Abstract
Over 13 months, 465 beavers, foxes, muskrats, otters, and raccoons were trapped in four counties in eastern Maryland and examined by molecular methods for microsporidia. A two-step nested PCR protocol was developed to amplify a 392-bp fragment of the internal transcribed spacer region of the rRNA gene of Enterocytozoon spp., with the use of primers complementary to the conserved regions of published nucleotide sequences. Fifty-nine PCR-positive samples were sequenced. Multiple alignments of these sequences identified 17 genotypes of Enterocytozoon spp. (WL1 to WL17); of these, 15 have not been reported before. Most of the genotypes were found in multiple species of wildlife and belonged to a major group consisting of all the previously described Enterocytozoon bieneusi genotypes from human and domestic animals. Some of the isolates from muskrats and raccoons formed two distinct groups. Results of this study indicate that fur-bearing mammals, especially those closely associated with surface water, can be a potential source of human-pathogenic E. bieneusi. However, there are also host-adapted Enterocytozoon genotypes in wildlife, which may represent species different from E. bieneusi and have no apparent public health significance. This is the first report of E. bieneusi in wildlife.
Microsporidia are obligate, intracellular parasites representing more than 1,200 species in 143 genera that infect invertebrate and vertebrate hosts. At least six genera including 14 species have been reported to infect humans (10). Enterocytozoon bieneusi is recognized as the most common microsporidian in humans, even though little is known of reservoirs or routes of infection (10, 15). In recent surveys of parasites of domestic animals, E. bieneusi was isolated from farm and companion animals including cattle, pigs, dogs, and a cat (1, 2, 5, 13, 15, 16). E. bieneusi was also isolated from simian immunodeficiency virus-infected immunodeficient macaques (3) and more recently from chickens (14). Spores of E. bieneusi have been detected in water from the Seine River in France, and PCR techniques have been developed for the detection of E. bieneusi in water samples (9, 19; J. M. Sparfel, C. Sarfati, O. Liguory, B. Caroff, N. Dumoutier, B. Gueglio, E. Billaud, F. Raffi, J. M. Molina, M. Miegeville, and F. Derouin, abstract, J. Eukaryot. Microbiol. 44:78S, 1997). Based on an epidemiological study in France, lake water contamination was suspected as the source of a possible outbreak of microsporidiosis involving 200 persons (4). Thus far, E. bieneusi is the only described species in the genus Enterocytozoon, although it is not clear whether the genus is truly monospecific.
The possibilities that some isolates of E. bieneusi might result in zoonotic infection under certain circumstances and that water might serve as a vehicle for dissemination of spores have led to the present study, in which fur-bearing animals, closely associated with surface waters, were trapped and their feces were examined for the presence of microsporidian spores. In this communication, the development of a two-step nested PCR protocol to amplify the internal transcribed spacer (ITS) region of the rRNA gene and nucleotide sequence characterizations of the amplified ITS fragments are described. Results of the study revealed the existence of extensive genetic diversity in E. bieneusi and related parasites, and some of the wildlife genotypes have zoonotic potential whereas others may represent species different from E. bieneusi. The data should be useful in understanding the taxonomy of Enterocytozoon spp., developing alternative molecular tools for the detection and differentiation of Enterocytozoon spp., and investigating outbreaks or endemic diseases caused by E. bieneusi.
MATERIALS AND METHODS
Isolates.
From January 2001 through January 2002 trappers licensed in the state of Maryland shipped feces from 85 beavers, 67 foxes, 239 muskrats, 19 otters, and 55 raccoons to the Animal Waste Pathogen Laboratory, Agriculture Research Service, U.S. Department of Agriculture, Beltsville, Md. Feces were obtained from animals trapped in streams and lowland areas of Caroline, Charles, Dorchester, and Talbot counties, with age and sex of animals recorded. The age of animals was estimated by body measurements (length and weight) and, in the case of otters, also by inspection of canine teeth. Feces collected directly from the colon of each animal were placed in an individual plastic specimen carton with a lid identifying the animal source, age, sex, date, and location of capture (Table 1). Feces were held under refrigeration without chemical preservatives for approximately 1 to 2 weeks before shipping.
