Abstract
Genomic libraries derived from environmental DNA (metagenomic libraries) are useful for characterizing uncultured microorganisms. However, conventional library-screening techniques permit characterization of relatively few environmental clones. Here we describe a novel approach for characterization of a metagenomic library by hybridizing the library with DNA from a set of groundwater isolates, reference strains, and communities. A cosmid library derived from a microcosm of groundwater microorganisms was used to construct a microarray (COSMO) containing ∼1-kb PCR products amplified from the inserts of 672 cosmids plus a set of 16S ribosomal DNA controls. COSMO was hybridized with Cy5-labeled genomic DNA from each bacterial strain, and the results were compared with the results for a common Cy3-labeled reference DNA sample consisting of a composite of genomic DNA from multiple species. The accuracy of the results was confirmed by the preferential hybridization of each strain to its corresponding rDNA probe. Cosmid clones were identified that hybridized specifically to each of 10 microcosm isolates, and other clones produced positive results with multiple related species, which is indicative of conserved genes. Many clones did not hybridize to any microcosm isolate; however, some of these clones hybridized to community genomic DNA, suggesting that they were derived from microbes that we failed to isolate in pure culture. Based on identification of genes by end sequencing of 17 such clones, DNA could be assigned to functions that have potential ecological importance, including hydrogen oxidation, nitrate reduction, and transposition. Metagenomic profiling offers an effective approach for rapidly characterizing many clones and identifying the clones corresponding to unidentified species of microorganisms.
Microorganisms contribute significantly to the earth's biological diversity, yet relatively few of the microorganisms present in nature have been cultured and characterized. It is generally accepted that less than 1% of bacteria and fungi present in most habitats have been cultivated for study in pure culture (2). Although direct analysis of environmental DNA samples by PCR is effective for showing the presence of uncultured microorganisms, biases in primer specificity and amplification of different targets prevent full recognition of microbial diversity (31, 37, 40, 41). Thus, new approaches to examination of community genomes are needed.
The use of large-insert genomic libraries is a powerful approach for isolating DNA sequences from complex mixtures of uncultured microorganisms. Direct cloning of DNA from environmental samples makes it possible to avoid some of the biases of cultivation and PCR. In addition, genomic fragments that are >100 kb long can be obtained, and they provide significant functional and taxonomic information about the organisms from which they were derived. Such metagenomic libraries have been used to identify novel genes from uncultivated species of archaea, bacteria, and viruses that are responsible for significant ecosystem processes (4, 5, 8, 12) and to isolate enzymes that are involved in biosynthesis of novel pharmaceuticals (7, 15, 24, 41, 42) or have other industrial uses (11, 16, 17, 26, 33).
Given the immense uncultivated and uncharacterized metabolic diversity in the environment, one would need to sequence relatively few bacterial artificial chromosome (BAC) or cosmid clones to discover fundamentally interesting sets of genes. If modern genomic techniques can be used to carry out more comprehensive surveys of metagenomic libraries, our understanding of natural genetic diversity should be greatly enhanced.
Screening a genomic library can be done a number of different ways. Typically, screening involves colony hybridization with a probe of interest, which yields information one gene at a time. Bioassays have been developed to screen libraries for genes involved in the production of specific enzymes (11, 16, 17, 26, 33) or natural products (15, 24, 42); however, this approach relies on the fortuitous expression of heterologous DNA by the library host strain. High-throughput end sequencing of BAC clones has been used to accelerate various single-genome projects (39), and it is currently being used to characterize some environmental DNA libraries (8).
Although the speed and effectiveness of brute-force sequencing are constantly improving, it is not yet practical to assemble a complete bacterial genome from a metagenome. There is still a need for new functional genomic approaches that systematically yield information about many of the elements in a metagenomic library. These new approaches should ideally allow us to identify the organism from which each clone came, to determine some functional characteristics of various clones, and to identify many more novel uncultivated bacteria.
We sought to develop a practical approach that would provide a large amount of information about the microbial community from a limited set of clones. The purpose of our approach was to classify many of our cosmids and to identify a few candidates for sequencing rather than to undertake a major sequencing and assembly project. Our method involves hybridization of the library with genomic DNA of various reference strains and bacterial isolates from the community under study. In addition, DNA derived from as-yet-uncultivated organisms can be identified by hybridization with metagenomic DNA.
Metagenomic profiling.
Metagenomic profiling is classification of clones based on hybridization of insert DNA to the genomes of bacterial isolates, reference strains, and environmental DNA.
DNA microarray technology has become an important tool for determining the gene contents of entire genomes and measuring the expression of genes (18). High-density arrays are effective for quantitative detection of genes in complex samples. Thus, microarrays are a promising technique for characterization of genes in environments such as soil and water (3, 9, 34, 44, 45). However, the use of microarrays has been limited to 16S rRNA markers or a relatively small set of functional genes, and no practical approach has been developed to specifically target the unculturable majority of the species in the environment.
