Abstract
Nuclear magnetic resonance (NMR) spectra were acquired from suspensions of clinically important yeast species of the genus Candida to characterize the relationship between metabolite profiles and species identification. Major metabolites were identified by using two-dimensional correlation NMR spectroscopy. One-dimensional proton NMR spectra were analyzed by using a staged statistical classification strategy. Analysis of NMR spectra from 442 isolates of Candida albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis resulted in rapid, accurate identification when compared with conventional and DNA-based identification. Spectral regions used for the classification of the five yeast species revealed species-specific differences in relative amounts of lipids, trehalose, polyols, and other metabolites. Isolates of C. parapsilosis and C. glabrata with unusual PCR fingerprinting patterns also generated atypical NMR spectra, suggesting the possibility of intraspecies discontinuity. We conclude that NMR spectroscopy combined with a statistical classification strategy is a rapid, nondestructive, and potentially valuable method for identification and chemotaxonomic characterization that may be broadly applicable to fungi and other microorganisms.
In clinical mycology, important emerging fungal pathogens in the genus Candida include Candida glabrata, C. tropicalis, C. parapsilosis, and C. krusei (Issatchenkia orientalis) (15, 20, 33, 35). Many strains within these species have reduced or variable susceptibility to antifungal drugs, and together they are more common than C. albicans as causes of candidemia in some centres (6, 15, 33, 34). Identification of Candida species can be problematic because of the paucity of phenotypic characters on which to base identification and the recognition that some characters are subject to intragroup variability or parallel evolution (12, 21, 37, 47). At present, the genus Candida remains an artificial assemblage of ascomycetous, mitosporic yeasts (28, 50).
Secondary metabolites and other compounds, such as proteins, lipids, or carbohydrates, have been utilized in chemotaxonomic approaches to the classification of fungi and lichenized fungi and for identification (11). Within the yeasts, classification to the level of genus has been achieved by using a monophasic approach, i.e., by analysis of profiles from particular groups of compounds, including fatty acids, carbohydrates, or polyols (1, 3, 13, 36). Identification of Candida isolates in routine laboratories is based on a polyphasic approach, which includes combinations of morphological characters, assimilation and fermentation profiles, identification of particular metabolites, and growth on differential media. These phenotypic methods are slow, may not distinguish between closely related species, and are sometimes unreliable, and not all clinically relevant species are included in the databases (10, 19, 23, 40, 42, 46, 49). Molecular methods of identification (including DNA-DNA reassociation, PCR fingerprinting, and DNA sequencing of small numbers of reference organisms) are generally more discriminatory than methods based on phenotypic characters. However, criteria for defining species boundaries based only on DNA sequences have not been agreed upon. Genotypic characters alone, which do not take into account expressed gene products of biological importance, may not provide a plausible and practical definition of species boundaries. As many genotypic and phenotypic characters as possible should be considered to further the understanding of species boundaries in the absence of sexuality.
NMR spectroscopy offers a high-throughput, rapid, polyphasic approach to establish the chemotype of microorganisms by providing information on a large range of metabolites rapidly and simultaneously. 1D proton (1H) NMR spectra of cell suspensions provide an overview of mobile hydrogen-containing compounds, which can be identified by multidimensional NMR correlation spectroscopy. Multivariate analysis and pattern recognition techniques detect differences in gross spectral characteristics (shape and pattern) of spectroscopic data from biological samples without the need to identify individual compounds (2, 14, 17, 24, 30, 44, 45). It was shown in a preliminary study that analysis of 1H NMR spectra by linear discriminate analysis is able to distinguish between selected species of streptococci and staphylococci with high accuracy (2).
We demonstrate here that application of a multistage, supervised SCS developed for analysis of NMR spectra (24, 44) results in accurate identification of isolates within the genus Candida.
MATERIALS AND METHODS
Abbreviations.
ATCC, American Type Culture Collection; CBS, Centraalbureau voor Schimmelcultures; COSY, correlation spectroscopy; FTIR, Fourier transform infrared; HSQC, heteronuclear single-quantum coherence; NMR, nuclear magnetic resonance; ORS, optimal region selection; TOCSY, total correlation spectroscopy; PBS-D2O, phosphate-buffered saline made up in deuterated water; SAB, Sabouraud's dextrose agar; SCS, statistical classification strategy; YPG, yeast extract-peptone-glucose broth; 1D, one dimensional; HMBC, heteronuclear multiple-bond correlation; and LDA, linear discriminate analysis.
Cultures.
Isolates were obtained from the culture collections at the Centre for Infectious Diseases and Microbiology (CIDM, University of Sydney at Westmead Hospital, Westmead, New South Wales, Australia), the ATCC (Manassas, Va.), and the CBS (Utrecht, The Netherlands). Recent clinical isolates were obtained from the CIDM Laboratory Services (CIDMLS), Institute of Clinical Pathology and Medical Research, Westmead Hospital. Type or neotype cultures of C. albicans (CBS 562), C. glabrata (CBS 138), C. krusei (CBS 573), C. parapsilosis (CBS 604), and C. tropicalis (CBS 94) were obtained from the CBS. Overall, 96% were clinical isolates (82% from eight hospitals in Australia and New Zealand, 9% from North and South America, 4% from Europe, and 1% from Asia) and 4% were of environmental origin. Isolates were stored either in autoclaved water at 25°C or in 10% glycerol in nutrient broth at −70°C. Identifications were made from duplicate cultures that had been incubated at 27°C for 42 to 48 h on SAB (Difco Laboratories, Detroit, Mich.). Specimens for MR spectroscopy were held at room temperature (20 to 30°C) for 1 to 4 h before use.
Identification.
Prior to storage, isolates had been identified biochemically (VITEK YBC or API ID32; BioMerieux, Marcy l'Etoile, France). All tests were carried out as specified by the manufacturers. To check for potential discrepancies arising from incorrect handling of stored cultures, random isolates were reidentified by conventional tests. Approximately 15% of isolates (n = 74) were also identified by PCR fingerprinting (23, 29). Conventional identification and PCR were performed routinely when there was disagreement between identification by conventional methods (VITEK/API) and statistical classification of NMR spectra.
PCR fingerprinting.
Briefly, genomic DNA was isolated and PCR was performed by using oligonucleotides of the minisatellite-specific core sequence of the wild-type phage M13 (5′-GAGGGTGGCGGTTCT-3′) (29) as a single primer. Reactions were carried out in a Perkin-Elmer thermal cycler (model 480) as follows: denaturation (35 cycles of 20 s at 94°C), annealing (1 min at 50°C), extension (20 s at 72°C), and final extension (6 min at 72°C). Products were separated by electrophoresis in 1.4% agarose gels in Tris-borate-EDTA buffer at 3 V cm−1. Amplification products were detected by staining with ethidium bromide and were visualized under UV light. PCR fingerprint profiles were manually evaluated. Type strains of all tested species were included as reference strains.
NMR spectroscopy.