TABLE 1.
Sources of Enterocytozoon-positive fecal samples and their genotype designation at the ITS region of the rRNA gene
| Specimen no. | Animal no. | Host | Age (mo) | Sexa | Genotype | Reference (genotype/host/GenBank accession no.) |
|---|---|---|---|---|---|---|
| 5496 | 45 | Raccoon | 30 | M | WL1 | This report |
| 5553 | 48 | Raccoon | 12 | F | WL1 | |
| 5972 | 53 | Raccoon | 30 | M | WL1 | |
| 3624 | 59 | Raccoon | 24 | M | WL1 | |
| 3608 | 57 | Raccoon | 30 | M | WL2 | This report |
| 3609 | 58 | Raccoon | 24 | M | WL2 | |
| 3603 | 56 | Raccoon | 30 | M | WL3 | This report |
| 3548 | 23 | Muskrat | 36 | F | WL4 | This report |
| 3570 | 25 | Muskrat | 30 | M | WL4 | |
| 3578 | 26 | Muskrat | 24 | M | WL4 | |
| 3633 | 27 | Muskrat | 24 | M | WL4 | |
| 3666 | 29 | Muskrat | 36 | F | WL4 | |
| 3713 | 30 | Muskrat | 30 | M | WL4 | |
| 3714 | 31 | Muskrat | 36 | M | WL4 | |
| 5999 | 41 | Muskrat | 24 | M | WL5 | This report |
| 5540 | 34 | Muskrat | 12 | F | WL6 | This report |
| 3599 | 1 | Beaver | 18 | M | WL7 | This report |
| 5514 | 4 | Beaver | 6 | F | WL8 | D/Human/AF101200 (5), |
| 5539 | 6 | Beaver | 12 | M | WL8 | PigEBITS9/Pig/AF34877 (2) |
| 5573 | 7 | Beaver | 6 | M | WL8 | |
| 6009 | 12 | Beaver | 24 | F | WL8 | |
| 3568 | 24 | Muskrat | 18 | M | WL8 | |
| 3665 | 28 | Muskrat | 36 | F | WL8 | |
| 5997 | 54 | Raccoon | 30 | M | WL8 | |
| 5973 | 14 | Fox | 30 | M | WL8 | |
| 5977 | 15 | Fox | 12 | F | WL8 | |
| 6003 | 11 | Beaver | 30 | M | WL9 | This report |
| 5489 | 32 | Muskrat | 12 | F | WL10 | This report |
| 6005 | 22 | Fox | 30 | M | WL11 | This report |
| 5505 | 3 | Beaver | 30 | F | WL12 | This report |
| 3667 | 43 | Otter | 24 | F | WL12 | |
| 5536 | 5 | Beaver | 12 | F | WL13 | EbpC/Pig/AF076042 (1), E/Pig/AF135832 (5) |
| 5969 | 8 | Beaver | 36 | F | WL13 | |
| 5971 | 9 | Beaver | 42 | M | WL13 | |
| 6000 | 10 | Beaver | 30 | F | WL13 | |
| 6011 | 13 | Beaver | 12 | M | WL13 | |
| 5980 | 17 | Fox | 24 | M | WL13 | |
| 5987 | 20 | Fox | 24 | M | WL13 | |
| 5993 | 21 | Fox | 24 | M | WL13 | |
| 5576 | 37 | Muskrat | 18 | F | WL13 | |
| 5966 | 40 | Muskrat | 12 | F | WL13 | |
| 6004 | 42 | Muskrat | 12 | M | WL13 | |
| 5502 | 44 | Otter | 24 | F | WL13 | |
| 5570 | 52 | Raccoon | 12 | M | WL13 | |
| 5527 | 33 | Muskrat | 18 | M | WL14 | This report |
| 5497 | 2 | Beaver | 30 | M | WL15 | This report |
| 5778 | 16 | Fox | 12 | F | WL15 | |
| 5985 | 18 | Fox | 18 | M | WL15 | |
| 5986 | 19 | Fox | 24 | F | WL15 | |
| 5541 | 35 | Muskrat | 12 | F | WL15 | |
| 5550 | 47 | Raccoon | 12 | M | WL15 | |
| 5568 | 50 | Raccoon | 12 | F | WL15 | |
| 5562 | 36 | Muskrat | 24 | F | WL16 | This report |
| 5580 | 38 | Muskrat | 12 | M | WL16 | |
| 5581 | 39 | Muskrat | 18 | F | WL16 | |
| 5567 | 49 | Raccoon | 42 | M | WL16 | |
| 6001 | 55 | Raccoon | 30 | M | WL16 | |
| 5517 | 46 | Raccoon | 36 | M | WL17 | This report |
| 5569 | 51 | Raccoon | 12 | F | WL17 |
M, male; F, female.
