfer initiation and, in conjunction with the helicase, couple it to the TraG protein for DNA transfer through a conjugation pore consisting of Trb proteins. Clearly, the existence of a potential oriT and its location, as well as the elucidation of the mechanism of conjugative transfer of these DNA segments, requires further experimental analysis.
As mentioned earlier, the Tn4371 bphR gene and its orthologue in the very closely related bph gene cluster in Achromobacter georgiopolitanum KKS102 were shown not to be involved in the regulation of the bph operon (21, 22). The presence of orthologues of this gene next to the traG-trb region in all of the DNA segments analyzed here suggests that these orthologues actually regulate the expression of the transfer genes. On plasmids Ti and RK2/RP4, the conjugal transfer genes are regulated in completely different ways: by quorum sensing for Ti (23) and cooperative interactions between TrbA and KorB for RK2/RP4 (L. Bingle, M. Zatyka, S. E. Manzoor, and C. M. Thomas, unpublished data). This most likely correlates with the different life-styles of the bacteria carrying either of those two plasmids. Regulation of the TraG-Trb transfer machinery genes by a LysR regulator would thus represent a third mode of regulation of very similar gene clusters, illustrating, once more, the fact that regulatory genes and the genes they regulate do not necessarily evolve together (5).
A clear modular picture emerges from the comparisons shown in Fig. 2. Very similar traG-trb operons and their potential LysR family regulator are separated from the conserved segment, including the repA and traF genes, by a set of completely unrelated ORFs. Preliminary results (S. Monchy and M. Mergeay, unpublished data) indicate that R. metallidurans CH34 contigs C636, C620, C573, and C373 are contiguous on the chromosome and thus likely to form one island, where the set of nonconserved genes is replaced by an IS1071 insertion sequence. On the other islands that contain C696 and C668 more sequencing is needed in order to determine whether a stretch of 10 to 15 kb of unpredictable nature indeed separates the repA-traF from the traG-trb region.
Other places in the islands seem to be prone to shorter insertions. Two of these, which flank the conserved region that covers Tn4371 orf RO00013 and orfRO00014, include truncated IS elements and serine recombinase encoding genes. These might be the footprints of rearrangements that led to the present structure.
Despite the fact that the mobility of the sequences discussed here has not yet been demonstrated experimentally (except for that of Tn4371), these sequences reside in a wide range of species in the β-proteobacteria (Ralstonia) and γ-proteobacteria (A. vinelandii and E. chrysanthemi), including soil and plant pathogenic strains, suggesting that the conjugation transfer machinery might confer a broad host range, as for the similar machinery of the IncP plasmids.
Tn4371-related sequences have been identified in other biphenyl- and chlorobiphenyl-degrading isolates by using DNA-DNA hybridization with appropriate Tn4371-borne gene probes (31a). A group of β-proteobacteria contained bph genes that are highly similar in organization to those of Tn4371 and KKS102. Three of these bacterial strains were able to transfer their bph genes to R. metallidurans in a mode similar to that of Tn4371; a 50-kb chromosomal fragment carrying the bph genes was, after insertion in RP4, transferred to CH34. The transferred DNA segment of two strains (1C3 and 4A4) showed an extended similarity with Tn4371, to the right of the bph catabolic genes and up to trbI. However, no homology was found with a Tn4371 int probe, which might be related to the fact that Int orthologues tend to be less conserved than Tra and Trb orthologues (except for TrbJ, see the legend to Fig. 4). Strains 1C3 and 4A4, although derived from the same geographical locations as strain A5, might carry Tn4371-related elements with a different int gene but the same bph gene cluster, again pointing toward a reassortment of building blocks.
Nomenclature.
Several generic terms have been proposed for elements that, like those discussed here, carry a site-specific recombinase (usually of the tyrosine recombinase family, but which a priori could just as well be serine recombinases) and a conjugative transfer machinery. The first recognized family of conjugative transposons was that of the gram-positive elements related to Tn916 (6). Their transfer machinery appears to be different from the type IV secretion system, although it clearly bears a similar function. A second family of integrated and conjugative elements was described in the genus Bacteroides (for a review, see reference 28), whose conjugative machinery is still different. Another family, the so-called integrative plasmids, was identified in Streptomyces spp. (29). These elements can sustain either an integrated or an autonomous plasmid state, as do the IncJ family of plasmids in enterobacteria (3). The Streptomyces elements seem to use a mode of conjugative transfer that differs from that of all other conjugative plasmids known today. DNA transfer here would involve a motor protein related to the SpoIIIE family of enzymes (24) and hence would be more related to the double-strand transfer or segregation of the chromosome in the prespore of sporulating gram-positive bacteria such as Bacillus subtilis (35). A generic term was proposed for those elements, ICE (for integrative and conjugative elements) (4). The generic term “genomic island” has been proposed for a wider range of large integrated mobile (or potentially mobile) elements. It is intended to include pathogenicity islands (PAI; see, for instance, reference 12), symbiotic islands (genes encoding components of the symbiosis machinery of nitrogen-fixing bacteria) (33) or, as in the case of Tn4371, catabolic islands, i.e., large mobile DNA segments, which encode enzymes that degrade a variety of toxic compounds (34). More general terms such as “fitness islands” or “ecological islands” have also been proposed (11), none of which is appropriate here since no experimental evidence is yet available to support a “fitness” or “ecological” role for the islands described. We thus called them “genomic islands.” This term, we think, should be more widely used for a general description of mobile entities located on chromosomes, especially in the annotation of bacterial genomes. Genomic islands should include elements that can move between strains either by conjugation, in which case they will be ICEs; by means of transduction by a phage, in which case they will be a defective prophage; or by transposition onto another mobile element, plasmid, or phage, in which case they would be a transposon. Provided their capacity to transfer by conjugation can be demonstrated (studies are currently under way for the E. chrysanthemi element), the elements described here would thus most likely fit among the ICEs.