Yeast colonies were gently removed from the SAB plate with a plastic inoculation loop and were suspended in 0.5 ml in PBS-D2O (PBS [pH 7.2, room temperature] made up in 99.5% D2O) (Australian Nuclear Science and Technology Organization, Lucas Heights, Australia) to a final concentration of 108 to 5 × 109 CFU ml−1. The suspension was immediately transferred to a 5-mm NMR tube (Wilmad Glass Co., Inc., Buena, N.J.). 1H NMR spectra were obtained at 37°C on a Bruker Avance 360-MHz NMR spectrometer by using a 5-mm {1H, 13C} inverse-detection dual-frequency probe. 1H NMR spectra were acquired with acquisition parameters as follows: frequency, 360.13 MHz; pulse angle, 90o (6 or 7 μs); repetition time, 2.3 s; 4,096 data points; 32 transients; and spectral width, 3,600 Hz. The samples were spun at 20 Hz to prevent the cells from settling in the NMR tube. The field was locked to D2O. Water suppression was performed by a selective excitation field gradient method (18). Spectra of cell suspensions were stable at 37°C for at least 2 h. Spectra were processed by using Bruker xwinnmr 2.6 software. Chemical shift calibration was performed by setting the center of the spectrum to 4.64 ppm (nominal position of the water resonance with respect to tetramethylsilane in PBS-D2O at 37°C). The viability and purity of cultures were confirmed by plating fungal cell suspensions (two isolates per species) after 2-h NMR experiments (1H NMR plus 1H and 1H COSY).
Signal assignment.
2D homo- and heteronuclear correlation spectra were acquired for 7 to 12 isolates per species to assign 1H NMR resonances to specific compounds. {1H, 1H} gradient COSY experiments were performed in magnitude mode. Acquisition parameters were as follows: spectral width in t2, 3,600 Hz; t2 time domain, 2,048; 256 increments of four or eight acquisitions each; and relaxation delay, 1 s. Sine-bell window functions were applied in the t1 dimension, and Gaussian-Lorentzian window functions were applied in the t2 dimension. Zero filling was used to expand the data matrix to 1,024 in the t1 dimension. TOCSY spectra with mixing times of 40 and 150 ms were acquired with 256 increments of 2,048 data points and 16 acquisitions. {1H, 13C} single-bond shift correlation spectra were obtained in the 1H detection mode by using an HSQC pulse sequence. The 1H NMR spectral width was 3,600 Hz, and the 13C NMR spectral width was 15,000 Hz. 13C decoupling during acquisition was achieved by GARP-1 (41). The evolution time (t1) was incremented to obtain 256 FIDs, each of 32 or 64 acquisitions and consisting of 2,048 data points. The relaxation delay was 2 s. A sine-bell function was applied in the t2 dimension, and a Gaussian-Lorentzian function was applied in the t1 dimension. Zero filling to 1,024 was used in the t1 dimension prior to Fourier transformation. {1H, 13C} gradient HMBC were acquired without proton decoupling by using the same parameters as for the HSQC experiments, except for a 13C spectral width of 20 kHz. Single-bond and long-range correlation experiments were usually optimized for 1JC,H of 140 Hz and nJC,H of 7 Hz, respectively. 1H and 13C chemical shifts in HSQC and HMBC were referenced by using the two ethanol cross-peaks at 3.65/58.3 and 1.18/17.6 ppm, which were present in all samples. 1D 1H NMR spectra were acquired before and after the 2D experiments to verify absence of metabolic changes during the time in the magnet.
Quantification of HSQC spectra.
Quantification of metabolites based on 1D 1H NMR spectra was not possible due to overlapping resonances. We applied a modified method by Bubb et al. that was based on the integration of fully relaxed {1H, 13C} HSQC cross-peaks (4). The concentrations of amino acid residues, carbohydrates, and alditols were estimated by comparison of volumes of resolved HSQC cross-peaks with those of p-aminobenzoic acid. To account for differences in relaxation times and 1J{1H, 13C} coupling constants between different compounds (for most of the cross-peaks the 1J{1H, 13C} coupling constants were found to be between 130 and 145 Hz), calibration factors were determined by comparison of the NMR-based concentrations with chemically determined trehalose, glycerol, and lysine concentrations (one isolate per species). To determine experimental errors, the procedure was repeated with a duplicate culture. The error of the method was found to be in the order of 30 to 40%, confirming previous reports (4). Despite the recognized limitations associated with the measurement of peak volumes in 2D NMR spectra, the concentration estimates are considered to be more meaningful than are qualitative descriptions of peak intensities (51).
Reproducibility of NMR spectra.
Short-term method variability was investigated by examination of duplicate cultures. Any effect of storage, i.e., minor variations in methods or cultures (e.g., differences between batches of culture media), were sought by retesting two isolates per Candida species up to five times over a 20-month period. The effect of specified culture conditions on classification of NMR spectra was investigated with duplicate cultures of two isolates per species by varying the following: temperature (25 to 40°C), pH (pH 5.0 or 7.2), incubation time (24 to 192 h), growth medium (SAB plates or YPG broth), and storage at room temperature (0 to 192 h).
Classification of NMR spectra.
An SCS was employed that was specifically designed for NMR and infrared spectra of biofluids and tissue biopsies, where databases contain many fewer spectra than data points in each spectrum (attributes) (31, 44). NMR data were prepared for statistical classification by using software developed in-house (Xprep; IBD, NRC, Winnipeg, Canada). Magnitude spectra, consisting of 4,096 data points over a spectral width of 10 ppm, were reduced to the most informative 1,500 points between 0.35 and 4.00 ppm. The spectra were normalized to unit area in this region. The correct alignment of the NMR spectra was checked visually by simultaneous and sequential display of all NMR spectra through use of the lipid resonance at 1.3 ppm.
The magnitude NMR spectra were analyzed by a genetic algorithm-based ORS in order to reduce the number of attributes and hence eliminate redundant information (31). Two or three maximally discriminatory subregions in the 1D NMR spectra of each group (species) were selected for development of LDA-based classifiers. The averages of these subregions were used to develop LDA-based pairwise classifiers for all combinations of the five Candida species (n = 10).
These classifiers were made robust by a bootstrap-based cross-validation method developed in-house (IBD, NRC) (8, 43). Specifically, about half the spectra from each class were selected at random and were used to train an LDA classifier (the “training set”). The remaining spectra were then tested against this classifier. This process was repeated 1,000 times (with random replacement), and the optimized LDA coefficients were saved. The ultimate classifier consisted of the weighted output of the 1,000 different bootstrap classifier coefficient sets. Each classifier yielded probabilities of class assignment for the individual spectra. For each sample x, the a posteriori probabilities pm(x) for all five classes (species) m = 1, 2,…, K were calculated according to
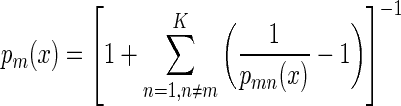 |
where pmn(x) is the class probability for x when submitted to the individual pair classifier Cmn. The individual probabilities of belonging to one of the five species were normalized by dividing them by the sum of all individual probabilities (e.g., p1[x] + p2[x] + p3[x] + p4[x] + p5[x]).
Class assignment was defined as crisp if the probability of belonging to one class was larger than the average between 1 and even probabilities ((1 + K−1)/2). Thus, for five classes, assignment was crisp if the probability was larger than 60%. Correct classification refers to assignment of a spectrum to the same species group as conventional classification, with a classification probability that is >60%. Indeterminate or fuzzy classification refers to assignment of a spectrum to any species group with a classification probability of ≤60%. Accuracy refers to the number of cultures correctly identified relative to the total number of cultures. Specificity refers to the number of cultures identified correctly as a given species relative to the number of cultures actually belonging to that species. Sensitivity refers to the number of correctly identified cultures from one species relative to the total number of cultures from that species.
Validation.
Classifiers were evaluated by using independent validation sets that consisted of NMR data from isolates that were not part of the training process (usually 130 to 200 cultures) and also included yeast species other than the five in the study. The initial classifiers were redeveloped and tested by using additional Candida isolates as they became available. Some spectra from the initial validation sets were later added to the training sets.