Upon arrival at the U.S. Department of Agriculture laboratory feces from each carton was transferred into a 50-ml centrifuge tube. Water was added to a final volume of 35 ml, the contents were mixed by vortexing, the slurry was poured through a 7.62-cm-diameter sieve with a pore size of 45 μm, and the sieved suspension was poured into another 50-ml centrifuge tube. Water was added to reach a final volume of 50 ml, and the tube was capped and centrifuged at room temperature (RT) for 10 min at 1,500 × g. Supernatant was decanted, the pellet was resuspended in 25 ml of water, and an additional 25 ml of aqueous CsCl2 (1.4 g/ml) was thoroughly mixed with the suspension. The tube was then centrifuged at RT for 15 min at 300 × g, 2 ml was aspirated from the surface and pipetted into a 15-ml centrifuge tube, 13 ml of deionized water was added, and the tube was centrifuged at RT for 15 min at 1,500 × g. The pellet was washed twice with deionized water, resuspended in 100 μl of deionized water, transferred to a 1.8-ml microcentrifuge tube, and stored at −70°C until 100 or more specimens had accumulated. Tubes were then shipped frozen overnight to the Centers for Disease Control and Prevention laboratory in Atlanta, Ga., for molecular studies.
DNA extraction.
Upon arrival at the Centers for Disease Control and Prevention laboratory, the frozen suspensions of cleaned fecal debris to be tested for the presence of spores of Enterocytozoon spp. were washed three times with distilled water. DNA was extracted from each washed fecal pellet after initial treatment with 1 M KOH for 15 min at 65°C, followed by neutralization with 25% HCl. The DNA lysate was then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) solution and purified using the QIAamp DNA stool kit (Qiagen Inc., Valencia, Calif.) according to the manufacturer's protocol, except that the DNA was resuspended in 100 μl of distilled water.
PCR amplification of the ITS region of the rRNA gene.
To amplify the ITS region of the rRNA gene, a two-step nested PCR protocol was developed, using primers complementary to the conserved published ITS nucleotide sequences of Enterocytozoon spp. downloaded from GenBank: human (AF101197 to AF101200, AF242475, AF242476 to AF242479, AF267145, and AF267147), pig (AF076040 to AF076043, AF135832 to AF135835, AF348469 to AF348477, and U61180), rhesus monkey (AF023245), cattle (AF135836 to AF135837, AF267143, and AF267144), cat (AF267141 to AF267142), llama (AF267146), and farm dogs and cats (AF118144 and AF059610). For the primary PCR, a PCR product of 410 bp was amplified using primers AL4037 (5′-GATGGTCATAGGGATGAAGAGCTT-3′) and AL4039 (5′-AATACAGGATCACTTGGATCCGT-3′). The PCR mixture consisted of 1.0 to 2.0 μl of DNA, 200 μM (each) deoxynucleoside triphosphates, 1× PCR buffer (Perkin-Elmer, Foster City, Calif.), 3.0 mM MgCl2, 5.0 U of Taq polymerase (GIBCO BRL, Frederick, Md.), and 200 nM (each) primers in a total of 100 μl of reaction mixture. The reactions were performed for 35 cycles (94°C for 45 s, 55°C for 45 s, and 72°C for 60 s) in a Perkin-Elmer GeneAmp PCR 9700 thermocycler, with an initial hot start (94°C for 3 min) and a final extension (72°C for 10 min). For the secondary PCR, a fragment of 392 bp was amplified using 2.5 μl of primary PCR and primers AL4038 (5′-AGGGATGAAGAGCTTCGGCTCTG-3′) and AL4040 (5′-AATATCCCTAATACAGGATCACT-3′). The conditions for the secondary PCR were identical to the primary PCR. The PCR products were analyzed by agarose gel electrophoresis and visualized after ethidium bromide staining. Each sample was analyzed by PCR at least twice.