Acknowledgments
We thank Jerome Gouzy and Christian Boucher for contributions to the transfer of the iANT package in Brussels and for providing access to unpublished results, Andrew Coulson for his help with the analysis of Tn4371 GC content, and Arlette Michaux and Annemie Ryngaert for technical assistance.
This work was supported by the Belgian Fonds de la Recherche Fondamentale Collective, the Fonds de la Recherche Scientifique Médicale, a TOURNESOL exchange grant from the Belgian CGRI and the French CNRS, and a grant from the Convention between the Université Libre de Bruxelles and SCK/CEN. A.T. is Directeur de Recherche from the Fonds National de la Recherche Scientifique (Belgium).
REFERENCES
- 1.Abremski, K. E., and R. H. Hoess. 1992. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 5:87-91. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Boltner, D., C. MacMahon, J. T. Pembroke, and A. M. Osborn. 2002. A conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 4a.Cabezon, E., J. I. Sastre, and F. de la Cruz.1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo, V., and J. Perez-Martin. 1996. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol. 19:1177-1184. [DOI] [PubMed] [Google Scholar]
- 6.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 8.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomis-Ruth, F. X., F. de la Cruz, and M. Coll. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res. Microbiol. 153:199-204. [DOI] [PubMed] [Google Scholar]
- 10.Goris, J., P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters, and P. Vandamme. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands, and bacterial pathogenicity: a Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentschel, U., and J. Hacker. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545-548. [DOI] [PubMed] [Google Scholar]
- 13.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biol. Sci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 14.Kalkum, M., R. Eisenbrandt, R. Lurz, and E. Lanka. 2002. Tying rings for sex. Trends Microbiol. 10:382-387. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, C., P. Rudnick, M. L. MacDonald, and T. Melton. 2001. Genus III, Azotobacter Beijerinck 1901. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 16.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 18.Maki, S., S. Takiguchi, T. Miki, and T. Horiuchi. 1992. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins: antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J. Biol. Chem. 267:12244-12251. [PubMed] [Google Scholar]
- 19.Merlin, C., D. Springael, M. Mergeay, and A. Toussaint. 1997. Organisation of the bph gene cluster of transposon Tn4371 encoding enzymes for the degradation of biphenyl and 4-chlorobiphenyl. Mol. Gen. Genet. 253:499-506. [DOI] [PubMed] [Google Scholar]
- 20.Merlin, C., D. Springael, and A. Toussaint. 1999. Tn4371: a modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway and RP4/Ti-like transfer. Plasmid 41:40-54. [DOI] [PubMed] [Google Scholar]
- 21.Mouz, S., C. Merlin, D. Springael, and A. Toussaint. 1999. A GntR-like negative regulator of transposon Tn4371 biphenyl degradation genes. Mol. Gen. Genet. 262:790-799. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsubo, Y., Y. Nagata, K. Kimbara, M. Takagi, and A. Ohta. 2000. Expression of the bph genes involved in biphenyl/PCB degradation in Pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene 256:223-228. [DOI] [PubMed] [Google Scholar]
- 23.Piper, K., S. Beck von Bodman, and S. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 24.Possoz, C., C. Ribard, J. Gagnat, J. L. Pernodet, and M. Guerineau. 2001. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 42:159-166. [DOI] [PubMed] [Google Scholar]
- 25.Reverchon, S., N. Hugouvieux-Cotte-Pattat, and J. Robert-Baudouy. 1985. Cloning of genes encoding pectolytic enzymes from a genomic library of the phytopathogenic bacterium, Erwinia chrysanthemi. Gene 35:121-130. [DOI] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, T. S. Mangeno, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and B. C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 28.Salyers, A. A., A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonet, J. M., F. Boccard, J. L. Pernodet, J. Gagnat, and M. Guerineau. 1987. Excision and integration of a self-transmissible replicon of Streptomyces ambofaciens. Gene 59:137-144. [DOI] [PubMed] [Google Scholar]
- 30.Springael, D., L. Diels, and M. Mergeay. 1994. Transfer and expression of PCB-degradative genes into heavy metal resistant Alcaligenes eutrophus strains. Biodegradation 5:343-357. [DOI] [PubMed] [Google Scholar]
- 31.Springael, D., S. Kreps, and M. Mergeay. 1993. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J. Bacteriol. 175:1674-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Springael, D., A. Ryngaert, C. Merlin, A. Toussaint, and M. Mergeay. 2001. Occurrence of Tn4371-related mobile elements and sequences in (chloro)biphenyl-degrading bacteria. Appl. Environ. Microbiol. 67:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Meer, J. R., R. Ravatn, and V. Sentchillo. 2001. The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch. Microbiol. 175:785-799. [DOI] [PubMed] [Google Scholar]
- 35.Wu, L. J., P. J. Lewis, R. Allmansberger, P. M. Hauser, and J. Errington. 1995. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 9:1316-1326. [DOI] [PubMed] [Google Scholar]
- 36.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and Z. M. Zatika. 2002. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.