RESULTS
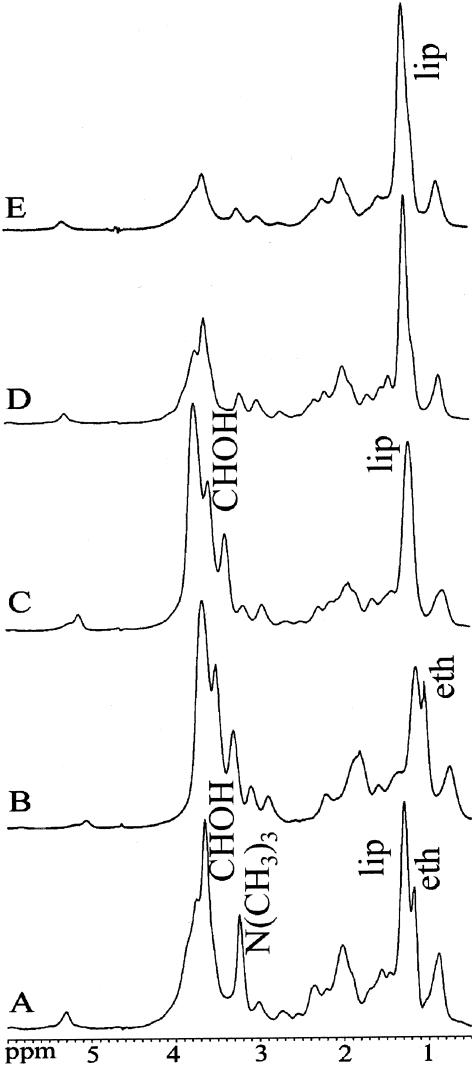
NMR spectra were obtained from 198 cultures of C. albicans (102 strains), 201 of C. glabrata (93 strains), 192 of C. parapsilosis (95 strains), 143 of C. krusei (79 strains) and 142 of C. tropicalis (73 strains). 1D NMR spectra representative of the five Candida species are shown in Fig. 1. Metabolites contributing to the major peaks were identified by 2D homo- and heteronuclear correlation spectra by using {1H, 1H} COSY, {1H, 1H} TOCSY, {1H, 13C} HSQC, and {1H, 13C} HMBC experiments. Spectra were dominated by resonances from lipids, polyols, carbohydrates, amino acid residues, ethanol, N(CH3)n groups (betaine and cholines), and, in some species, acetate. Metabolites consistently present in NMR spectra from the respective Candida species and with the potential to distinguish between them are summarized in Table 1. Since absolute quantification of metabolites was not possible due to overlapping resonances, the relative amount of each was estimated from the respective cross-peak intensities in the {1H, 13C} HSQC spectra (Table 1). Characteristic differences were noted in the relative amount of lipids, trehalose, ethanol, and acetate and the polyol profile.
FIG. 1.
Representative 1H NMR spectra from suspensions of Candida species in PBS-D2O. (A) C. albicans, (B) C. krusei, (C) C. glabrata, (D) C. parapsilosis, and (E) C. tropicalis. Signal assignment was performed by using {1H, 1H} COSY and {1H, 13C} HSQC spectra. Abbreviations: CHOH, carbohydrate and polyol residues; eth, ethanol; lip, lipids; and N(CH3)3, betaine- and choline-containing metabolites (mainly glycerophosphocholine). Signal assignment was made from COSY spectra except for polyols and carbohydrate residues, when {1H, 13C} correlation NMR spectra (HMBC and HSQC) were used. 1H NMR spectra were obtained after processing in xwinnmr (multiplication with an exponential function resulting in a line broadening of 1 Hz, Fourier transformation, first- and second-order phase correction, and polynomial baseline correction).
TABLE 1.
Metabolites of Candida species that were identified by 2D correlation NMR spectrad
| Compound | HSQC cross-peak | Metabolite concn (mM) for:
|
||||
|---|---|---|---|---|---|---|
| C. albicans | C. glabrata | C. krusei | C. parapsilosis | C. tropicalis | ||
| Lipidsa | 1.29/30.5 | 25-50 | 10-30 | 10-35 | 20-40 | 30-80 |
| Ala | 1.49/17.2 | 1-3 | 1-3 | 1-3 | 1-3 | 1-3 |
| Asp | 2.69/37.5 | 1-3 | 1-3 | 1-3 | <1 | <1 |
| Glu | 2.34/34.7 | 1-5 | 1-3 | 1-3 | 2-5 | 2-5 |
| Ile | 1.99/36.7 | ND | <1c | <1c | ND | ND |
| Leu | 1.7/40.6 | ND | <1c | ND | ND | ND |
| Lys | 3.02/40.5 | 1-10 | 1-5 | 3-10 | 1-5 | 2-8 |
| Val | 0.98/17.6 | ND | ND | <1c | ND | ND |
| Thr | 1.33/21.0 | ND | ND | <1 | ND | ND |
| Pheala | 3.29/37.7 | <1c | ND | <1 | <1 | <1c |
| Arg | 3.25/41.3 | ND | ND | ND | ND | <1 |
| Gaba | 3.04/39.9 | ND | ND | ND | ND | <1c |
| Trehalose | 5.19/94.0 | ND | 5-30 | 2-15b | Low | ND |
| Glucose | 5.23/92.9 | <1 | ND | ND | ND | <1 |
| Uridine | 7.87/142.6 | <1c | <1 | ND | ND | <1c |
| Ethanol | 3.65/58.3 | 15-40 | 10-40 | 10-30 | 5-25 | 5-15 |
| Acetate | 1.91/23.8 | 2-8 | 5-20 | 5-10 | <1 | <1 |
| Ribitol | 3.84/73.2 | Possibly | ND | ND | ND | ND |
| Glycerol | 3.78/72.3 | 5-15 | 2-10 | 5-15 | 5-10 | 8-20 |
| Arabitol | 3.90/71.0 | 5-10 | ND | 2-5b | 2-8 | 1-2c |
| Mannitol | 3.87/64.1 | ND | ND | <1 | ND | ND |
| Betaine | 3.27/54.1 | 1-3 | 1-3 | <1 | 1-3 | 1-3 |
| Dulcitol | 3.83/63.7 | ND | ND | ND | ND | 1-3 |
| GPC | 3.23/54.8 | <1c | 1-3 | ND | 2-5 | ND |
Lipid quantitation has not been corrected for the length of the fatty acid chain. The concentration refers to concentration of the represented CH2 units.
Of the five C. krusei isolates studied by {1H, 13C} HSQC spectra, one contained arabitol but no trehalose and the remaining four contained trehalose but no arabitol.
ND, not detected in all isolates.
The concentration of these compounds was determined relative to 10 mM p-aminobenzoic acid in the cell suspension and was based on the indicated {1H, 13C} HSQC cross-peak volumes, corrected for the representing number of carbon atoms. Concentration ranges were determined from at least three isolates per species. Organisms (5 × 108 to 2 × 109 CFU) were suspended in 0.5 ml of PBS/D2O to avoid sedimentation of cells in the NMR tube. Concentrations below 1 mM could not be determined reliably. The estimated error for concentration estimates of all other metabolites is of the order of 30 to 40% and confirms previous reports (4). Therefore, detectable cross-peaks of very low intensity were labeled as <1 mM.
Statistical classification strategy.