Nucleotide sequencing and phylogenetic analysis.
With the use of Microcon PCR centrifugal filter devices (Millipore, Bedford, Mass.), the secondary PCR products with the expected size were purified and sequenced in both directions using the Big Dye Terminator Cycle Sequencing Ready Reaction kit on an ABI 3100 automated sequencer (Perkin-Elmer). Sequence accuracy was confirmed by sequencing of two separate PCR products. Multiple alignment of the nucleotide sequences was performed using the Wisconsin Package Version 9.0 program (Genetics Computer Group, Madison, Wis.). To assess the extent of genetic diversity and evolutionary relationships among Enterocytozoon genotypes, phylogenetic analysis was performed on the aligned sequences. In this analysis, the published ITS nucleotide sequences representing various E. bieneusi genotypes (1, 2, 3, 5, 13, 14, 15, 16) were aligned with ITS sequences obtained from beavers, muskrats, raccoons, and foxes. Based on the evolutionary distances calculated by the Kimura two-parameter model, a neighbor-joining tree (18) was constructed using the TreeconW program (20). Bootstrap analysis was used to assess the reliability of grouping using 1,000 pseudoreplicates (8). Since bootstrap values may be conservative estimates of the reliability of the clades, values above 70% are reported (7). Maximum likelihood analysis was used to validate the phylogenetic relationship inferred from the neighbor-joining analysis, using the program Phylip implemented in the phylogenetic package DAMBE (http://aix1.uottawa.ca/∼xxia/). The most divergent sequence (Enterocytozoon sp. from a dog; GenBank accession no. AF059610) was used as an outgroup in the phylogenetic analysis.
Nucleotide sequence accession number.
Nucleotide sequences of the ITS region of the rRNA gene of Enterocytozoon spp. from beavers, muskrats, raccoons, and foxes representing different genotypes (WL1 to WL17) were deposited in the GenBank database under accession numbers AY237209 to AY237225.
RESULTS
In order to identify microsporidian infection in all the 465 different fur-bearing wild mammalian hosts (85 beavers, 67 foxes, 239 muskrats, 19 otters, and 55 raccoons), a two-step nested PCR was performed at least twice to amplify the ITS region (392 bp) of the rRNA gene with Enterocytozoon-specific primers. Of the 465 wildlife isolates characterized, 59 were found to be PCR positive.
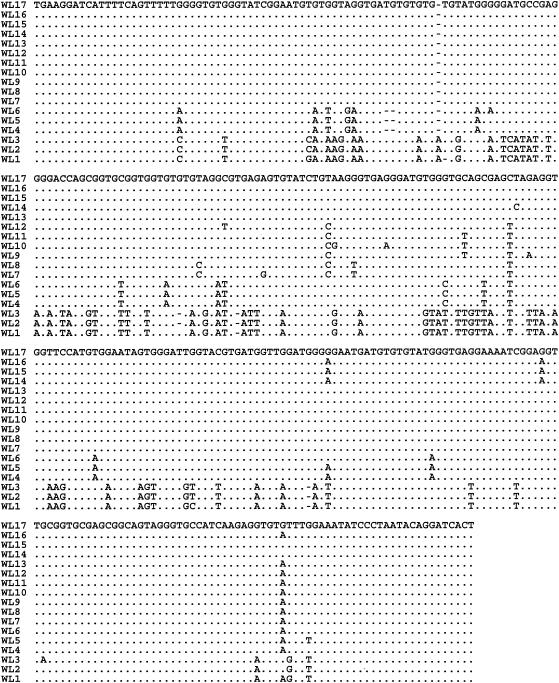
Nucleotide sequences of the ITS were generated for all 59 PCR-positive isolates (9 isolates from foxes, 13 isolates from beavers, 20 isolates from muskrats, 15 isolates from raccoons, and 2 isolates from otters; Table 1). Infected animals varied between 6 and 42 months in age, with more male than female animals being infected (35 versus 24, respectively). The extent of genetic diversity in Enterocytozoon spp. was measured by multiple alignments of the nucleotide sequences of the ITS region of the rRNA gene (Fig. 2). The analysis revealed distinct sequences for some of the isolates from muskrats and raccoons. The analysis further revealed the presence of at least 17 distinct genotypes of Enterocytozoon spp. (WL1 to WL17) within these wildlife isolates.