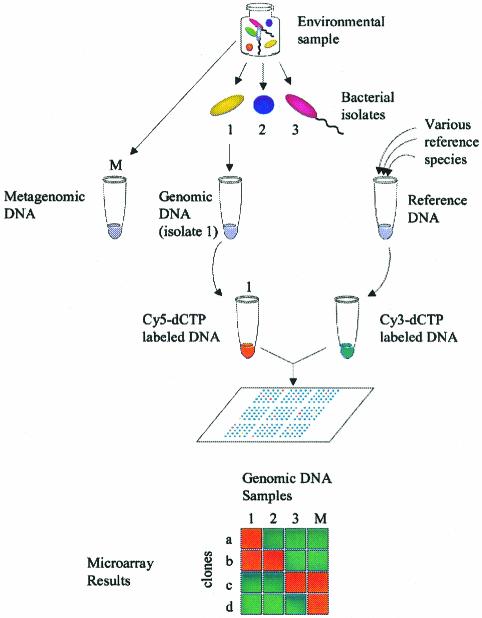
We used a microarray platform to screen a metagenomic library with whole microbial genomes and community genomes (Fig. 1). The microarray (COSMO) contained ∼1-kb PCR products amplified from the inserts of 672 cosmids along with a set of controls (16S ribosomal DNA [rDNA] probes). From an environmental sample (the same sample from which the library was derived), numerous bacterial isolates were obtained. Genomic DNA was purified from each environmental isolate. In addition, metagenomic DNA was purified directly from the mixed population, which was done without cultivation of bacteria. Each test genome was labeled with Cy5-dCTP and probed with COSMO. In order to subtract any signal that may have come from nonspecific hybridization, in each experiment we used two-color hybridization, in which each test genome was compared to a reference sample of common bacterial DNA. The reference DNA consisted of a pooled sample of genomic DNA from 14 species of bacteria (which effectively diluted the strain-specific genes and enriched common sequences). The reference DNA was given a different label (Cy3), and equal amounts of test and reference DNA were combined and hybridized to COSMO. We refer to this approach as comparative genomic hybridization (CGH). CGH was repeated for all environmental isolates, as well as for the metagenomic DNA sample(s). Positive results were determined based on a Cy5/Cy3 ratio greater than 1 (>0 on a log2 scale). As a result, we obtained a profile for each clone in the metagenomic library (i.e., a graphical representation of its hybridization to one or more species of bacteria). Clones that were specific to a test strain or community hybridized only to those DNA samples. Clones that contained a conserved sequence within their corresponding PCR amplicons hybridized to the genomes of multiple species.
FIG. 1.
Metagenomic profiling. Genomic DNA is purified from various bacterial isolates, as well as from a mixed population. A reference sample of common bacterial DNA is created by pooling the genomic DNA of many strains. In each CGH experiment, the genome of each strain or community is analyzed in comparison to the reference DNA. Some examples of informative results are shown. A clone of environmental DNA may correspond to only bacterial strain 1 (clone a), multiple strains (clone b), the metagenome and a bacterial isolate (clone c), or only the metagenome (clone d).
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
To evaluate our approach, we needed a microbial community that could be manipulated in the laboratory. A stable community was developed from an inoculum of biofilm material collected from the Snake River Plain Aquifer in southeastern Idaho. Biofilm was collected from the basalt aquifer by suspending a basket containing 100 ml of ceramic beads in an open borehole at a depth of 73 m adjacent to a zone of high groundwater flow. After 80 days, the basket was retrieved, and the beads were immersed in 100 ml of sterile phosphate-buffered saline. Cells were collected by gently vortexing the submerged beads for 15 min. One milliliter of a cell suspension was used to inoculate triplicate flasks containing 100 ml of minimal succinate medium and 100 ml of glass beads. Minimal succinate medium contained (per liter of deionized water) 6.0 g of K2HPO4, 3.0 g of KH2PO4, 1.0 g of (NH4)2SO4, and 4.0 g of succinic acid. The pH of the medium was adjusted to 7.5 with 10 M NaOH. One milliliter of sterile 1 M MgSO4 and 1.0 ml of sterile 45 mM CaCl2 were added after autoclaving. Microcosms were developed by incubating the flasks without shaking for 1 week at 30°C. After incubation, the cells were suspended by gently vortexing the flask and decanting the medium into two 50-ml polypropylene tubes, and cells were collected by centrifugation and resuspended in 1.0 ml of phosphate-buffered saline. Approximately 1010 cells were used for construction of a cosmid library, and the remaining cells were stored in glycerol at −80°C.
To obtain community DNA that were to be used for microarray target samples, a second enrichment of the microcosm was carried out under the same conditions by using a 0.5% inoculum of frozen stock. After incubation, two different fractions of cells were collected (pellicle and planktonic). First, the pellicle was removed with a sterile spatula and placed in a 50-ml polypropylene tube, and then the remaining cells were suspended by gently vortexing the flask, decanting the medium into two 50-ml polypropylene tubes, and collecting the cells by centrifugation. DNA from the samples described above was probed with COSMO in order to identify clones corresponding to microbial strains that were enriched in the mixed culture but that we failed to isolate in pure culture. We used the unisolated fraction of the microcosm to represent the uncultured microorganisms in the environment.
