Abstract
A presumed antimicrobial enzyme system, the Curvularia haloperoxidase system, was examined with the aim of evaluating its potential as a sanitizing agent. In the presence of hydrogen peroxide, Curvularia haloperoxidase facilitates the oxidation of halides, such as chloride, bromide, and iodide, to antimicrobial compounds. The Curvularia haloperoxidase system caused several-log-unit reductions in counts of bacteria (Pseudomonas spp., Escherichia coli, Serratia marcescens, Aeromonas salmonicida, Shewanella putrefaciens, Staphylococcus epidermidis, and Listeria monocytogenes), yeasts (Candida sp. and Rhodotorula sp.), and filamentous fungi (Aspergillus niger, Aspergillus tubigensis, Aspergillus versicolor, Fusarium oxysporum, Penicillium chrysogenum, and Penicillium paxilli) cultured in suspension. Also, bacteria adhering to the surfaces of contact lenses were killed. The numbers of S. marcescens and S. epidermidis cells adhering to contact lenses were reduced from 4.0 and 4.9 log CFU to 1.2 and 2.7 log CFU, respectively, after treatment with the Curvularia haloperoxidase system. The killing effect of the Curvularia haloperoxidase system was rapid, and 106 CFU of E. coli cells/ml were eliminated within 10 min of treatment. Furthermore, the antimicrobial effect was short lived, causing no antibacterial effect against E. coli 10 min after the system was mixed. Bovine serum albumin (1%) and alginate (1%) inhibited the antimicrobial activity of the Curvularia haloperoxidase system, whereas glucose and Tween 20 did not affect its activity. In conclusion, the Curvularia haloperoxidase system is an effective sanitizing system and has the potential for a vast range of applications, for instance, for disinfection of contact lenses or medical devices.
Bacteria and other microorganisms attach readily to surfaces in both natural and man-made ecosystems. If they are left undisturbed, and if nutrients are supplied, biofilms may form (9, 10). Microorganisms attached as single cells or in monolayers and more mature microbial biofilms are the cause of problems in several areas, for example, in the food industry, where bacteria colonize processing equipment and subsequently contaminate (or recontaminate) the products (22). There are reports of clones of pathogenic bacteria, such as Listeria monocytogenes, which have persisted in food-processing environments for several years (12, 27). Even more severe problems are seen in the medical area, where surface-adhered microorganisms cause infections from contaminated contact lenses (42) or medical devices, such as catheters or ear tubing (4, 28, 34). Effective cleaning and surface disinfection is essential to control these hazards.
A vast range of agents or compounds that can inactivate microorganisms are therefore used across many sectors, from the pharmaceutical industry to the food industry to water distribution lines. Such sanitizers are used both for surface disinfection and for water purification (15, 21, 25). Due to the environmental impact, there is a growing concern about the disposal of sanitizers and chemically synthesized disinfectants. This has led to increased research efforts to identify and evaluate natural antimicrobial compounds as novel sanitizers (14, 30). Natural antimicrobial systems are widespread, occurring in mammals, cold-blooded animals, plants, and microorganisms (3). Examples of such compounds are iron-chelating substances, such as lactoferrin (41), small basic peptides (16, 17, 20), and enzymes with antimicrobial activity. Examples of the last are lactoperoxidase, secreted from various mammalian glands (14, 43), and lysozyme, found in body fluids, such as tears, saliva, and human milk (26).
Peroxidases other than lactoperoxidase may also exhibit antimicrobial activity. Thus, preliminary studies have demonstrated that a haloperoxidase isolated from the filamentous fungus Curvularia verruculosa has antimicrobial activity (18). Curvularia haloperoxidase (Novozymes A/S, Bagsværd, Denmark), examined in this study, is not commercially available but is produced by recombinant technology that allows production on an industrial scale. The enzyme oxidizes halides, such as bromide, chloride, and iodide, in the presence of hydrogen peroxide, and it is believed, albeit not experimentally verified, that reactive oxygen species with antimicrobial effects are produced. Commercial sanitizers based on, for example, hydrogen peroxide alone have several side effects, such as corrosion. The Curvularia haloperoxidase system uses 100-fold-lower concentrations of H2O2 and would therefore be expected to be less corrosive. The present study was undertaken with the aim of evaluating the Curvularia haloperoxidase system as a sanitizing agent. The stability of the system, as well as effects of potential interfering agents, were studied to evaluate potential areas of application. Finally, the system was tested on bacteria adhering to contact lenses to determine its potential as a surface disinfectant.
(This work was carried out as part of an industrial Ph.D. study by Eva Holm Hansen.)
MATERIALS AND METHODS
Microorganisms and growth conditions.