FIG. 2.
Genetic variation in the ITS region of the rRNA gene of Enterocytozoon spp. Seventeen distinct genotypes (WL1 to WL17) based on these sequences were evident. Dots denote sequence identity to WL17. Dashes denote deletions.
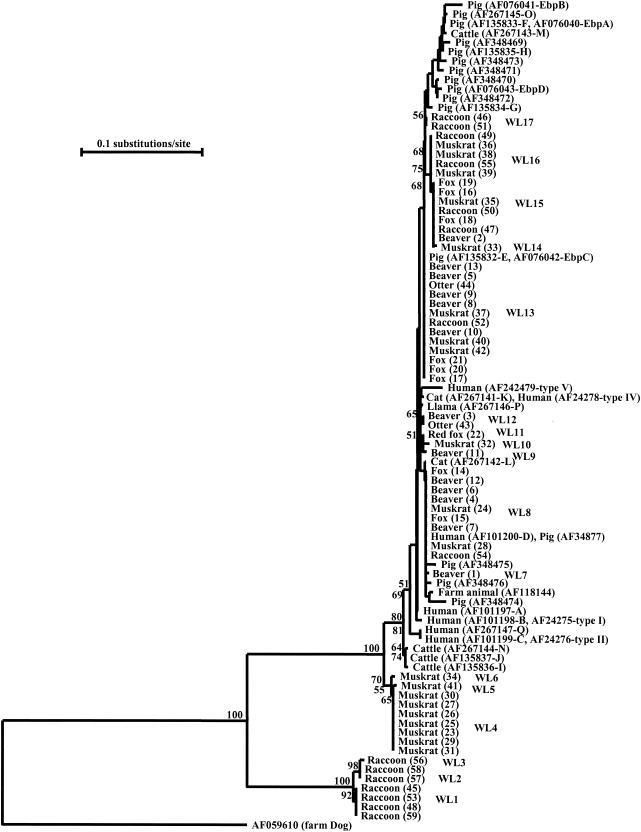
Phylogenetic analysis was performed to understand the genetic relationship among Enterocytozoon genotypes. A neighbor-joining tree was constructed from aligned ITS sequences of various Enterocytozoon spp. isolates from wildlife (present study) and all the published E. bieneusi ITS nucleotide sequences from humans and domestic animals (Table 2; Fig. 1). Three major clusters were evident from these combined data. The first major cluster consisted of all previously reported isolates of E. bieneusi from human, cattle, cat, llama, rhesus, pig, and farm dog and cat sources (1, 2, 5, 11, 13, 15, 16, 17; P. Deplazes, A. Mathis, C. Muller, and R. Weber, abstract, J. Eukaryot. Microbiol. 43:93S, 1996) as well as some isolates from beavers, muskrats, foxes, raccoons, and otters. The second major cluster was formed by some isolates from muskrats. The third major cluster was represented by some raccoon isolates. All major clusters were well supported by bootstrap analysis (Fig. 1).
TABLE 2.