From the original microcosm and all subsequent enrichments, bacterial species were isolated by plating serial dilutions of liquid cultures onto plates containing tryptic soy agar. Bacterial strains were identified by selecting colonies with unique morphology that appeared during a 7-day incubation at 30°C. Ten different strains were obtained (see Fig. 4) along with the Pseudomonas aeruginosa and Staphylococcus aureus reference strains. To determine the identity of each isolate, the 16S rRNA gene was amplified by PCR from a genomic DNA template by using eubacterial primers 27F (20) and 907R (29). 16S amplicons were sequenced by using the 27F primer.
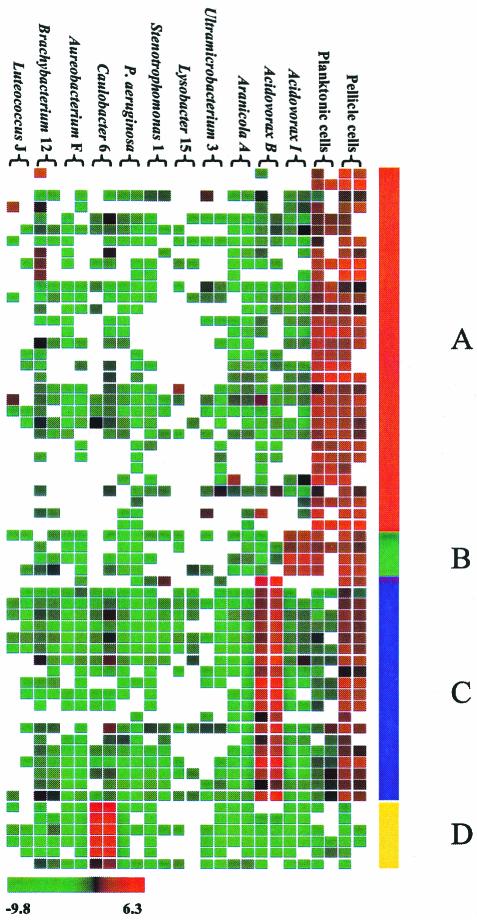
FIG. 4.
Identification of clones corresponding to uncultivated organisms. CGH experiments were performed by using metagenomic DNA from different cell fractions, including free-swimming (planktonic) cells and cells accumulating at the surface (pellicle cells). When duplicate experiments were added to the original data set, it was possible to identify uncultivated DNA (A). In addition, probes classified by single-genome CGH could be used to track the distribution of an organism in the mixed population (B, C, and D).
Library construction.
A cosmid library was constructed from the genomic DNA of the original mixed bacterial enrichment. Bacterial cells were embedded in agarose, and genomic DNA was purified by agarose-embedded cell lysis, followed by partial digestion of the agarose plugs with Sau3AI as described by Stein et al. (35). The metagenomic library was constructed by using a SuperCos I cosmid kit (Stratagene) according to the manufacturer's protocols. Clones were picked randomly into 96-well plates containing Luria-Bertani medium (44) supplemented with 100 mg of ampicillin per liter and 0.1 volume of 10× Hogness buffer (40% glycerol, 36 mM K2HPO4, 13 mM KH2PO4, 20 mM trisodium citrate, 10 mM MgSO4 in deionized water). After overnight incubation, the library was stored at −80°C. Plasmids were purified by using a REAL prep 96 kit (Qiagen) and a BioRobot 3000 (Qiagen) liquid-handling system. When we examined XhoI restriction digests of 10 clones by agarose gel electrophoresis, we observed no duplicate clones, and we determined that the average insert size of the cosmids was ∼40 kb. The results presented below verify that COSMO represented much of the microcosm's diversity. We did not attempt to characterize the species represented in the cosmid library prior to fabrication of COSMO. The task of amplifying 16S rRNA genes from a pool of cosmids was confounded by the presence of contaminating Escherichia coli genomic DNA in the plasmid preparation. (While the manuscript was being reviewed, Liles et al. [21] published a new procedure for eliminating E. coli 16S rDNA from pools of purified BACs.)
Array fabrication.
All microarray experiments were performed with COSMO, a DNA microarray containing end fragments of 672 cosmids selected randomly from the metagenomic library plus a set of reference genes (16S rDNA markers from several microcosm isolates [Fig. 2]). Cosmid end fragments were produced by a thermal asymmetric interlaced PCR method (22). Briefly, the thermal asymmetric interlaced PCR method involved sequential cycles of linear amplification of insert DNA from the T7 end of the vector with nested primers, followed by exponential amplification of the specific product by random priming, which resulted in PCR products that were 200 to 2,000 bp long. High-throughput PCR was performed with an MBS 384S thermocycler system (ThermoHybaid). 16S rRNA gene controls were also amplified by PCR. The quality and quantity of DNA were confirmed by agarose gel electrophoresis. PCR products were purified by using 384-well filter plates (Millipore) and were resuspended in 15 μl of 1× Spotting Solution Plus (Telechem) to obtain a final DNA concentration of 100 to 200 ng/μl. Each sample was spotted in duplicate on SuperAmine slides (Telechem) by using a Microgrid arrayer (BioRobotics). Slide cross-linking, washing, and blocking steps were carried out by using the manufacturer's protocols (http://arrayit.com/PDF/Super_Microarray_Substrates.pdf).