Two or three maximally discriminatory subregions were identified in the spectra of each class (species) by the genetic algorithm-based ORS process (31, 44). These regions were used to develop the LDA-based pairwise classifiers for all combinations of the five Candida species. The initial classifiers, developed on 162 cultures, resulted in a classification (identification) accuracy of 97% compared with biochemical methods of identification. However, identification of isolates of an independent validation data set (n = 145) resulted in only 88% agreement between VITEK/API and NMR/SCS. In eight cases, at least one of the duplicate cultures was classified by NMR/SCS as indeterminant (“fuzzy”). The 20 isolates yielding discrepant results (Table 2) were recultured from stock, and their identity was determined by biochemical analysis and PCR. Four cultures were mixed, one was not identified by biochemical analysis or PCR, and two yielded concordant results between PCR and NMR. One isolate, belonging to Cryptococcus neoformans, was classified by NMR as C. glabrata. One C. dubliniensis isolate was indeterminate or was classified as C. albicans. In five cases, one or both of the duplicates were misclassified by NMR/SCS as an alternate Candida species. The classifiers were redeveloped from an expanded data set (training set) of 355 cultures and were validated by using new cultures of Candida species from within and outside those in the training set. Classification accuracy in the validation set was improved (Fig. 2). Discrepant or indeterminate results (n = 19) arose from species that were not part of the training set used for classifier development. These included clinically unusual Candida species (n = 9), C. dubliniensis (n = 4), C. neoformans (n = 1), and Saccharomyces cerevisiae (n = 5). Six of these 19 isolates were not identified by VITEK/API.
TABLE 2.
Classification results for cultures that were misidentified at some stage of the classifier developmenta
| Stage | Isolate | Classification by VITEK/API | Classification by NMR/SCS | Classification by PCR fingerprinting after recultivation | Validation with final classifier |
|---|---|---|---|---|---|
| Development | WM 204 | C. albicans | Fuzzy (1) | C. albicans | C. albicans |
| of first classifiers | WM 923 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis |
| (n = 162) | WM 1007 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis |
| WM 929 | C. glabrata | C. krusei (2) | C. glabrata, atypical PCR | C. krusei | |
| Validation | WM 1029 | C. albicans | C. albicans (1), fuzzy (1) | C. dubliniensis | ND |
| of first classifiers | WM 1146 | C. glabrata | C. tropicalis (2) | C. tropicalis | C. tropicalis |
| (n = 145) | WM 1047 | C. glabrata | C. krusei (1) | Mixed, A, C. tropicalis; B, unidentified | A, C. tropicalis; B, ND |
| WM 1050 | Unidentified | C. krusei (2) | Unidentified | ND | |
| WM 1051 | Unidentified | C. albicans (1), C. glabrata (1) | Mixed, A, unidentified; B, Trichosporon beigelii | ND | |
| WM 1052 | Unidentified | C. glabrata (1), C. krusei (1) | Mixed, A, C. glabrata; B, unidentified | A, C. glabrata; B, ND | |
| WM 1070 | C. albicans | C. parapsilosis (1), fuzzy (1) | Mixed, A, C. parapsilosis; B, C. lusitaniae | A, C. parapsilosis; B, ND | |
| WM 1102 | C. glabrata | C. krusei (2) | C. glabrata | C. glabrata | |
| WM 1105 | C. krusei | C. parapsilosis (1) | C. krusei | C. krusei | |
| WM 1107 | C. glabrata | C. parapsilosis (1) | C. glabrata, atypical PCR | Fuzzy | |
| WM 1106 | C. glabrata | C. krusei (1), fuzzy (1) | C. krusei | C. krusei | |
| WM 1110 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis | |
| WM 1119 | Filobasidiella neoformans | C. glabrata (2) | F. neoformans | ND | |
| WM 1151 | C. parapsilosis | C. glabrata (1) | C. parapsilosis | C. parapsilosis | |
| WM 1154 | C. krusei | Fuzzy (1) | C. krusei | C. krusei | |
| WM 1155 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis | |
| WM 1156 | C. parapsilosis | C. glabrata (2) | C. parapsilosis | C. parapsilosis | |
| WM 1166 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis | |
| WM 1170 | C. albicans | Fuzzy (1) | C. albicans | C. albicans | |
| WM 1223 | C. albicans | Fuzzy (1) | C. albicans | C. albicans | |
| Development | WM 921 | C. albicans | Fuzzy (1) | C. albicans | C. albicans |
| of second classifiers | WM 933 | C. albicans | C. tropicalis (1) | C. albicans | Fuzzy |
| (n = 355) | WM 1180 | C. albicans | Fuzzy (1) | C. albicans | C. albicans |
| ATCC 90030 | C. glabrata | Fuzzy (1) | C. glabrata | C. glabrata | |
| WM 1054 | C. krusei | C. parapsilosis (1) | C. krusei | C. glabrata | |
| WM 1006 | C. parapsilosis | C. tropicalis (1) | C. parapsilosis | C. parapsilosis | |
| WM 972 | C. parapsilosis | Fuzzy (1) | C. parapsilosis | C. parapsilosis | |
| WM 1007 | C. parapsilosis | C. albicans (1) | C. parapsilosis, type 3 | Fuzzy | |
| WM 1121 | C. glabrata | C. tropicalis (1) | C. glabrata | C. glabrata | |
| WM 1193 | C. albicans | C. tropicalis (1) | C. albicans | C. albicans | |
| WM 929 | C. glabrata | C. krusei (2) | C. glabrata, atypical PCR | C. krusei | |
| 1218666 | C. krusei | Fuzzy (1) | C. krusei | C. krusei | |
| Validation | WM 1210 | Y. lipolytica | C. parapsilosis (1), C. tropicalis | ND | ND |
| of second classifiers | WM 1207 | C. albicans | C. tropicalis (1), fuzzy (1) | C. albicans | Fuzzy |
| (n = 229) | WM 1191 | C. albicans | C. tropicalis (1) | C. albicans | C. albicans |
| WM 1173 | C. albicans | Fuzzy (1) | C. albicans | C. albicans | |
| 1059299 | C. glabrata | Fuzzy (1) | C. glabrata | C. glabrata | |
| JH23 | S. cerevisiae | C. glabrata (1), C. krusei (1) | ND | ND | |
| WM 01.177 | Unidentified | C. glabrata (1), fuzzy (1) | Unidentified | ND | |
| WM 01.176 | C. famata | C. parapsilosis (1), fuzzy (1) | C. parapsilosis | C. parapsilosis | |
| 7208673 | C. glabrata | Fuzzy (2) | C. glabrata | C. glabrata | |
| 7227274 | C. glabrata | Fuzzy (2) | C. glabrata | C. glabrata | |
| JH45 | S. cerevisiae | C. tropicalis (1), C. glabrata (1) | ND | ND | |
| WM 1124 | S. cerevisiae | C. glabrata (2) | ND | ND | |
| WM 1227 | S. cerevisiae | C. glabrata (2) | ND | ND | |
| WM 01.179 | Unidentified | C. krusei (1), fuzzy (1) | Kluyveromyces marxianus var. marxianus | ND | |
| WM 01.180 | C. zeylanoides | C. glabrata (1), C. krusei (1) | Unidentified | ND | |
| WM 01.181 | C. zeylanoides | C. krusei (1), fuzzy (1) | C. glabrata, atypical PCR | Fuzzy | |
| WM 258 | Unidentified | C. tropicalis (1), fuzzy (1) | C. nitrativorans | ND | |
| WM 599 | Y. lipolytica | C. tropicalis (2) | Y. lipolytica | ND | |
| WM 01.14 | Unidentified | C. parapsilosis (1), fuzzy (1) | C. parapsilosis | C. parapsilosis/PICK> | |