Published E. bieneusi ITS genotypes
| GenBank accession no. | Host(s) | Genotype | Refer- ence(s) |
|---|---|---|---|
| AF101197 | Human | A | 1, 5, 17 |
| AF101198, AF242475 | Human | B, type I | 1, 5, 11, 17 |
| AF101199, AF242476 | Human | C, type II | 1, 5, 11, 17 |
| AF101200, AF023245, AF348477 | Human, macaque, pig | D, PigEBITS9 | 2, 3, 5 |
| AF267147 | Human | Q | 5 |
| AF242477 | Human | Type III | 11 |
| AF242478, AF267141 | Human, cat | K, type IV | 5, 11 |
| AF242479 | Human | V | 11 |
| AF135836 | Cattle | I | 5 |
| AF135837 | Cattle, chicken | J | 5, 14, 15 |
| AF267143 | Cattle | M | 5 |
| AF267144 | Cattle | N | 5 |
| AF267142 | Cat | L | 5 |
| AF118144 | Cat | EbfelA | 13 |
| AF059610 | Farm dog and cat | 13 | |
| AF267146 | Llama | P | 5 |
| AF076040, AF135833 | Pig | EbpA, F | 1, 2, 5 |
| AF135834 | Pig | G | 5, 15 |
| AF135835 | Pig | H | 5, 15 |
| AF267145 | Pig | O | 5 |
| AF076041 | Pig | EbpB | 1 |
| AF076042, AF135832 | Pig | EbpC, E | 1, 5, 15 |
| AF076043 | Pig | EbpD | 1 |
| AF348469 | Pig | PigEBITS1 | 2 |
| AF348470 | Pig | PigEBITS2 | 2 |
| AF348471 | Pig | PigEBITS3 | 2 |
| AF348472 | Pig | PigEBITS4 | 2 |
| AF348473 | Pig | PigEBITS5 | 2 |
| AF348474 | Pig | PigEBITS6 | 2 |
| AF348475 | Pig | PigEBITS7 | 2 |
| AF348476 | Pig | PigEBITS8 | 2 |
FIG. 1.
Phylogenetic relationships of Enterocytozoon spp. inferred from the neighbor-joining analysis of the ITS region of the rRNA gene. Numbers on branches are percent bootstrap values from 1,000 replications.
Within the first major cluster, some isolates (WL8 genotype) from muskrats (isolates 24 and 28), beavers (isolates 4, 6, 7, and 12), foxes (isolates 14 and 15), and raccoon (isolate 54) were identical to the published E. bieneusi ITS sequences from a human (genotype D) and a pig (GenBank accession no. AF101200 and AF34877, respectively) and formed a major clade. Similarly, the ITS sequences (WL13 genotype) from some beavers (isolates 5, 8, 9, 10, and 13), muskrats (isolates 37, 40, and 42), foxes (isolates 17, 20, and 21), and one raccoon (isolate 52) and one otter (isolate 44) were identical to E. bieneusi sequences from pigs (genotypes EbpC and E, GenBank accession no. AF076042 and AF135832, respectively) and formed a second major clade. The third major clade (WL15 genotype) in the cluster contained some E. bieneusi isolates from raccoons (isolates 47 and 50) and foxes (isolates 16, 18, and 19) and one isolate each from muskrat (isolate 35) and beaver (isolate 2). The fourth major clade (WL16 genotype) was formed by some E. bieneusi isolates from raccoons (isolates 49 and 55) and muskrats (isolates 36, 38, and 39). The remaining genotypes were scattered at several places within the first major cluster (Fig. 1).
The second major cluster was formed by muskrat isolates only, and three distinct muskrat Enterocytozoon genotypes (WL4, WL5, and WL6) were evident in this cluster. The nucleotide sequences (WL4 genotype) of seven muskrat isolates (isolates 23, 25, 26, 27, 29, 30, and 31) were identical but differed from the second muskrat genotype (WL6 genotype; isolate 34) at two positions (G to A at position 148 and A to G at position 286) and from the third muskrat genotype (WL5 genotype; isolate 41) at one position (G to T at position 363) (Fig. 2).
The third major cluster was formed by isolates of raccoon only. Three distinct genotypes (WL1, WL2, and WL3) of raccoon Enterocytozoon spp. (isolates 45, 48, 53, 56, 57 58, and 59) were observed within the third major cluster and differed from one another at four nucleotide positions (C or G at position 124, T or C at position 266, A or G at position 323, and G or A at position 360) (Fig. 2).