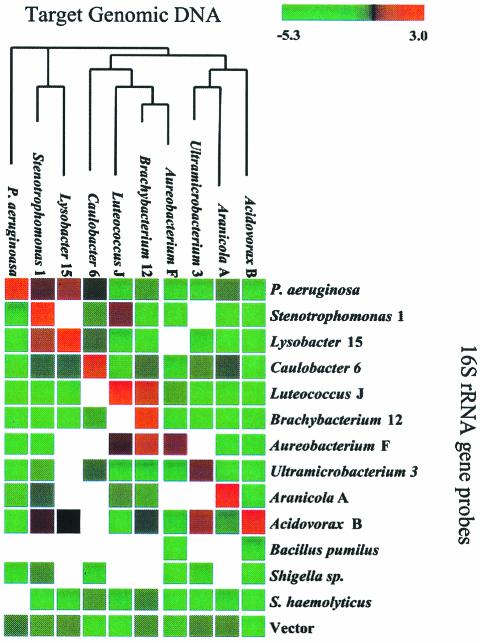
FIG. 2.
Microarray validation with 16S rDNA controls. The data are the results obtained with rRNA probes in 10 different experiments (one replicate per strain). The results of each experiment are presented as a column of color-coded values. The colors, ranging from green to red, correspond to the log2 of the Cy5/Cy3 ratio (see key at the top). Preferential hybridization of the test strain to a rRNA gene probe is indicated by a positive log2 ratio (red). Results with a signal-to-noise ratio of less than 3 are indicated by blank squares. Species are sorted horizontally and vertically according to their phylogenetic relationships (as determined by Clustal W alignment of rRNA sequences) in order to show the amount of cross-hybridization that occurs between the sequences of related strains. The last four probes shown are additional negative controls.
DNA preparation, labeling, and hybridization.
Genomic target DNA was purified from bacterial isolates and mixed cultures as described by Wilson (43). The reference sample was prepared by mixing equal amounts of genomic DNA from the 10 species of bacterial isolates that were used in this study and from the reference organisms E. coli, P. aeruginosa, S. aureus, and Bacillus subtilis. Fluorescently labeled target DNA was made as described by Pollack et al. (30). Briefly, 2 μg of target DNA was digested completely with MspI and purified by ethanol precipitation. Prior to labeling, 10 ng of salT (14) (a rice gene used as an internal standard) was added to the sample. Target DNA was labeled by incorporation of Cy5-dCTP (for test DNA) or Cy3-dCTP (for reference DNA) (Amersham Pharmacia) by random primer synthesis (BioPrime labeling kit; Invitrogen). Labeled target DNA was purified with CHROMA SPIN+ TE-30 gel filtration columns (Clontech). Test DNA and reference DNA were combined with 40 μg of human Cot-1 DNA and 100 μg of salmon sperm genomic DNA and reduced to a volume of 5 μl by using Microcon YM-30 concentrators (Millipore); 20 μl of 1.25× unihyb hybridization buffer (Telechem) was added to the target DNA mixture, and the target was preannealed to blocking DNA by boiling the preparation for 1.5 min, followed by 30 min of incubation at 37°C. Twenty-five microliters of probe was used per slide. Hybridization was performed for 8 h at 65°C. Posthybridization washing was performed by using the slide manufacturer's protocol.
Data analysis.
Arrays were scanned with an Axon 4000 scanner, and fluorescence measurements were obtained by using Genepix Pro 3.0 software (Axon). Data sets were filtered for spots with a signal-to-noise ratio greater than 3.0 by using Microsoft Excel. Results are reported below as log2 of the Cy5/Cy3 ratio. Data analysis and graphical display were done with Expression NTI (Informax Inc.) in the following manner: (i) all clones that failed to produce a positive result (log2, >0) in duplicate experiments with at least one target genome were removed from the data set, (ii) clones were clustered by complete linkage of genes by using the correlation coefficient, and (iii) experiments were sorted according to the phylogenetic relationships of test strains as determined by Clustal W alignment of 16S rRNA sequences.
Analysis of cosmid end sequences.
The inserts of select clones were sequenced from both ends of the multiple cloning site by using the T3 and T7 primers. The plasmid template was purified from 500-ml cultures of E. coli by using a large-construct kit (Qiagen). A sequencing reaction mixture (total volume, 20 μl) was prepared by combining 1 μg of plasmid DNA with 0.32 μl of a 10 μM primer T3 or T7 stock solution in Tris-EDTA buffer and 8.0 μl of a Big Dye mixture. PCR was performed with a PTC-100 thermocycler (MJ Research) for 80 cycles (94°C for 30 s, 47°C for 15 s, and 60°C for 4 min). Reaction mixtures were purified with DTR gel filtration columns (Edge Biosystems). Nucleotide sequences (average size, 450 bp) were determined with an ABI 3100 DNA sequencer.