The microorganisms (with strain numbers and origins) used in this study are listed in Table 1. Among the tested strains were three Pseudomonas putida efflux pump mutants (DOT-T1E-PS30, DOT-T1E-PS34, and DOT-T1E-82) (39). DOT-T1E-PS30 was a double mutant and lacked functional TtgDEF and TtgGHI pumps, while DOT-T1E-PS34 and DOT-T1E-82 were triple mutants lacking functional TtgABC, TtgDEF, and TtgGHI pumps. The sensitivities of these mutants to the Curvularia haloperoxidase system were compared to that of the wild-type parent strain, DOT-T1E. All microorganisms were cultured at 25°C. Gram-negative bacteria were cultured in tryptone soy broth (Oxoid CM129), while gram-positive bacteria were cultured in brain heart infusion (BHI) (Oxoid CM225) for 24 h. As an exception, Listeria monocytogenes was cultured at 37°C in BHI supplemented with 3 g of glucose liter−1. Yeasts were grown for 48 h in yeast extract-peptone-dextrose (YPD) broth (containing [per liter] 5.0 g of yeast extract, 10.0 g of peptone, and 10.0 g of glucose, pH 5.5). Different media were used for sporulation of filamentous fungi. Aspergillus spp. and Penicillium spp. were grown for 5 to 10 days on Czapek yeast extract agar (35) and thereafter for 4 to 7 days on malt extract agar (MEA) modified after the method of Blakeslee (5). MEA was modified by adding (per liter of MEA) 1 ml of trace metal solution consisting of 10 g of ZnSO4 · 7H2O (Merck 8883) liter−1 and 5 g of CuSO4 · 5H2O (Merck 2790) liter−1. Fusarium sp. was grown for 7 and then for 8 days on Specieller Nährstoffarmer agar (31) under UV light.
TABLE 1.
Log CFU of selected bacteria, yeasts, and filamentous fungi ml−1 before and after treatment with Curvularia haloperoxidase system (enzyme, halide, and H2O2)
| Organism | Species | Strain no./origin or reference | Treatment
|
Log CFU ml−1 ± SD
|
|||
|---|---|---|---|---|---|---|---|
| Enzyme (mg liter−1) | H2O2 (mM) | Halide (mM) | Before treatment | After treatmente | |||
| Gram-negative bacteria | P. aeruginosa | PaO1 | 0.125 | 0.250 | 0.667c | 6.1 ± 0.0 | 4.4 ± 0.1*** |
| P. putida | 2442 | 0.125 | 0.250 | 0.667c | 5.1 ± 0.0 | 3.1 ± 0.2*** | |
| P. putida | 2442 | 1.000 | 1.000 | 5.000c | 5.2 ± 0.1 | <1.0*** | |
| P. putida | DOT-T1E (39) | 0.050 | 0.100 | 0.250c | 5.8 ± 0.0 | 2.6 ± 0.3** | |
| P. putida | DOT-T1E-PS30 (39) | 0.050 | 0.100 | 0.250c | 6.0 ± 0.0 | 3.7 ± 0.0*** | |
| P. putida | DOT-T1E-PS34 (39) | 0.050 | 0.100 | 0.250c | 6.1 ± 0.2 | 3.8 ± 0.0** | |
| P. putida | DOT-T1E-82 (39) | 0.050 | 0.100 | 0.250c | 5.8 ± 0.2 | 3.0 ± 0.5** | |
| E. coli | ATCC 23282 | 0.125 | 0.250 | 0.667c | 5.7 ± 0.0 | 1.2 ± 0.2*** | |
| S. marcescens | ATCC 14041 | 0.125 | 0.250 | 0.667c | 6.2 ± 0.0 | <1.0*** | |
| A. salmonicidaa | Jno 3175/88 | 0.125 | 0.250 | 0.667c | 5.7 ± 0.0 | <1.0*** | |
| S. putrefaciensa | A6/isolated from whole, gutted cod; 0°C | 0.125 | 0.250 | 0.667c | 5.5 ± 0.0 | <1.0*** | |
| Gram-positive bacteria | S. epidermidis | DSM 20042 | 0.125 | 0.250 | 0.667c | 5.6 ± 0.0 | <1.0*** |
| L. monocytogenes | O32 (Campden Food and Drink Research Association)/Scott A; human case | 0.125 | 0.250 | 0.667c | 6.2 ± 0.0 | 1.9 ± 1.2* | |
| Yeasts | Rhodotorula sp.a | B33-1/fish-processing factory | 1.000 | 1.000 | 5.000c | 4.4 ± 0.0 | 4.4 ± 0.0NS |
| Rhodotorula sp.a | B33-1/fish-processing factory | 0.100 | 0.100 | 0.100c | 4.4 ± 0.1 | <1.0*** | |
| Candida sp.a | B114-3/fish-processing factory | 1.000 | 1.000 | 5.000c | 4.3 ± 0.0 | 4.2 ± 0.0* | |
| Candida sp.a | B114-3/fish-processing factory | 0.100 | 0.100 | 0.100d | 4.4 ± 0.0 | <1.0*** | |
| Filamentous fungi | A. nigerb | IBT 14491/Lechuigillia cave in New Mexico | 1.000 | 1.000 | 5.000c | 5.0 ± 0.1 | 4.9 ± 0.0NS |
| A. nigerb | IBT 14491/Lechuigillia cave in New Mexico | 0.050 | 0.050 | 0.050d | 5.4 ± 0.1 | 4.9 ± 0.1** | |
| A. nigerb | IBT 3250/fermentation plant | 0.050 | 0.050 | 0.050d | 5.1 ± 0.1 | 2.5 ± 0.1*** | |
| A. tubigensisb | IBT 22444/grape | 0.050 | 0.050 | 0.050d | 5.0 ± 0.1 | 1.9 ± 0.1*** | |
| A. tubigensisb | IBT 4579 | 0.050 | 0.050 | 0.050d | 5.5 ± 0.1 | 3.1 ± 0.0*** | |
| A. versicolorb | IBT 23325/salt-evaporating plant | 0.050 | 0.050 | 0.050d | 5.1 ± 0.0 | 5.2 ± 0.0NS | |
| F. oxysporumb | IBT 2478/37°C hot water | 1.000 | 1.000 | 5.000c | 5.3 ± 0.1 | 5.3 ± 0.1NS | |
| F. oxysporumb | IBT 2478/37°C hot water | 0.050 | 0.050 | 0.050d | 5.3 ± 0.0 | <1.0*** | |
| P. chrysogenumb | IBT 23469/soil | 0.050 | 0.050 | 0.050d | 5.0 ± 0.1 | <1.0*** | |
| P. paxillib | IBT 16202/optical instrument in Panama | 0.050 | 0.050 | 0.050d | 5.2 ± 0.1 | <1.0*** | |
From culture collection at Danish Institute for Fisheries Research, Department of Seafood Research, DK-2800 Kgs. Lyngby, Denmark.