Results of the neighbor-joining analysis were confirmed by maximum likelihood analysis. Thus, three major clusters were also seen in the maximum likelihood tree. As in the neighbor-joining analysis, the cluster consisting of genotypes WL1, WL2, and WL3 formed the basal branch and the cluster consisting of genotypes WL4, WL5, and WL6 formed the next branch (data not shown). Even within the major cluster containing all E. bieneusi sequences from humans and domestic animals, the three genotypes from cattle (I, J, and N) and two genotypes from humans (C and Q) branched out first in both the neighbor-joining and maximum likelihood analyses (Fig. 1 and data not shown).
DISCUSSION
To date, based on the nucleotide sequence of the ITS, 31 distinct genotypes of E. bieneusi parasites have been reported from humans and domestic animals (1, 2, 5, 11, 13, 15, 16, 17; Deplazes et al., J. Eukaryot. Microbiol. 43:93S, 1996). Most of the genotypes identified before were from humans and pigs, and some of the genotypes were given different names by different researchers (Table 2). Results of the present study show for the first time the presence of Enterocytozoon spp. in wildlife associated with surface water. A total of 17 genotypes were found, with 15 of them being new. In this study, sequence characterization of isolates has been used to assess the public health significance of Enterocytozoon spp. from wildlife.
One E. bieneusi genotype reported in pigs and humans (GenBank accession no. AF101200 and AF34877, respectively) was identical to some of the isolates from muskrats, beavers, and foxes and one isolate from a raccoon that formed the genotype WL8. Similarly, the ITS sequences from some beavers, muskrats, raccoons, foxes, and one otter belonging to the genotype WL13 were identical to the E. bieneusi ITS sequences previously reported from pigs (GenBank accession no. AF076042 and AF135832). Furthermore, three more genotypes (WL12, WL15, and WL16) contained isolates from multiple species of wildlife. These findings indicate that some E. bieneusi genotypes can infect a broad range of animals and that these parasites are genetically related to or even identical to those infecting humans and domestic animals.
Some animals appear to have host-specific Enterocytozoon genotypes. For example, isolates in the cluster formed by genotypes WL4, WL5, and WL6 were all from muskrats, and isolates in the cluster formed by genotypes WL1, WL2, and WL3 were all from raccoons. These Enterocytozoon genotypes not only have ITS sequences very divergent from those of E. bieneusi genotypes but also are consistently placed in clusters outside the major E. bieneusi clades in both neighbor-joining and maximum likelihood analyses. Thus, they may represent species different from the human-pathogenic E. bieneusi. This observation needs to be confirmed by sequence and phylogenetic analyses of the small-subunit rRNA gene and other genetic loci. The identification of host-adapted Enterocytozoon spp. in this study was unlikely due to the broader specificity of primers used. Three previously identified bovine genotypes (I, J, and N) might also be host-adapted genotypes, as they formed a group separate from most E. bieneusi isolates (Fig. 1).
The transmission routes and the sources of human infection with E. bieneusi have not been identified directly. Spores of E. bieneusi have been frequently found in water in the United States and France (6; Sparfel et al., J. Eukaryot. Microbiol. 44:78S, 1997), but the human-infective potentials and sources of these spores are not known. Results of phylogenetic analysis have shown a close relatedness of human E. bieneusi with genotypes in farm animals (5). Indeed, two genotypes pathogenic for humans have been found in pigs (2, 12), suggestingthat zoonotic infection is possible. In the present study, E. bieneusi was found in a number of wild mammals (beavers, foxes, muskrat, otters, and raccoons) in four counties of eastern Maryland. Most of the E. bieneusi genotypes from these mammals were genetically related to human-pathogenic E. bieneusi, and one genotype has been found in humans. Thus, fur-bearing mammals living in or near surface water can be a source of contamination with human-pathogenic E. bieneusi.
In summary, results of the present study indicate that microsporidiosis due to Enterocytozoon spp. is prevalent in wild fur-bearing mammals and that extensive genetic polymorphism exists in Enterocytozoon spp. from wild mammals. The ITS region of the rRNA gene is a good genetic marker for the analysis of molecular evolutionary and taxonomic relationships of Enterocytozoon spp. The molecular data suggest that E. bieneusi from wildlife can be a potential source of microsporidian contamination in water, which in turn can be a risk to public health.