The potential functions and phylogenetic affinities of cosmid end sequences were determined by performing a nucleotide and translated-protein search of GenBank by using the Basic Local Alignment Search Tool (BLAST) (1). The potential function of a given sequence was determined by examination of all homologous sequences with expect values of <1e-2. A particular function was assigned if (i) a consensus was apparent among the best hits and (ii) there was no disagreement between the consensus of the nucleotide results and the translated-protein results. We also listed the species corresponding to the nucleotide best hit; when no significant nucleotide matches were observed, we listed the species corresponding to the protein best hit (Table 1).
TABLE 1.
Potential genes observed in cosmid end sequencesa
| Clone | Gene in T7 end | Gene in T3 endb |
|---|---|---|
| 2H6 | Transposase (Burkholderia fungorum) | HP (Chloroflexus aurantiacus) |
| 3C2 | ABC transporter (Pseudomonas aeruginosa) | HP (Burkholderia fungorum) |
| 3F7 | NS | ABC transporter (Ralstonia metallidurans) |
| 3G10 | Replication initiator protein dnaA (Ralstonia metallidurans) | HP (Sinlorhizobium meliloti) |
| 3H9 | Unknown (Azotobacter vinelandii) | HP (Desulfitobacterium sp.) |
| 5D6 | Denitrifying NorE and NorF genes (Pseudomonas stutzeri) | NS |
| 5D8 | 50S ribosomal subunit protein L32 (Ralstonia solanacearum) | HP (Desulfovibrio desulfuricans) |
| 5D11 | Short-chain dehydrogenase (Rhizobium sp. strain NGR234) | HP (Halobacterium sp. strain NR) |
| 5G2 | Putative [NiFe] hydrogenase (Streptomyces averimitilis) | NS |
| 6A7 | Transposase and tRNA synthase (Ralstonia metallidurans) | Probable serine hydroxymethyl transferase (Microbulbifer degradans) |
| 6A12 | Insertion sequence IS1051-X (Xanthomonas oryzae) | NS |
| 6D10 | Transposase (Ralstonia metallidurans) | Cytochrome c-like protein (Burkholderia fungorum) |
| 6F10 | Probable organic solvent resistance protein (Ralstonia solanacearum) | Sensory transduction histidine kinase (Magnetospirillum magnetotacticum) |
| 7A12 | Integrase-like protein (Xanthomonas axonopodis) | Peptidyl-prolyl cis-trans isomerase (Ralstonia solanacearum) |
| 7B11 | Cation-transporting ATPase (Mycobacterium tuberculosis) | HP (Geobacter metallireducens) |
| 7H7 | HP (Novosphingobium aromaticivorans) | HP (Halobacterium sp. strain NRC-1) |
Sequences were read from the T7 and T3 ends of each clone. Each result is a consensus of all significant results from nucleotide and translated protein searches of the GenBank database. The species or strain corresponding to the most significant result at the nucleotide level is indicated in parentheses.
HP, hypothetical protein; NS, no significant homology.
RESULTS
Data validation.
A single self-versus-self hybridization was performed by hybridizing COSMO with two samples of Acidovorax B genomic DNA that had been prepared and labeled separately, one with Cy5 and the other with Cy3. Seventy-six probes corresponding to 43 different cosmids passed our filtering criteria (see Materials and Methods). The values for the Cy5/Cy3 ratio ranged from 0.77 to 1.19 (mean, 1.00; standard deviation, 0.12) (raw data not shown). Based on this experiment, the performance of different probes and dyes should have caused less than a 1.3-fold deviation from the mean.
The sensitivity and specificity of our microarray analysis were evident from the results obtained for controls and reference genes (Fig. 2) in our CGH experiments with individual genomes (see below). 16S rRNA genes from several microcosm isolates were obtained by PCR and included in COSMO as controls. Each rRNA gene served as a positive control for its corresponding species and as a negative control for distantly related species. Additional negative controls are shown last in the figures. The results indicated that our hybridization conditions allowed identification of strain-specific genes. In all but one case, genomic DNA of each strain hybridized preferentially to its corresponding rRNA probe (yielding the highest ratio). In most cases, some hybridization to a related strain was observed, but the level of hybridization was lower. In the case of Ultramicrobacterium 3, genomic DNA hybridized nearly equally to the Ultramicrobacterium 3 and Acidovorax B rRNA probes. No positive results were obtained with the Bacillus sp., Shigella sp., and Staphylococcus sp. negative controls or with the cosmid vector.
Hybridization with individual genomes.