From culture collection at BioCentrum, Section of Food Biotechnology and Mycology, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark.
Bromide used as halide.
Iodide used as halide.
*, P < 0.05 comparing levels before and after treatment using Student's t test; **, P < 0.01 comparing levels before and after treatment using Student's t test; ***, P < 0.001 comparing levels before and after treatment using Student's t test; NS, nonsignificant comparing levels before and after treatment using Student's t test.
Harvesting of cells and conidia.
Bacteria and yeast cells were harvested (3,360 × g; 10 min), washed once in 0.05 M HEPES (Sigma H-3375; pH 7) or 0.05 M MES (morpholineethanesulfonic acid) (Sigma M-8258; pH 6) buffer and resuspended in HEPES or MES buffer. HEPES and MES buffer were used for evaluating the microbiocidal effect of Curvularia haloperoxidase with bromide and iodide, respectively, as the halide. Conidia from filamentous fungi on MEA and Specieller Nährstoffarmer agar plates were suspended in MES buffer, and hyphae were removed by filtering the suspension through sterile glass wool.
Enumeration of microorganisms.
Plate counting was used to determine cell numbers following exposure to the Curvularia haloperoxidase system. Tenfold serial dilutions were prepared in sterile physiological saline with 0.1% peptone. To inhibit any carryover of the enzyme antimicrobial effect, the first 10-fold dilution was prepared in sterile physiological saline with 0.1% peptone and 0.5% bovine serum albumin (BSA) (Sigma A-7906), since BSA abolished the antimicrobial effect (see data below). Appropriate dilutions were surface plated, and all plates were incubated at 25°C. Gram-negative bacteria, gram-positive bacteria, and yeasts were enumerated on tryptone soy agar (Oxoid CM131), BHI agar (BHI broth with 15 g of agar liter−1), and YPD agar (containing [per liter] 5.0 g of yeast extract, 10.0 g of peptone, 10.0 g of glucose, and 20 g of agar, pH 5.5), respectively. BHI agar supplemented with glucose (3 g liter−1) was used for enumeration of L. monocytogenes cells, and plates were incubated at 37°C. Tryptone soy agar, BHI agar, and YPD agar plates were incubated for 2 to 3 days. Conidia of filamentous fungi were plated on MEA and incubated for 4 to 5 days.
Curvularia haloperoxidase antimicrobial effect.
Harvested bacteria and yeast cells were diluted to an optical density of 0.4 at 450 nm and diluted 100 times in HEPES or MES buffer, yielding bacterial counts of ∼106 CFU ml−1 and yeast counts of 104 CFU ml−1. The conidium solution was diluted to ∼5 × 105 conidia ml−1 in HEPES or MES buffer. Separate solutions of Curvularia haloperoxidase, hydrogen peroxide (Merck 1.07209), and halide in the form of KBr (Sigma P-9881) or KI (Merck 1.05043) were made six times stronger than the final concentration used in the experiment. Stock solutions of Curvularia haloperoxidase and halide were prepared in HEPES when bromide was used as the halide or in MES buffer when iodide was used. The hydrogen peroxide solution was mixed in Milli-Q water. Volumes of 200 μl of cell or conidium suspension, 100 μl of Curvularia haloperoxidase, 100 μl of hydrogen peroxide, 100 μl of halide, and 100 μl of buffer were mixed in a sterile Eppendorf tube. The final concentrations varied between 0.125 and 1.000 mg of enzyme liter−1, 0.250 and 1.000 mM hydrogen peroxide, 0.667 and 5 mM bromide, and 0.050 and 0.100 mM iodide. All enzyme experiments were performed at 25°C, because disinfection of contact lenses is used in this study as an example of surface disinfection, and contact lenses are disinfected at room temperature. Experiments were performed in duplicate, counts of bacteria were log transformed, and levels before and after treatment were compared using Student's t test.
Four different kinds of experiments were performed with Curvularia haloperoxidase to investigate (i) the antimicrobial spectrum, (ii) killing kinetics, (iii) the stability of the antimicrobial effect, and (iv) the effects of interfering compounds. These experiments were performed as follows.
(i) Antimicrobial spectrum.
Suspensions of bacteria, yeasts, and filamentous fungi were incubated with the Curvularia haloperoxidase system for 20 min at 25°C under agitation (270 rpm), and cell numbers were determined by plate counting.
(ii) Killing kinetics.
The total volume of enzyme, hydrogen peroxide, halide, and cell suspension was increased to 10 ml by mixing 3.34 ml of cell suspension, 1.66 ml of hydrogen peroxide solution, and 5.0 ml of combined enzyme and bromide solution. The hydrogen peroxide solution was prepared in Milli-Q water at six times the concentration used in the final mixture, and the combined enzyme and bromide solution was prepared in HEPES buffer at two times the concentration used in the final mixture. Suspensions of cells, hydrogen peroxide, and bromide plus enzyme were mixed in a 25-ml glass flask and incubated at 25°C under agitation (200 rpm). Samples were withdrawn every 2 to 5 min to determine cell numbers.