| WM 1070 | Unidentified | C. tropicalis (1), fuzzy (1) | Mixed, A, C. parapsilosis; B, C. tropicalis | A, C. parapsilosis; B, C. tropicalis | |
| WM 1009 | F. neoformans | C. glabrata (1), fuzzy (1) | F. neoformans | ND | |
| WM 1124 | S. cerevisiae | C. glabrata (2) | S. cerevisiae | ND | |
| WM 01.195 | C. tropicalis | Fuzzy (1) | Mixed, A, C. tropicalis; B, unidentified | A, C. tropicalis; B, ND | |
| WM 01.199 | C. krusei | C. krusei (1), C. tropicalis (1) | Unidentified | ND | |
| WM 01.211 | ND | C. tropicalis (2) | Y. lipolytica | ND | |
| WM 01.205 | C. tropicalis | C. albicans (1), C. tropicalis (1) | Mixed, A, Y. lipolytica; B, unidentified | ND | |
| Development | WM 221 | C. albicans | Fuzzy | C. albicans | |
| of final classifiers | WM 1207 | C. albicans | Fuzzy | C. albicans | |
| (n = 444) | WM 02.03 | C. albicans | Fuzzy | C. albicans | |
| WM 929 | C. glabrata | Fuzzy | C. glabrata, atypical PCR | ||
| WM 01.181 | C. glabrata | Fuzzy | C. glabrata, atypical PCR | ||
| WM 1142 | C. glabrata | Fuzzy | C. glabrata | ||
| WM 1146 | C. glabrata | Fuzzy | C. glabrata | ||
| WM 932 | C. krusei | Fuzzy | C. krusei | ||
| WM 990 | C. krusei | C. glabrata | C. krusei | ||
| WM 993 | C. krusei | Fuzzy | C. krusei | ||
| WM 1154 | C. krusei | Fuzzy | C. krusei | ||
| 00-027-1760 | C. krusei | C. parapsilosis | C. krusei | ||
| 99-237-2930 | C. krusei | Fuzzy | C. krusei | ||
| WM 1003 | C. parapsilosis | Fuzzy | C. parapsilosis | ||
| WM 1007 | C. parapsilosis | Fuzzy | C. parapsilosis, type 3 | ||
| WM 01.18 | C. parapsilosis | Fuzzy | C. parapsilosis, type 3 | ||
| WM 01.56 | C. parapsilosis | Fuzzy | C. parapsilosis, type 3 | ||
| WM 930 | C. tropicalis | C. albicans | C. tropicalis | ||
| WM 981 | C. tropicalis | C. parapsilosis | C. tropicalis | ||
| Validation | WM 02.57 | C. glabrata | C. tropicalis (1) | C. glabrata | |
| of final classifiers | WM 02.05 | C. albicans | C. parapsilosis (1) | C. albicans | |
| (n = 133) | WM 02.78 | C. krusei | C. tropicalis (1) | C. krusei | |
| WM 01.203 | C. tropicalis | C. glabrata (1) | C. tropicalis | ||
| WM 02.08 | C. krusei | Fuzzy (1) | C. krusei | ||
| WM 02.86 | C. tropicalis | Fuzzy (1) | C. tropicalis |
The number in parentheses indicates the number of misidentified cultures (all isolates were cultured as duplicates except for the development and test of the final classifiers. All isolates were recultured and tested against the final classifiers except for isolates that were identified as species that were not part of this study. n indicates the total number of cultures in the respective data set. In cases of mixed cultures, the purified cultures were PCR fingerprinted. Purified cultures were labeled as A, B,.... “Unidentified” refers to indeterminate identification by VITEK and API tests and by PCR fingerprinting when the respective pattern did not match with one of the five studied Candida species or species studied by Latouche et al. (23). The comment “mixed” refers to the fact that no pure culture was obtained after recultivation of the isolate from storage. The purified isolates are named A and B. ND indicates that the respective test was not done.
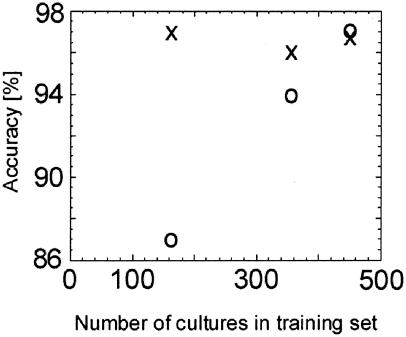
FIG. 2.
Accuracy of the classification of Candida species with increasing numbers of isolates. Classifiers were developed as isolates became available. A first set of pairwise classifiers was developed on 162 cultures. The accuracy of the classifiers on the training set is indicated by crosses. These classifiers were then tested against an independent validation set of cultures (circles). Only the number of cultures used for the development of classifiers (training set) is indicated. For number of cultures in the validation set, see Table 2. Accuracy was determined based on correct identification (compared to PCR fingerprinting). Isolates belonging to species not included in this study but part of the validation data sets were not considered.
Development and validation of the final classifiers.
Mixed cultures and those not identified by PCR or biochemical testing were excluded from the training sets. Single rather than duplicate cultures were used, since discrepancies between duplicates in the training set were rare (<1% [Table 2]). The final classifiers were trained on 444 spectra. The two most discriminatory spectral regions determined by ORS resulted in high classification accuracy (Table 3). An increase in the number of regions used to develop the classifiers improved the accuracy of classification by not more than 1 to 2% (data not shown). All isolates for which classification by SCS/NMR was indeterminate or for which there was disagreement between VITEK/API and SCS/NMR are summarized in Table 2.
TABLE 3.
Performance of final pairwise classifiers on a training and validation set of NMR spectra from cell suspensions of Candida spp.a
| Pairwise classifiers | Chemical shift range (ppm) | Training data set
|
Validation data set
|
Indeterminate sample | No. (%) of isolates with indeterminate classification for:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Sensitivity (%) | Specificity (%) | No. of isolates | Sensitivity (%) | Specificity (%) | Training data set | Validation data set | |||
| C.alb vs C.glab | 2.18-2.25; 3.72-3.79 | 102:93 | 100 | 100 | 33:26 | 100 | 100 | C.alb | 3 (3) | 0 (0) |
| C.alb vs C.krus | 1.62-1.77; 3.70-3.73 | 102:79 | 100 | 100 | 33:21 | 100 | 100 | C.glab | 4 (4) | 0 (0) |
| C.alb vs C.para | 1.52-1.58; 2.17-2.24 | 102:95 | 100 | 100 | 33:27 | 97 | 100 | C.krus | 3 (3) | 1 (5) |
| C.alb vs C.trop | 1.48-1.58; 2.97-3.14 | 102:73 | 100 | 99 | 33:23 | 100 | 100 | C.para | 3 (3) | 0 (0) |
| C.glab vs C.krus | 1.74-1.76; 2.30-2.37 | 93:79 | 100 | 98 | 26:21 | 100 | 100 | C.trop | 0 (0) | 1 (4) |
| C.glab vs C.para | 2.30-2.37; 3.67-3.77 | 93:95 | 100 | 100 | 26:27 | 100 | 100 | |||
| C.glab vs C.trop | 3.24-3.30; 3.74-3.79 | 93:73 | 100 | 100 | 26:23 | 96 | 96 | |||
| C.krus vs C.para | 2.05-2.12; 2.48-2.56 | 79:95 | 98 | 100 | 21:27 | 100 | 100 | |||
| C.krus vs C.trop | 1.46-1.51; 3.92-4.04 | 79:73 | 100 | 100 | 21:23 | 95 | 100 | |||
| C.para vs C.trop | 3.76-3.82; 3.95-4.01 | 95:73 | 100 | 97 | 27:23 | 100 | 100 | |||
The left part of the table shows the 1H NMR chemical shift regions used for the individual pairwise classifiers and the sensitivity and specificity of the pairwise classifiers on the training set and a validation set. The numbers of isolates refer to the species indicated in column 1. The right part of the table lists the number of isolates in the two data sets that were not confidently (class assignment P > 60%) assigned to one of the species (fuzzy classification). C.alb, C. albicans; C.glab, C. glabrata; C.krus, C. krusei; C.para, C. parapsilosis; and C.trop, C. tropicalis.