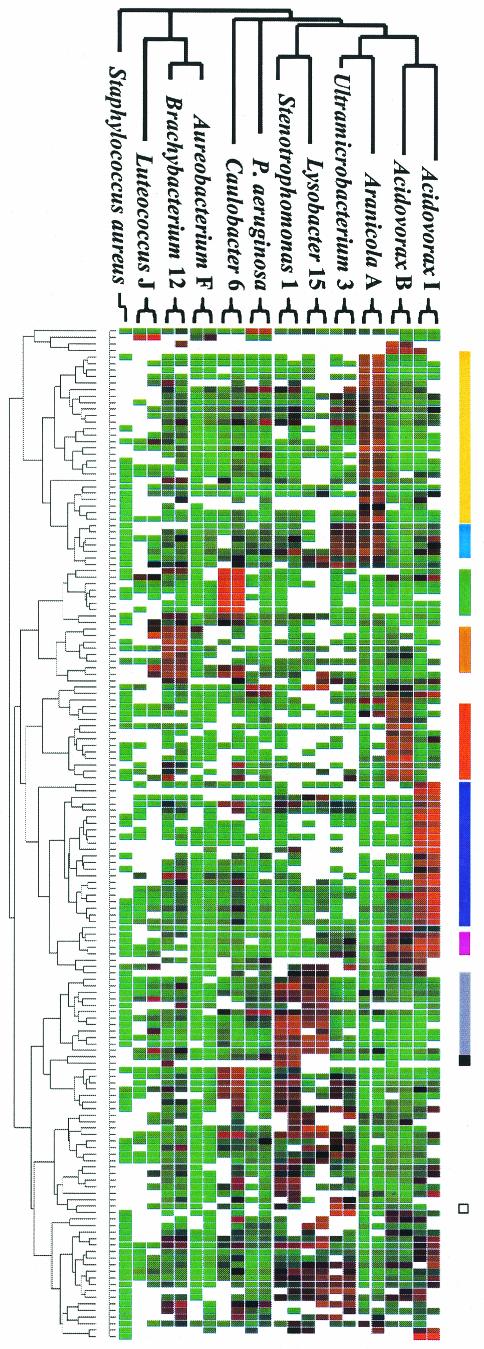
Various reference strains and microcosm isolates were used individually as target DNA in experiments to locate clones related to these organisms. The clustered microarray results for various microcosm isolates and reference strains revealed distinct classes of DNA that corresponded to individual strains or groups of bacteria (Fig. 3). Numerous strain-specific clones were apparent. The data also revealed examples in which clones hybridized to multiple related organisms (indicative of conserved genes). Based on these patterns, we classified some clones as members a particular species, genus, or branch (Fig. 3).
FIG. 3.

Classification of clones based on hybridization of COSMO with individual genomes. Twelve different strains were analyzed with COSMO in duplicate. Experiments were sorted horizontally according to the phylogenetic relationships of the test strains. Clones were clustered (by rows) according to the correlation coefficients of the microarray data across 23 experiments. S. aureus was included to show that cross-hybridization between very distant relatives may not be significant, but one replicate was removed in order to minimize its effect on clustering. Based on the clustered data, distinct classes of clones are apparent. The colored bars on the right indicate the clones with reproducible hybridization patterns specific for one test strain (strain-specific clones) and clones that hybridized to multiple related species (conserved clones).
At the bottom of Fig. 3, some results appear to be scrambled (i.e., distinct patterns are not easily distinguished). The profiles observed for this group of clones are not consistent with profiles of strain-specific and conserved clones. In general, these clones appeared to cross-hybridize between species to a greater extent. A variety of different patterns were observed, and for the sake of simplicity they were not labeled.
Classifying uncultured DNA.
In the experiment described above (Fig. 3), 156 probes (clones) passed our filtering process. The remaining 524 clones failed to produce a positive log2 ratio with any test strain or failed to produce any significant signal. Some of these clones may have corresponded to organisms present in the microcosm that we failed to isolate in pure culture. We considered the unisolated organisms in our microcosm to be analogous to uncultured microbes in the environment. To identify cosmids derived from such organisms, we performed a similar experiment using genomic DNA extracted directly from the mixed bacterial population as the test DNA (the same reference DNA was used). The results of two metagenomic CGH experiments were added to the data set prior to clustering (Fig. 4). The results identified a number of clones that were present in the community and not in our catalog of isolates (Fig. 4A). Such clones were classified as uncultured.
A single experiment, such as the one described above, yielded a spectrum of ratios. It is likely that the clones that yielded a log2 ratio of 1 corresponded to a different organism than the clones that yielded a log2 ratio of 6. The uncultured class could be separated into subgroups if there were clear differences in the abundance of different genes in the community, but it was not obvious where to draw the line between one organism and another.