(iii) Stability of antimicrobial effect.
Curvularia haloperoxidase, bromide, and hydrogen peroxide were mixed and left at room temperature. At intervals between 0 and 45 min after the components were mixed, cells of Escherichia coli suspended in HEPES were added, and the mixture was incubated for 20 min at 25°C under agitation (270 rpm). Cell numbers were determined by plate counting.
(iv) Interfering compounds.
The following suspensions were prepared in Milli-Q water: 6% BSA (Sigma A-7906), 6% alginic acid (Fluka 05550), 0.6% gum xanthan (Sigma G-1253), 6% Tween 20 (Merck 8.22184), and 6% glucose (Merck 1.08337). A volume of 100 μl of one of these suspensions or dilutions thereof was added to 200 μl of E. coli cell suspension, 100 μl of Curvularia haloperoxidase solution, 100 μl of hydrogen peroxide solution, and 100 μl of bromide solution. Cell numbers were measured by plate counting 20 min after mixing the ingredients.
Disinfection of contact lenses.
Cells of Serratia marcescens or Staphylococcus epidermidis were harvested as described above and suspended in 10 ml of HEPES buffer to ∼106 CFU ml−1. The bacteria were allowed to attach by immersing contact lenses (73% water, group II, nonionic; sterile UV multifocal; Rythmic, Essilor, France) in the bacterial suspension for 5 h at 25°C with agitation (125 rpm). The lenses were washed for 1 min in 10 ml of HEPES after exposure to the cell suspension to remove loosely attached cells. Thereafter, the lenses were incubated with the Curvularia haloperoxidase system or the constituents of the Curvularia haloperoxidase system for 20 min at 25°C without agitation. The proportions of enzyme-H2O2-bromide-HEPES were 1:1:1:7 ml. Solutions of enzyme, hydrogen peroxide, and bromide were therefore prepared 10 times stronger than the desired final concentration in the mixture. The final concentrations used were 0.125 mg of enzyme liter−1, 0.667 mM bromide, and 0.250 mM hydrogen peroxide. For control experiments, the volume of the omitted constituents was replaced with HEPES buffer. The numbers of attached live cells on the lenses were determined after treatment by using indirect conductance measurements (2, 6, 11, 19, 33) with the Flexi 2000 instrument (Malthus Instruments Ltd., West Sussex, United Kingdom). In brief, the lenses were placed in Malthus glass tubes containing 3 ml of tryptone soy broth, and an insert with 0.5 ml of 0.1 N KOH was placed in each tube. Electrodes were immersed in the KOH solution, and the glass tubes were incubated in a water bath at 25°C. The conductance of the KOH solution was measured continuously, and detection times, e.g., the time from the start of the experiment until a significant change occurred in the conductance, were registered. Multiplication of microbial cells on the contact lenses results in CO2 development, and as CO2 is absorbed in the KOH, conductance decreases. The time from the start of the measurement until a significant change occurs in conductance is inversely linearly proportional to the initial number of cells. A standard curve relating detection times to cell numbers was prepared for each of the two bacteria from a 10-fold dilution series of the bacterial cell suspension used for the attaching experiment.
RESULTS
Antimicrobial spectrum of Curvularia haloperoxidase system.
The antimicrobial activity of the Curvularia haloperoxidase system was tested against selected bacteria, yeasts, and filamentous fungi (Table 1).
(i) Bacteria.
The Curvularia haloperoxidase system clearly had a bactericidal effect against both gram-negative and gram-positive bacteria, and the numbers of all tested bacteria were considerably reduced after 20 min of treatment using 0.125 mg of enzyme liter−1, 0.667 mM bromide, and 0.250 mM hydrogen peroxide. Using Student's t test, the reduction in numbers of Pseudomonas aeruginosa, P. putida 2442, E. coli, S. marcescens, Aeromonas salmonicida, Shewanella putrefaciens, and S. epidermidis cells were significant, with a P value of <0.001, and the reduction in L. monocytogenes was significant with a P value of <0.05.
The extent of the bactericidal effect was dependent on the concentration of the Curvularia haloperoxidase system, resulting in increased activity with increasing concentrations. P. putida 2442 was reduced from 5.1 ± 0.0 to 3.1 ± 0.2 log CFU ml−1 when exposed to 0.125 mg of enzyme liter−1, 0.667 mM bromide, and 0.250 mM hydrogen peroxide. No P. putida cells could be detected (<1 CFU ml−1) after 20 min when the doses were increased to 1 mg of enzyme liter−1, 5 mM bromide, and 1 mM hydrogen peroxide.
P. putida 2442 appeared more resistant to the Curvularia haloperoxidase system than E. coli, S. marcescens, A. salmonicida, S. putrefaciens, L. monocytogenes, and S. epidermidis when the same concentrations of the enzyme system were used. Approximately 105 cells ml−1 of P. putida 2442 were reduced to 103 cells ml−1 by a treatment which caused reductions of the other bacteria to 10 cells ml−1 or below. To determine if the widespread occurrence of multidrug efflux pump systems in Pseudomonas spp. were the cause of the relative resistance, we evaluated the effects of the Curvularia haloperoxidase system on three P. putida efflux pump mutants, DOT-T1E-PS30, DOT-T1E-PS34, and DOT-T1E-82 (39). The sensitivities of these mutants to the Curvularia haloperoxidase system were compared to that of the wild-type parent strain, DOT-T1E. During treatment, the wild-type strain was reduced from 5.8 ± 0.0 to 2.6 ± 0.3 log CFU ml−1 (Table 1). The pump mutants were as sensitive as the wild-type parent strain; for example, DOT-T1E-82 was reduced from 5.8 ± 0.2 to 3.0 ± 0.5 log CFU ml−1.