The final validation set contained recent clinical yeast isolates whose identity had not been confirmed at the time of validation (Tables 2 and 3). Isolates belonging to species not included in this study were subsequently excluded from the final independent validation set. The accuracy of identification in both the final training set and the independent validation set was similarly high (Fig. 2). NMR spectra of previously indeterminate or misclassified isolates were tested against the final classifiers. The identification agreed with PCR-based identification in 39 of the 49 instances of previously indeterminate or misidentified isolates (Table 2).
Reproducibility and effect of specific culture conditions on identification by SCS/NMR.
The reproducibility of the method was evaluated by repeated testing of five cultures of two isolates per species. The results obtained under standard culture conditions were highly reproducible, resulting in only one fuzzy classification out of 50 cultures. The effect of different culture conditions was tested by using duplicate cultures of two isolates per species. Variation in the time of incubation (24, 49, 72, 96, and 192 h) revealed that, with the exception of one C. albicans culture that was indeterminately classified, identifications based on 24- to 72-h cultures were correct. Four cultures (two of C. albicans) were indeterminate, and two cultures were misclassified after incubation for 96 h. Seven cultures were indeterminate, and four were misclassified after incubation for 192 h. Two isolates of C. parapsilosis and one of C. albicans failed to grow at 40°C, but cultures incubated at 25°C and 30°C were identified correctly. Two cultures of C. krusei and one of C. parapsilosis were indeterminate after growth at 25°C. One culture of C. krusei and one of C. tropicalis were misidentified, and two cultures of C. krusei and one of C. glabrata were indeterminate after incubation at 35°C. One culture of C. krusei and one of C. tropicalis were misidentified after incubation at 40°C. Storage for up to 24 h at room temperature did not influence the classification results. Storage for an additional 96 h resulted in indeterminate or misclassification of 30% of all cultures and up to 50% of those of C. krusei and C. glabrata. The growth medium significantly influenced the NMR spectral characteristics. Cultures in YPG displayed quantitatively different metabolite profiles with generally larger amounts of ethanol and acetate than in those cultured on SAB plates (data not shown). This resulted in up to 50% of C. albicans and C. glabrata isolates being indeterminate or misclassified.
DISCUSSION
Our data confirm the potential of 1H NMR spectroscopy combined with analysis by an SCS (NMR/SCS) for a high-throughput, rapid, nondestructive identification of yeast species. Using the artificial genus Candida as the model for establishing proof of concept, the overall accuracy of identification of an independent validation set of isolates using the final classifiers was 97%. NMR/SCS was at least as accurate as conventional phenotypic identification (23). As with all phenotypic methods, reproducibility and accuracy of identification are dependent on the use of standardized growth medium and conditions. However, the NMR/SCS method does exhibit tolerances of short- and long-term procedural variations. Use of different batches of a specified culture medium, incubation at 25 to 30°C for 24 to 72 h, heterogeneity within colonies scraped from agar plates, and storage at room temperature for up to 24 h before NMR spectroscopy did not significantly affect reproducibility or accuracy of identification. Species-specific differences were noted following substantial changes in culture conditions, most likely due to different metabolic rates and responses to such changes (e.g., large amounts of ethanol were produced by C. albicans after prolonged incubation, and C. krusei produced larger amounts of α,α-trehalose when cultured at higher temperatures).
Statistical classification strategy.
1H NMR spectra of living cells contain information derived simultaneously from all mobile chemicals (metabolites and cell components) that are present in the cell suspension. The advantage of NMR spectroscopy compared with other biochemical methods of identification is that this information is retrieved rapidly from a single test. The need to assign the respective NMR spectra to particular metabolites is circumvented by analyzing variations in the signal pattern rather than identification of individual compounds. The SCS was specifically designed for the analysis of NMR and infrared spectra from biological fluids and tissues (44), where individual spectra contain many more data points (attributes) than the number of spectra (sample size) (reviewed in reference 24). The genetic algorithm-based ORS for attribute reduction used in NMR/SCS has the advantage over principal component analyses applied by others for analysis of NMR, FTIR, and Raman spectra (9, 17, 26, 27) that the selected features retain their spectral identity (correspond to spectral subregions). The most discriminatory spectral regions identified by ORS point to particular metabolites that contributed to a successful discrimination. These spectral regions (metabolites) are consistent for all isolates within the particular species. Thus, NMR/SCS also provides a means of rapidly screening for stable phenotypic markers (metabolite profiles) that can be used for classification. This is of particular value in yeast species with few distinctive phenotypic characters (28). SCS proved to be a robust method for identification of Candida isolates to the species level in the present study, based on analysis of only two spectral regions selected by the ORS. The smaller the number of attributes utilized for a successful analysis, the more robust it is, as both theoretical and empirical evidence suggests that the sample-to-attribute ratio must be greater than 10 to 1 (31).
The robustness and reliability of the NMR/SCS analysis were proven by testing against additional, independent isolates that were not included in the development of the initial classifiers. As expected, the accuracy of identification increased with increasing numbers of isolates in the training set (Fig. 2). Classifiers were considered robust when the accuracy and crispness of identification using an independent validation set approached those obtained on the training set. Convergence of the accuracy for the training and validation sets was achieved with 444 cultures in the training set (Fig. 2). This may vary for different classification problems and is potentially problematic for species where the number of known isolates is small. The larger the training set, the likelier it is that the randomly selected members (isolates) of a particular class (species) represent the most of the data space (phenotype range) occupied by all members of that species. The increased reliability of the classifiers with more isolates was confirmed by the improved results on isolates that were initially classified as indeterminate or misclassified (Table 2).
Metabolite profiles.
Although NMR/SCS is based on biochemical information in NMR spectra, identification of individual compounds is not necessary for successful classification. We confirmed by 2D correlation NMR spectroscopic techniques that many of the most discriminatory regions utilized for development of the pairwise classifiers in the SCS correspond to the polyol/carbohydrate region of the NMR spectra (3.5 to 4.0 ppm) and to a less specific region (1.5 to 2.5 ppm) exhibiting resonance of lipids, amino acid residues, acetate, and other O- and N-acetyl compounds. Differences in polyol, lipid, and carbohydrate composition of material from purified yeast extracts, cell walls, or capsular components have been used previously to classify fungi at a taxonomic level above species (1, 3, 13, 36). Table 1 shows that more than a single metabolite or group of compounds differ quantitatively between species. The advantage of NMR spectroscopic studies based on suspensions of whole cells is that all NMR-visible metabolites can be included in the analysis, which increases the discriminatory power of the method and results in a rapid sample throughput. Analysis of large databases is essential for any chemotaxonomic approach to overcome natural variations between isolates of the same taxon (5). Although the NMR/SCS approach selects out spectral regions and metabolites with the most discriminatory power, it still uses only a small portion of the entire NMR spectrum. The polyphasic approach of NMR spectroscopy, based on an overview of secondary metabolites, primary metabolite pools, and other compounds (including carbohydrates, lipids, and amino acids), provided a more discriminating and rapid method for classification of fungi to the species level than did monophasic chemotypic methods. In addition, the utilization of live cells rather than cell extracts minimizes manual sample handling. Alternative, nondestructive, and polyphasic spectroscopic techniques being investigated for microbial identification (FTIR and near infrared multichannel Raman spectroscopy) are less robust and cannot be used to identify particular metabolites (26, 49). Furthermore, these techniques require preprocessing of plate cultures before spectroscopy, which adds at least an extra 6 h to the identification time (26, 27).
Characterization of indeterminate and misclassified isolates.