In an attempt to resolve the uncultivated community in slightly better detail, the microcosm cells were fractionated into two types, pellicle cells and free-swimming cells, which were analyzed separately. All uncultured clones (Fig. 4A) were found to have the same even distribution between planktonic and pellicle cells; therefore, we were not able to determine if these clones corresponded to multiple species. However, we observed that clones corresponding to some of the cultivable species had distinct distribution patterns. Acidivorax I was apparently distributed throughout the microcosm (Fig. 4B), Acidivorax B was present primarily in the pellicle (Fig. 4C), and Caulobacter 6 was not abundant in either fraction (Fig. 4D). These clusters were labeled and presented as examples of clones from different species that, in principle, could be distinguished based solely on community analysis.
Sequence analysis of uncultivated DNA.
In order to identify functional characteristics of uncultured microorganisms from the microcosm, insert DNA from each of 17 random clones from cluster A (Fig. 4A) was sequenced from the T7 and T3 primer sites that flanked the insert. A nucleotide and translated-protein search of GenBank was performed with each cosmid end sequence (Table 1). Among 34 sequences, 18 functional genes were identified, 10 hits were obtained with genes of unknown function, and 6 sequences yielded no significant result. The great majority of the sequences were found to be significantly similar to genes from members of the Proteobacteria, including seven genes from Ralstonia sp. Some of the sequences could be assigned to functions having ecological importance, including a putative [NiFe] hydrogenase, nitrate reduction, and several transposases. Four different insertion sequence elements (ISs) were observed in five clones, 2H6, 6A12, 6A7, 6D10, and 7A12 (sequences from 6A7 and 6D10 were from different positions of the same gene). All ISs that we identified occurred precisely in the T7 end fragment.
DISCUSSION
The experiments described above illustrate a useful technique for rapidly classifying DNA from a metagenomic library and, in the process, identifying conserved and divergent genome fragments from a particular community, genus, strain, or species and DNA corresponding to strains that have not been isolated in pure culture.
Hybridizing COSMO with single genomes and cluster analysis of the microarray results were effective for visualizing groups of clones that have unique patterns of hybridization to different bacterial genomes (Fig. 3). We identified clones that hybridized to a single strain, as well as clones that hybridized to related species. In this manner, we classified clones as strain specific or specific to broader phylogenetic group.
Many of the clones in the scrambled section of Fig. 3 have much broader specificity, and some cross-hybridize only between two distantly related strains. The latter finding may be of interest to researchers who wish to identify genes that have been transferred horizontally between species. However, before such claims are made based on hybridization between more divergent species, the correlation between homology and signal intensity must be examined more carefully.
Identification of clones corresponding to cultivable strains is also useful for eliminating clones that do not need to be examined in a culture-independent manner. This may in fact be the primary objective, in which case it would not be necessary to probe genomes one at a time, as we did. The cultivable genomes could be combined into pools in order to accelerate the process.
The eventual identification of clones corresponding to uncultivated microorganisms is the most valuable aspect of this approach. We identified clones that hybridized uniquely to total community DNA. These clones corresponded to one or more species of bacteria that were enriched in mixed cultures but that we were unable to obtain by isolation on tryptic soy agar. End sequencing of 17 cosmids resulted in identification of numerous sequences that had nucleotide similarity to the genes of Ralstonia spp. In addition, we identified a putative hydrogenase and genes involved in reduction of nitrate. Some species of bacteria, including Ralstonia spp. (6, 19, 32), have the ability to couple hydrogen oxidation to nitrate reduction. The putative hydrogenase gene and the norE homologue were not found in the same clone, and we could not confirm that they are linked any way. Such proof would require isolating both sets of genes on the same fragment of DNA. However, knowledge of the metabolic genes present in the uncultured population provides information that should be important for selectively culturing such organisms if they are indeed present.
Among the clones that hybridized only to community DNA, a remarkably high frequency of ISs was observed. We do not believe that this is a unique characteristic of the uncultivated species. ISs occur in a wide range of genera (25). A variety of ISs can occur in a single genome, and a single type of IS may have a copy number of >20 (38). We believe that the five occurrences of four different IS-like elements precisely in a clone's T7 end fragment (the probe actually printed on the array) may have reflected a greater sensitivity of our method for high-copy-number sequences.
Of course, in a mixed population, our method detects the most abundant genes. A common limitation of microarray studies is sensitivity. The detection limit for microarray analysis of soil samples is on the order of ∼50 ng of a single bacterial genome (10); therefore, in a typical experiment one would detect only organisms that constitute at least 1/40 of the population. However, new techniques for uniform amplification of genomic DNA (27) and enrichment of unique sequences (23, 28) may be applied to environmental samples in order to access the single-copy genes of less abundant species.
We hope to further develop applications of the COSMO microarray for environmental samples. We demonstrate here that in addition to simple identification of an uncultured subset of clones, additional classes of organisms can be defined by comparing different environmental samples from the same habitat (Fig. 4B to D). Clones that have a unique distribution in the two fractions may constitute a separate phylogenetic class. By comparing the metagenomic profiles of a field site to a map of metabolic activities (13) or microbial species (36), clones whose distribution correlates with a biological process may be identified. In this manner, ecologically important genes for which there is not a specific probe or assay may be identified.