(ii) Yeasts.
The number of cells of Rhodotorula sp. was unaffected by treatment with 1 mg of enzyme liter−1, 5 mM bromide, and 1 mM hydrogen peroxide. A slight decrease (P < 0.05) was seen when Candida sp. was treated with this combination. When bromide was replaced with iodide, there was a marked effect against these strains (P < 0.001). Rhodotorula sp. was reduced from 4.4 ± 0.1 to <1.0 log CFU ml−1, and Candida sp. was reduced from 4.4 ± 0.0 to <1.0 log CFU ml−1 during treatment with 0.1 mg of enzyme liter−1, 0.1 mM iodide, and 0.1 mM hydrogen peroxide for 20 min.
(iii) Filamentous fungi.
Filamentous fungi, like yeasts, were not affected by the Curvularia haloperoxidase system when bromide was used as the halide. The numbers of conidia of Aspergillus niger and Fusarium oxysporum were unaffected by treatment with 1 mg of enzyme liter−1, 5 mM bromide, and 1 mM hydrogen peroxide. When bromide was replaced with iodide, there was a marked effect against these strains. Except for the reduction in numbers of A. niger (IBT 14491) conidia, which was significant with a P value of <0.01, the reductions of the numbers of conidia of all other tested strains were significant with a P value of <0.001 (Table 1). Treatment with 0.05 mg of enzyme liter−1, 0.05 mM iodide, and 0.05 mM hydrogen peroxide for 20 min reduced the number of F. oxysporum conidia from 5.3 ± 0.0 to <1.0 log CFU ml−1. Using the same concentration of the Curvularia haloperoxidase system, A. niger (IBT 14491) conidia were reduced from 5.4 ± 0.1 to 4.9 ± 0.1 log CFU ml−1.
Conidia from other strains of filamentous fungi were also treated with 0.05 mg of enzyme liter−1, 0.05 mM iodide, and 0.05 mM hydrogen peroxide. Aspergillus tubigensis IBT 22444 and A. tubigensis IBT 4579 were reduced by this treatment from 5.0 ± 0.1 and 5.5 ± 0.1 to 1.9 ± 0.1 and 3.1 ± 0.0 log CFU ml−1, respectively. By use of the same doses, Penicillium chrysogenum and Penicillium paxilli were reduced from 5.0 ± 0.1 and 5.2 ± 0.1, respectively, to <1.0 log CFU/ml. In contrast, treatment of Aspergillus versicolor conidia with 0.05 mg of enzyme liter−1, 0.05 mM iodide, and 0.05 mM hydrogen peroxide for 20 min did not affect the number of conidia.
Killing kinetics.
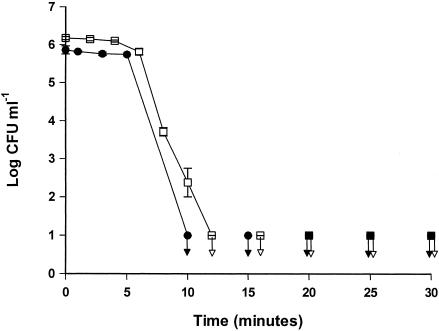
In order to assess the killing kinetics of the Curvularia haloperoxidase system, cells of E. coli and S. marcescens were treated in separate experiments with 0.125 mg of enzyme liter−1, 0.667 mM bromide, and 0.250 mM hydrogen peroxide. The initial numbers of E. coli and S. marcescens cells were 5.9 ± 0.1 and 6.2 ± 0.0 log CFU ml−1, respectively (Fig. 1). During the first 4 to 5 min of treatment, the numbers of both E. coli and S. marcescens cells remained constant, but thereafter the numbers declined. Cells of E. coli or S. marcescens could not be detected after 10 to 12 min of treatment.
FIG. 1.
Numbers of E. coli (•) and S. marcescens (□) cells during treatment with the Curvularia haloperoxidase system at 25°C. The arrows indicate samples below the detection level. The error bars represent standard deviations of duplicate determinations.
Stability of antimicrobial effect.
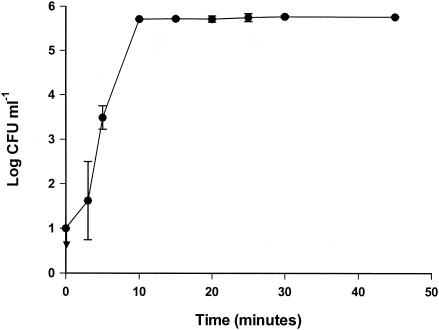
The Curvularia haloperoxidase system, consisting of 0.125 mg of enzyme liter−1, 0.667 mM bromide, and 0.250 mM hydrogen peroxide, was mixed, and the stability of the system was investigated. The bactericidal effect of the Curvularia haloperoxidase system was not stable. Suspending E. coli cells immediately after mixing the Curvularia haloperoxidase system caused complete killing (Fig. 2), and E. coli cells could not be detected after treatment. Five minutes after the Curvularia haloperoxidase system was mixed, the bactericidal effect against E. coli was reduced compared to the bactericidal effect exerted immediately after mixing. Only 3.5 ± 0.3 out of 5.8 ± 0.0 log CFU ml−1 survived treatment with the Curvularia haloperoxidase system when the system was left for 5 min before the addition of the cells. The bactericidal effect against E. coli was lost when the cells were suspended 10 min after the Curvularia haloperoxidase system was mixed, and all 5.8 ± 0.0 log CFU ml−1 survived.