Acknowledgments
This work was supported in part by funds from the U.S. Environmental Protection Agency.
REFERENCES
- 1.Breitenmoser, A. C., A. Mathis, E. Burgi, R. Weber, and P. Deplazes. 1999. High prevalence of Enterocytozoon bieneusi in swine with four genotypes that differ from those identified in humans. Parasitology 118:447-453. [DOI] [PubMed] [Google Scholar]
- 2.Buckholt, M. A., J. H. Lee, and S. Tzipori. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalifoux, L. V., A. Carville, D. Pauley, B. Thompson, A. A. Lackner, and K. G. Mansfield. 2000. Enterocytozoon bieneusi as a cause of proliferative serositis in simian immunodeficiency virus-infected immunodeficient macaques (Macaca mulatta). Arch. Pathol. Lab. Med. 124:1480-1484. [DOI] [PubMed] [Google Scholar]
- 4.Cotte, L., M. Rabodonirina, F. Chapuis, F. Bailly, F. Bissuel, C. Raynal, P. Gelas, F. Persat, M. Piens, and C. Trepo. 1999. Waterborne outbreak of intestinal microsporidiosis in persons with and without immunodeficiency virus infection. J. Infect. Dis. 180:2003-2008. [DOI] [PubMed] [Google Scholar]
- 5.Dengjel, B., M. Zahler, W. Hermanns, K. Heinritzi, T. Spillmann, A. Thomschke, T. Loscher, R. Gothe, and H. Rinder. 2001. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 39:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowd, S. E., C. P. Gerba, and I. L. Pepper. 1998. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encepalitozoon intestinalis, and Vittaforma corneae in water. Appl. Environ. Microbiol. 64:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efron, B., E. Halloran, and S. Holmes. 1996. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 93:13429-13434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on the phylogenies: an approach using bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, S., O. Liguory, O., M. Santillana-Hyat, E. Guillot, C. Sarfati, N. Dumoutier, J. Molina, and F. Derouin. 2000. Detection of microsporidia in surface water: a one-year follow-up study. FEMS Immunol. Med. Microbiol. 29:95-100. [DOI] [PubMed] [Google Scholar]
- 10.Franzen, C., and A. Muller. 1999. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 12:243-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liguory, O., F. David, C. Sarfati, F. Derouin, and J. M. Molina. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansfield, K. G., A. Carville, D. Shvetz, J. MacKey, S. Tzipori, and A. A. Lackner. 1997. Identification of an Enterocytozoon bieneusi-like microsporidian parasite in simian-immunodeficiency-virus-inoculated macaques with hepatobiliary disease. Am. J. Pathol. 150:1395-1405. [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis, A., A. C. Breitenmoser, and P. Deplazes. 1999. Detection of new Enterocytozoon genotypes in fecal samples of farm dogs and a cat. Parasite 6:189-193. [DOI] [PubMed] [Google Scholar]
- 14.Reetz, J., H. Rinder, A. Thomschke, H. Manke, M. Schwebs, and A. Bruderek. 2002. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). Int. J. Parasitol. 32:785-787. [DOI] [PubMed] [Google Scholar]
- 15.Rinder, H., A. Thomschke, B. Dengjel, R. Gothe, T. Loscher, and M. Zahler. 2000. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185-188. [DOI] [PubMed] [Google Scholar]
- 16.Rinder, H., A. Thomschke, A. Essig, K. Heinritzi, R. Gothe, and M. Zahler. 1999. Mikrosporidien bei Schweinen mit lebensbedrohlicher Diarrho. Tieraerztl. Prax. 27:231-234. [Google Scholar]
- 17.Rinder, H., S. Katzwinkel-Wladarsch, and T. Loscher. 1997. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol. Res. 83:670-672. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Sorel, N., E. Guillot, M. Thellier, I. Accoceberry, A. Datry, L. Mesnard-Rouiller, and M. Miegeville. 2003. Development of an immunomagnetic separation-polymerase chain reaction (IMS-PCR) assay specific for Enterocytozoon bieneusi in water samples. J. Appl. Microbiol. 94:273-279. [DOI] [PubMed] [Google Scholar]
- 20.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]