It is of interest that classification using the final classifiers resulted in correct identification of 80% of previously misclassified isolates. Apart from the fact that the isolates used in our analysis still represent only a small fraction of the entire data space, additional factors that may have contributed to incorrect or indeterminate identification are the (i) presence of mixed cultures for seven isolates and (ii) intraspecific discontinuity among seven isolates (Table 2).
Mixed isolates were in most cases misidentified by both VITEK/API as well as NMR/SCS, with marginally better results for NMR-based identification, presumably because one of the cultures was dominating the NMR spectrum. NMR/SCS identification of the purified and recultured isolates resulted in the assignment to the correct species in all cases.
Intraspecies discontinuity has been reported in a number of Candida species (22, 25, 32, 38, 39). A small number of isolates, analyzed by NMR/SCS, were indeterminate or misclassified throughout the study. Most of these isolates showed both NMR spectra and genotypes (PCR and partial actin gene sequences) different from the majority. For example, three distinct groups have been recognized for C. parapsilosis (25, 38, 39). Among the less common types II and III, type III is more different from type I, consistent with allocation of a different biotype code in the API 20C identification kit (38). Some C. parapsilosis type II (WM 1.57) and type III (WM 01.18, WM 1007, and WM 1.56) isolates included in this study were identified by partial sequence analysis of the actin gene (7). Whereas only one culture of WM 1.57 was at some stage indeterminately classified by NMR/SCS, all cultures of the more distantly related type III were repeatedly indeterminate or misclassified (Table 2), indicating unique NMR spectra and therefore “chemotypes” that were distinct. Similarly, isolates of C. glabrata (WM 929, WM 01.181, and WM 1107) with an unusual PCR fingerprinting pattern (data not shown) were more often classified as indeterminate or misclassified by NMR/SCS.
Identification of unusual clinical isolates.
Some isolates in the blinded validation sets were found subsequently to belong to species not included in this study (Table 2). They were either indeterminately classified or misclassified as closely related species. For example, isolates of C. dubliniensis were consistently identified as C. albicans, consistent with their close genetic relationship (48). The type strain of C. dubliniensis (CBS 7987) was also classified as C. albicans when compared against the classifiers (data not shown). In other cases metabolic similarities resulted in consistent indeterminate identification or misidentification. For example, Yarrowia lipolytica, a species with high lipid content, was misidentified as C. tropicalis, the species in the training set with the highest lipid content. C. neoformans was consistently identified as C. glabrata. Both are characterized by a very high trehalose content (16) (Table 2).
Identification of species outside those used for classifier development requires an additional data analysis approach based for example on a distance analysis. This was applied successfully for analysis of complex proteomic data (R. L. Somorjai et al., unpublished data) and is under development for microbial applications.
The presented data suggest that NMR/SCS will provide a rapid chemotypic method for microbial identification that can be fully automated. Species characterized by paucity of phenotypic characters were investigated in this study, and the accuracy of NMR/SCS exceeded that of traditional identification systems.
Acknowledgments
We acknowledge C. Halliday for assistance with cultures from the Australian candidemia study, W. Meyer for making available isolates from the culture collection of the Molecular Mycology Laboratory at Westmead Hospital, and D. Yurdakul and J. Watzl for some NMR measurements.
This work was supported by the National Health and Medical Research Council of Australia (grant 153805).
REFERENCES
- 1.Botha, A., and J. L. F. Kock. 1993. Application of fatty acid profiles in the identification of yeasts. Int. J. Food Microbiol. 19:39-51. [DOI] [PubMed] [Google Scholar]
- 2.Bourne, R., U. Himmelreich, A. Sharma, C. Mountford, and T. Sorrell. 2001. Identification of Enterococcus, Streptococcus, and Staphylococcus by multivariate analysis of proton magnetic resonance spectroscopic data from plate cultures. J. Clin. Microbiol. 39:2916-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brondz, I., and I. Olsen. 1990. Multivariate analysis of cellular carbohydrates and fatty acids of Candida albicans, Torulopsis glabrata, and Saccharomyces cerevisiae. J. Clin. Microbiol. 28:1854-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubb, W. A., L. C. Wright, M. Cagney, R. T. Santangelo, T. C. Sorrell, and P. W. Kuchel. 1999. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: identification of possible virulence factors. Magn. Reson. Med. 42:442-453. [DOI] [PubMed] [Google Scholar]
- 5.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronooxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, D. C., M. G. Rinaldi, K. A. Haynes, J. H. Rex, R. C. Summerbell, E. J. Anaissie, A. Li, and D. J. Sullivan. 1998. Importance of Candida species other than Candida albicans as opportunistic pathogens. Med. Mycol. 36:156-165. [PubMed] [Google Scholar]
- 7.Daniel, H. M., T. C. Sorrell, and W. Meyer. 2001. Partial sequence analysis of the actin gene and its potential for studying the phylogeny of Candida species and their teleomorphs. Int. J. Syst. E vol. Microbiol. 51:1593-1606. [DOI] [PubMed] [Google Scholar]
- 8.Efron, B. and R. Tibshirani. 1993. An introduction to the bootstrap. Monographs on statistics and applied probability; 57. Chapman & Hall, New York, N.Y.
- 9.Flury, B. D. 1994. Developments in principal component analysis, p. 14-33. In W. J. Krzanowski (ed.), Descriptive multivariate analysis. Clarendon Press, Oxford, United Kingdom.
- 10.Fricker-Hidalgo, H., S. Orenga, B. Lebeau, H. Pelloux, M. P. Brenier-Pinchart, P. Ambroise-Thomas, and R. Grillot. 2001. Evaluation of Candida ID, a new chromogenic medium for fungal isolation and preliminary identification of some yeast species. J. Clin. Microbiol. 39:1647-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisvad, J. C., P. D. Bridge, and D. K. Arora. 1998. Chemical fungal taxonomy. Marcel Dekker, Inc., New York, N.Y.