FIG. 2.
Stability of Curvularia haloperoxidase system. The Curvularia haloperoxidase system was mixed and left for 0 to 45 min. Cells of E. coli were added, and the cell numbers were measured after 20 min. The arrow indicates a sample below the detection level. The error bars represent standard deviations of duplicate determinations.
Interfering compounds.
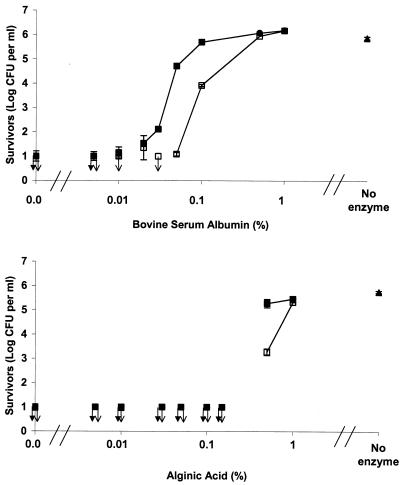
The interferences of different compounds with the bactericidal effect of the Curvularia haloperoxidase system were evaluated. BSA interfered with the antimicrobial effect, and the degree of interference depended on the concentration of BSA, as well as the concentration of the Curvularia haloperoxidase system. A concentration of 0.005% BSA did not interfere with the bactericidal effect of the haloperoxidase system (Fig. 3). Using a low concentration of the Curvularia haloperoxidase system, a BSA concentration of 0.01% increased the number of surviving E. coli cells from <1.0 to 1.2 ± 0.2 log CFU ml−1. Increasing the BSA concentration from 0.01 to 0.1% increased the number of surviving E. coli cells further, and all E. coli cells survived at a BSA concentration of 0.5% with a low enzyme concentration. Using a higher enzyme concentration, a smaller fraction of E. coli cells survived than in samples with similar BSA contents treated with a low enzyme concentration. Nevertheless, a BSA content of 1% caused all E. coli cells to survive even when a high enzyme concentration was used.
FIG. 3.
Interference of protein and alginic acid with the bactericidal effect of the Curvularia haloperoxidase system against E. coli. □, low enzyme concentration (treatment with 0.125 mg of haloperoxidase liter−1, 0.667 mM KBr, and 0.25 mM H2O2); ▪, high enzyme concentration (1.0 mg of haloperoxidase liter−1, 5.0 mM KBr, and 1.0 mM H2O2). The arrows indicate samples below the detection level. The error bars represent standard deviations of duplicate determinations.
Similarly to BSA, alginic acid interfered with the bactericidal effect of the Curvularia haloperoxidase system against E. coli (Fig. 3). Alginic acid in concentrations from 0 to 1% was used in killing experiments with E. coli, and cells were treated with the same concentrations of the Curvularia haloperoxidase system used to investigate protein interference. Alginic acid in concentrations up to 0.15% did not interfere with the bactericidal effect of the Curvularia haloperoxidase system. Adding 0.5% alginic acid caused a substantial number of E. coli cells to survive the treatment with both low and high enzyme concentrations. Similar to the protein interference, the interference of alginic acid was also dependent on the concentration of the Curvularia haloperoxidase system. Using a low enzyme concentration, 5.2 ± 0.2 log CFU ml−1 survived the treatment when 0.5% alginic acid was added compared to 3.3 ± 0.1 log CFU ml−1 with a high enzyme concentration and the same concentration of alginic acid; 1% alginic acid caused the survival of 5.5 ± 0.0 and 5.3 ± 0.0 out of 5.8 ± 0.0 log CFU ml−1 with low and high enzyme concentrations, respectively.
Glucose and Tween 20 in concentrations up to 1% did not affect the bactericidal effect of the Curvularia haloperoxidase system against E. coli.
Disinfection of contact lenses with the Curvularia haloperoxidase system.
S. marcescens and S. epidermidis adhered readily to sterile contact lenses immersed for 5 h in a suspension of S. marcescens or S. epidermidis of ∼6 log CFU ml−1. After loosely attached cells were removed by washing the lenses in HEPES buffer, the log CFU counts per lens were between 4 and 5. The lenses were treated for 20 min with 0.125 mg of enzyme liter−1, 0.250 mM hydrogen peroxide, and 0.667 mM bromide, reducing the numbers of both S. marcescens and S. epidermidis cells on the lens surfaces. After treatment with the Curvularia haloperoxidase system, the numbers of S. marcescens cells per lens were 1.2 ± 0.9 compared to 4.0 ± 0.4 log CFU on lenses placed in HEPES buffer for 20 min. The numbers of S. epidermidis cells were reduced by the treatment from 4.9 ± 0.1 log CFU on lenses placed in HEPES buffer to 2.7 ± 0.7 log CFU on the treated lenses. For both strains, control treatments with enzyme or a combination of bromide and hydrogen peroxide did not affect cell numbers on the lenses, showing that the bactericidal effect was attributable only to the three-component system of enzyme, bromide, and hydrogen peroxide.