Systematic organization of a metagenomic library is essential for performing a more comprehensive study of a microbial community. We must continue to utilize modern techniques of genome analysis and adapt them to the study of complex mixed genomes, keeping in mind that the purpose of a genome-wide study is to accelerate the discovery of genes that are important for specific processes.
A study of the microbial metagenome need not seek truly comprehensive knowledge concerning all microbial genomes if it is possible to first negotiate the genomic landscape of a given site and find what is relevant and interesting. A practical approach to metagenomics is (i) to quickly identify familiar genes, (ii) to identify the unknowns, and (iii) to attempt to classify them based on knowledge such as where certain genes are present and what apparent linkages there are between different genes. Once a set of clones that is linked to a biological process is identified, the specific genes involved may be identified from the complete DNA sequences of the clones. Metagenomic profiling is an appropriate technique for these tasks. We believe that microarray studies in combination with DNA sequence analysis will be important tools for enhancing our understanding of earth's microbial diversity.
Acknowledgments
We thank Cornelia Sawatzky for assistance with the preparation of the manuscript and Alan Caplan for supplying control DNA for the microarray.
This work was funded by a predoctoral fellowship from the Inland Northwest Research Alliance. Additionally, the project was supported by NIH grant P20 RR16454 from the BRIN Program of the National Center for Research Resources.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 5.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 6.Bernhard, M., E. Schwartz, J. Rietdorf, and B. Friedrich. 1996. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J. Bacteriol. 178:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, S., and J. Clardy. 2000. Long-chain N-acetyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 122:12903-12904. [Google Scholar]
- 8.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, J. C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, J. C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers, H., J. Pernthaler, F. O. Glockner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, R., I. Thompson, and M. Bailey. 1995. Metabolic profiling as a means of characterizing plant-associated microbial communities. FEMS Microbiol. Ecol. 16:9-18. [Google Scholar]
- 14.Garcia, A. B., J. Engler, S. Iyer, T. Gerats, M. Van Montagu, and A. B. Caplan. 1997. Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol. 115:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henne, A., R. Daniel, R. A. Schmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyce, E. A., K. Chan, N. R. Salama, and S. Falkow. 2002. Redefining bacterial populations: a post-genomic reformation. Nat. Rev. Genet. 3:462-473. [DOI] [PubMed] [Google Scholar]
- 19.Kleihues, L., O. Lenz, M. Bernhard, T. Buhrke, and B. Friedrich. 2000. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J. Bacteriol. 182:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 21.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 23.Lucito, R., J. West, A. Reiner, J. Alexander, D. Esposito, B. Mishra, S. Powers, L. Norton, and M. Wigler. 2000. Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res. 10:1726-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacNeil, I. A., C. L. Tiong, C. Minor, P. R. August, T. H. Grossman, K. A. Loiacono, B. A. Lynch, T. Phillips, S. Narula, R. Sundaramoorthi, A. Tyler, T. Aldredge, H. Long, M. Gilman, D. Holt, and M. S. Osburne. 2001. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 3:301-308. [PubMed] [Google Scholar]
- 25.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majernik, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+ (Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makrigiorgos, G. M., S. Chakrabarti, Y. Zhang, M. Kaur, and B. D. Price. 2002. A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat. Biotechnol. 20:936-939. [DOI] [PubMed] [Google Scholar]
- 28.Mavrodi, D. V., O. V. Mavrodi, B. B. McSpadden-Gardener, B. B. Landa, D. M. Weller, and L. S. Thomashow. 2002. Identification of differences in genome content among phlD-positive Pseudomonas fluorescens strains by using PCR-based subtractive hybridization. Appl. Environ. Microbiol. 68:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 30.Pollack, J. R., C. M. Perou, A. A. Alizadeh, M. B. Eisen, A. Pergamenschikov, C. F. Williams, S. S. Jeffrey, D. Botstein, and P. O. Brown. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 31.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romermann, D., J. Warrelmann, R. A. Bender, and B. Friedrich. 1989. An rpoN-like gene of Alcaligenes eutrophus and Pseudomonas facilis controls expression of diverse metabolic pathways, including hydrogen oxidation. J. Bacteriol. 171:1093-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoner, D. L., M. C. Geary, L. J. White, R. D. Lee, J. A. Brizzee, A. C. Rodman, and R. C. Rope. 2001. Mapping microbial biodiversity. Appl. Environ. Microbiol. 67:4324-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urasaki, A., Y. Sekine, and E. Ohtsubo. 2002. Transposition of cyanobacterium insertion element ISY100 in Escherichia coli. J. Bacteriol. 184:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venter, J. C., H. O. Smith, and L. Hood. 1996. A new strategy for genome sequencing. Nature 381:364-366. [DOI] [PubMed] [Google Scholar]
- 40.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G. C. and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiol. 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G. Y., E. Graziani, B. Waters, W. Pan, X. Li, J. McDermott, G. Meurer, G. Saxena, R. J. Andersen, and J. Davies. 2000. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2:2401-2404. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, K. 1992. Preparation of genomic DNA from bacteria, p. 2-10-2-11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 44.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]