- 12.Fuson, G. B., C. W. Price, and H. J. Phaff. 1980. Deoxyribonucleic acid base sequence relatedness among strains of Pichia ohmeri that produce dimorphic ascospores. Int. J. Syst. Bacteriol. 30:217-219. [Google Scholar]
- 13.Gorin, P. A. J., and J. F. T. Spencer. 1970. Proton magnetic resonance spectroscopy—an aid in identification and chemotaxonomy of yeast. Adv. Appl. Microbiol. 13:25-89. [Google Scholar]
- 14.Hagberg, G. 1998. From magnetic resonance spectroscopy to classification of tumours. A review of pattern recognition methods. NMR Biomed. 11:148-156. [DOI] [PubMed] [Google Scholar]
- 15.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmelreich, U., T. E. Dzendrowskyj, C. Allen, S. Dowd, R. Malik, B. P. Shehan, P. Russell, C. E. Mountford, and T. C. Sorrell. 2001. Cryptococcomas distinguished from gliomas with MR spectroscopy: an experimental rat and cell culture study. Radiology 220:122-128. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, E., A. W. Nicholls, J. C. Lindon, S. Ramos, M. Spraul, P. Neidig, S. C. Connor, J. Connelly, S. J. P. Damment, J. Haselden, and J. K. Nicholson. 1998. Development of a model for classification of toxin-induced lesions using 1H NMR spectroscopy of urine combined with pattern recognition. NMR Biomed. 11:235-244. [DOI] [PubMed] [Google Scholar]
- 18.Hwang, T.-L., and A. J. Shaka. 1995. Water suppression that works: excitation sculpting using arbitrary waveforms and pulse field gradients. J. Magn. Reson. 112:275-279. [Google Scholar]
- 19.Kitch, T. T., M. R. Jacobs, M. R. McGinnis, and P. C. Appelbaum. 1996. Ability of RapID Yeast Plus System to identify 304 clinically significant yeasts within 5 hours. J. Clin. Microbiol. 34:1069-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50:243-260. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P. 1984. Synonymy of the yeast genera Hansenula and Pichia demonstrated through comparisons of deoxyribonucleic acid relatedness. Antonie Leeuwenhoek 50:209-217. [DOI] [PubMed] [Google Scholar]
- 22.Lachance, M.-A., H. J. Phaff, W. T. Starmer, A. Moffitt, and L. G. Olson. 1986. Interspecific discontinuity in the genus Clavispora Rodrigues de Miranda by phenetic analysis, genomic deoxyribonucleic acid reassociation, and restriction mapping of ribosomal deoxyribonucleic acid. Int. J. Syst. Bacteriol. 36:524-530. [Google Scholar]
- 23.Latouche, G. N., H.-M. Daniel, O. C. Lee, T. G. Mitchell, T. C. Sorrell, and W. Meyer. 1997. Comparison of use of phenotypic and genotypic characteristics for identification of species of the anamorph genus Candida and related teleomorph yeast species. J. Clin. Microbiol. 35:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lean, C. L., R. L. Somorjai, I. C. P. Smith, P. Russell, and C. E. Mountford. 2002. Accurate diagnosis and prognosis of human cancers by proton MRS and a three-stage classification strategy. Annu. Rep. NMR Spectrosc. 48:71-111. [Google Scholar]
- 25.Lehmann, P. F., D. Lin, and B. A. Lasker. 1992. Genotypic identification and characterization of species and strains within the genus Candida using random amplified polymorphic DNA. J. Clin. Microbiol. 30:3249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maquelin, K., L.-P. Choo-Smith, T. van Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining, and G. J. Puppels. 2000. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal. Chem. 72:12-19. [DOI] [PubMed] [Google Scholar]
- 27.Maquelin, K., L.-P. Choo-Smith, H. P. Endtz, H. A. Bruining, and G. J. Puppels. 2002. Rapid identification of Candida species by confocal Raman microspectroscopy. J. Clin. Microbiol. 40:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, S. A., R. W. Payne, and D. Yarrow. 1998. Candida Berkout, p. 454-573. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts: a taxonomic study. Elsevier, Amsterdam, The Netherlands.
- 29.Meyer, W., G. N. Latouche, H. M. Daniel, M. Thanos, T. G. Mitchell, D. Yarrow, G. Schõnian, and T. Sorrell. 1997. Identification of pathogenic yeasts of the imperfect genus Candida by PCR-fingerprinting. Electrophoresis 18:1558-1559. [DOI] [PubMed] [Google Scholar]
- 30.Mountford, C. E., R. L. Somorjai, P. Malycha, L. Gluch, C. Lean, P. Russell, B. Barraclough, D. Gillett, U. Himmelreich, B. Dolenko, A. E. Nikulin, and I. C. P. Smith. 2001. Diagnosis and prognosis of breast cancer by magnetic resonance spectroscopy of fine-needle aspirates analysed using a statistical classification strategy. Br. J. Surg. 88:1234-1240. [DOI] [PubMed] [Google Scholar]
- 31.Nikulin, A., B. Dolenko, T. Bezabeh, and R. Somorjai. 1998. Near-optimal region selection for feature space reduction: novel preprocessing methods for classifying MR spectra. NMR Biomed. 11:209-217. [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa, A., H. Tomomatsu, T. Sugita, R. Ikeda, and T. Shinoda. 1996. Taxonomic position of clinical isolates of Candida famata. J. Med. Vet. Mycol. 34:411-419. [DOI] [PubMed] [Google Scholar]
- 33.Pfaller, M. A. 1995. Epidemiology of candidiasis. J. Hosp. Infect. 30(Suppl.):329-338. [DOI] [PubMed] [Google Scholar]
- 34.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, R. J. Hollis, and The SENTRY Participant Group. 2000. Bloodstream infections due to Candida species: SENTRY Antimicrobial Surveillance Program in North America and Latin America, 1997-1998. Antimicrobial Agents Chemother. 44:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, R. J. Hollis, and the SENTRY Participants Group. 2002. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfyffer, G. E., B. U. Pfyffer, and D. M. Rast. 1986. The polyol pattern, chemotaxonomy, and phylogeny of the fungi. Sydowia 39:160-201. [Google Scholar]
- 37.Price, C. W., G. B. Fuson, and H. J. Phaff. 1978. Genome comparison in yeast systematics: delimitation of species within the genera Schwanniomyces, Saccharomyces, Debaryomyces, and Pichia. Microbiol. Rev. 42:161-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy, B., and S. A. Meyer. 1998. Confirmation of the distinct genotype groups within the form species Candida parapsilosis. J. Clin. Microbiol. 36:216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnitzler, N., R. Lütticken, and G. Haase. 1999. Rapid identification of Candida glabrata by using a dipstick to detect trehalase-generated glucose. J. Clin. Microbiol. 37:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaka, A. J., P. B. Barker, and R. Freeman. 1985. Computer-optimized decoupling scheme for wideband applications and low-level operation. J. Magn. Reson. 64:547-552. [Google Scholar]
- 42.Slifkin, M. 2000. Tween 80 opacity test responses of various Candida species. J. Clin. Microbiol. 38:4626-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somorjai, R., B. Dolenko, A. Nikulin, N. Pizzi, G. Scarth, P. Zhilkin, W. Halliday, D. Fewer, N. Hill, I. Ross, M. West, I. C. Smith, S. M. Donnelly, A. C. Kuesel, and K. M. Briere. 1996. Classification of 1H MR spectra of human brain biopsies: the influence of preprocessing and computerized consensus diagnosis on classification accuracy. J. Magn. Res. Imaging 6:437-444. [DOI] [PubMed] [Google Scholar]
- 44.Somorjai, R. L., B. Dolenko, A. Nikulin, P. Nickerson, D. Rush, A. Shaw, M. Glogowski, J. Rendell, and R. Deslauriers. 2002. Distinguishing normal from rejecting renal allografts: application of a three-stage classification strategy to MR and IR spectra of urine. Vib. Spectrosc. 28:97-102. [Google Scholar]
- 45.Spraul, M., M. Hofmann, M. Ackermann, A. W. Nicholls, S. J. P. Damment, J. N. Haselden, J. P. Shockor, J. K. Nicholson, and J. C. Lindon. 1997. Flow injection proton nuclear magnetic resonance spectroscopy combined with pattern recognition methods: implications for rapid structural studies and high throughput biochemical screening. Anal. Commun. 34:339-341. [Google Scholar]
- 46.Stager, C. E., and J. R. Davis. 1992. Automated systems for identification of microorganisms. Clin. Microbiol. Rev. 5:302-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starmer, W. T., H. J. Phaff, M. Miranda, and M. W. Miller. 1978. Pichia amethionina, a new heterothallic yeast associated with the decaying stems of cereoid cacti. Int. J. Syst. Bacteriol. 28:433-441. [Google Scholar]
- 48.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. A. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 49.Tintelnot, K., G. Haase, M. Seibold, F. Bergmann, M. Staemmler, T. Franz, and D. Naumann. 2000. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J. Clin. Microbiol. 38:1599-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viljoen, B. C., and J. L. F. Kock. 1989. A taxonomic study of the yeast genus Candida Berkhout. Syst. Appl. Microbiol. 12:91-102. [Google Scholar]
- 51.Weiss, G. H., J. E. Kiefer, and J. A. Ferretti. 1992. Accuracy and precision in the estimation of internuclear distances for structure determinations. J. Magn. Reson. 97:227-234. [Google Scholar]