DISCUSSION
The present study demonstrated that the Curvularia haloperoxidase system has a rapid antimicrobial effect against a broad spectrum of bacteria, yeasts, and filamentous fungi. Compared to other, monocomponent sanitizers, such as hydrogen peroxide (1), much lower concentrations are used and the exposure time is shorter. The concentration and contact time of hydrogen peroxide is important because of its corroding effect. Disinfection with 3% hydrogen peroxide caused corrosion of equipment used in ocular science, and after 144 disinfection soakings, the equipment was inaccurate because of the corrosion (7). When hydrogen peroxide is used as the sole antibacterial principle, exposure for 15 to 60 min using 0.1 to 0.6% concentrations has been required to kill urinary tract pathogens, such as E. coli, S. epidermidis, and S. marcescens (40). In comparison, using the Curvularia haloperoxidase system, a concentration of 0.25 mM (∼0.0009%) hydrogen peroxide was used to kill these three species (Table 1). The concentration of hydrogen peroxide used in our study is, depending on species and strain, 110 to 670 times lower than the concentration used by Schaeffer et al. (40). It should be mentioned that the initial number of cells in our study (∼106 CFU ml−1) is lower than that in the study by Schaeffer et al. (40) (∼108 CFU ml−1), and the Curvularia haloperoxidase system may require higher concentrations of components if aimed at higher cell concentrations. Alasri et al. (1) showed that 0.31% hydrogen peroxide reduced E. coli (ATCC 8739) by 5 log reductions during 30 min of treatment, whereas 4.5 log reductions were obtained by treating E. coli with the Curvularia haloperoxidase system using 0.25 mM (∼0.0009%) hydrogen peroxide. Since exposure time is also an important parameter when considering side effects of chemical sanitizers, it is advantageous that the killing effect of the Curvularia system is extremely rapid (Fig. 1). Also, the short-lived effect is beneficial, as the bioactive compounds disappear rapidly (Fig. 2) and thereby limit the introduction of potentially harmful chemical agents, for example, into the eye after disinfection of contact lenses.
The Curvularia haloperoxidase system has a broad spectrum and exerts its killing effect against bacteria, yeasts, or filamentous fungi. It is likely, albeit not experimentally verified, that Curvularia haloperoxidase oxidizes halides, such as bromide, chloride, and iodide, in the presence of hydrogen peroxide and produces reactive oxygen species with an antimicrobial effect. Other peroxidase systems, such as the lactoperoxidase system, are well described (38, 43). The principal electron donor for lactoperoxidase is the pseudohalide thiocyanate (SCN−). In the presence of hydrogen peroxide, lactoperoxidase oxidizes SCN− to thiocyanogen (SCN)2, which hydrolyzes rapidly to hypothiocyanous acid (HOSCN) or hypothiocyanate (OSCN−). (SCN)2 and HOSCN can oxidize protein sulfhydryl groups to sulfenyl thiocyanate derivates. It is believed that Curvularia haloperoxidase, like lactoperoxidase, oxidizes halides, for example, bromide, to its hypohalide hypobromite. These hypohalides have an oxidizing effect against proteins in the microorganism, causing the antimicrobial effect. The fact that protein, such as BSA, interferes with the antimicrobial effect of the system supports this hypothesis about the mode of action of the Curvularia haloperoxidase system.
The interfering effect of protein clearly limits the application of the Curvularia haloperoxidase system, which would not be suitable as an antimicrobial agent in environments rich in protein. Other compounds were also investigated for their potential interference because of the relevance to assessing applicability. Tween 20 was investigated instead of lipid because of its solubility and showed no interference with the antimicrobial effect. No interference was observed when the carbohydrate glucose was present during treatment. Alginate protected E. coli from the antimicrobial effects of the Curvularia haloperoxidase system. Exopolysaccharides are present in the matrix of a microbial biofilm (10), and their potential interference is therefore relevant when an antimicrobial system is considered as a surface sanitizer. Alginate is an extracellular polysaccharide produced by a variety of gram-negative bacteria, including Azotobacter vinelandii, Pseudomonas fluorescens, and P. aeruginosa (32). Structurally, alginate is a linear glycoronan composed of (1 → 4)-linked residues of β-d-mannuronic acid and its C-(5)-epimer α-l-guluronic acid, and physically it has the property of forming viscous solutions and gels (8). Learn et al. (24) have investigated hypochlorite scavenging by P. aeruginosa alginate, and apparently the uronic acid core and the O-acetyl groups of pseudomonal alginate have the ability to scavenge hypochlorite. Furthermore, they demonstrated that a mucoid P. aeruginosa was more resistant to killing by hypochlorite than a nonmucoid mutant, and the addition of purified alginate to the nonmucoid mutant protected the organism by a greater resistance to killing by hypochlorite. As mentioned, we hypothesize that hypohalides are produced by the Curvularia haloperoxidase system, and the interference of alginate with the antimicrobial effect is thus to be anticipated based on the study by Learn et al. (24). Therefore, it is unlikely that the Curvularia haloperoxidase system can be used for the inactivation of mature biofilms.
In contrast, the Curvularia haloperoxidase system proved excellent as a surface sanitizer when used on bacteria adhering to contact lenses (Table 2). The U.S. Food and Drug Administration requires that contact lens care products not only be antibacterial but also eliminate yeasts and some filamentous fungi (13). Since the Curvularia haloperoxidase system also has pronounced microbiocidal effect against yeasts and filamentous fungi (Table 1), it qualifies as a potential contact lens disinfectant. Miller et al. (29) have assessed the antimicrobial activity of lens care products and found that two out of five products examined did not meet the Food and Drug Administration's requirements for antimicrobial effect against yeasts and filamentous fungi. Although it is rare, eye infections can also be caused by protozoa, such as Acanthamoeba, and occurrences are often associated with contact lens use (23, 36, 37). The effect of the Curvularia haloperoxidase system against protozoa has not been tested. The antibacterial effect on thin layers of adhered bacteria and the rapid and unstable effect, as well as the overall low concentration of compounds, point to a vast range of other applications, for instance, as a sanitizer of equipment used in ocular science or medical devices, such as pacemakers and catheters. The exact mechanism of action of the Curvularia haloperoxidase system is unknown, and such knowledge will be crucial for predicting any potential side effects and for evaluating other areas of application.
TABLE 2.
Curvularia haloperoxidase killing of S. marcescens and S. epidermidis cells attached to the surfaces of contact lenses
| Treatment | Log CFU/lensa
|
|
|---|---|---|
| S. marcescens | S. epidermidis | |
| Control | ||
| HEPES | 4.0 ± 0.4 | 4.9 ± 0.1 |
| KBr + H2O2 | 4.3 ± 0.5 | 4.6 ± 0.1 |
| Curvularia haloperoxidase | 3.9 ± 0.3 | 4.7b |
| Treatment | ||
| Curvularia haloperoxidase system | 1.2 ± 0.9* | 2.7 ± 0.7* |
*, p < 0.05 comparing log CFU/lens on lenses treated with the Curvularia haloperoxidase system and on lenses treated with control treatments, using Student's t test.
Only a single test.
Acknowledgments
This study was financed by Novozymes A/S and the Danish Academy of Technical Sciences.
REFERENCES
- 1.Alasri, A., C. Roques, G. Michel, C. Cabassud, and P. Aptel. 1992. Bactericidal properties of peracetic acid and hydrogen peroxide, alone and in combination, and chlorine and formaldehyde against bacterial water strains. Can. J. Microbiol. 38:635-642. [DOI] [PubMed] [Google Scholar]
- 2.Bagge, D., M. Hjelm, C. Johansen, I. Huber, and L. Gram. 2001. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl. Environ. Microbiol. 67:2319-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. G., R. G. Board, and H. C. Sparks. 2001. Natural antimicrobial systems and their potential in food preservation of the future. Biotechnol. Appl. Biochem. 8:103-147. [PubMed] [Google Scholar]
- 4.Berry, J. A., J. F. Biedlingmaier, and P. J. Whelan. 2000. In vitro resistance to bacterial biofilm formation on coated fluoroplastic tympanostomy tubes. Otolaryngol. Head Neck Surg. 123:246-251. [DOI] [PubMed] [Google Scholar]
- 5.Blakeslee, A. F. 1915. Linder's roll tube method of separation cultures. Phytopathology 5:68-69. [Google Scholar]
- 6.Bolton, F. J. 1990. An investigation of indirect conductimetry for detection of some food-borne bacteria. J. Appl. Bacteriol. 69:655-661. [DOI] [PubMed] [Google Scholar]
- 7.Chronister, C. L. 1997. Structural damage to Schiotz tonometers after disinfection with solutions. Optom. Vision Sci. 74:164-166. [DOI] [PubMed] [Google Scholar]
- 8.Clementi, F. 1997. Alginate production by Azotobacter vinelandii. Crit. Rev. Biotechnol. 17:327-361. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137-140. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappinscott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 11.Dezenclos, T., M. Ascon-Cabrera, D. Ascon, J. M. Lebeault, and A. Pauss. 1994. Optimisation of the indirect impedancemetry technique; a handy technique for microbial growth measurement. Appl. Microbiol. Biotechnol. 42:232-238. [Google Scholar]
- 12.Fonnesbech Vogel, B., H. H. Huss, B. Ojeniyi, P. Ahrens, and L. Gram. 2001. Elucidation of Listeria monocytogenes contamination routes in cold-smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 67:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Food and Drug Administration. 1997. Guidance for industry: premarket notification (510(k)) guidance document for contact lens care products. U.S. Department of Health and Human Services, Center for Devices and Radiological Health, Washington, D.C.
- 14.Fuglsang, C. C., C. Johansen, S. Christgau, and J. AdlerNissen. 1995. Antimicrobial enzymes: applications and future potential in the food industry. Trends Food Sci. Technol. 6:390-396. [Google Scholar]
- 15.Gerba, C. P., D. C. Johnson, and M. N. Hasan. 1997. Efficacy of iodine water purification tablets against Cryptosporidium oocysts and Giardia cysts. Wild. Environ. Med. 8:96-100. [DOI] [PubMed] [Google Scholar]
- 16.Giacometti, A., O. Cirioni, F. Barchiesi, M. S. Del Prete, and G. Scalise. 1999. Antimicrobial activity of polycationic peptides. Peptides 20:1265-1273. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, R. E. W., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 18.Johansen, C., and Novo Nordisk A/S. June 2001. Antimicrobial composition containing a haloperoxidase, a hydrogen peroxide source, a halide source, and an ammonium source. U.S. patent 6,251,386.
- 19.Johansen, C., P. Falholt, and L. Gram. 1997. Enzymatic removal and disinfection of bacterial biofilms. Appl. Environ. Microbiol. 63:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen, C., A. Verheul, L. Gram, T. Gill, and T. Abee. 1997. Protamine-induced permeabilization of cell envelopes of gram-positive and gram-negative bacteria. Appl. Environ. Microbiol. 63:1155-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kallen, B. A. J., and E. Robert. 2000. Drinking water chlorination and delivery outcome—a registry-based study in Sweden. Reprod. Toxicol. 14:303-309. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, C. G., and S. K. Anand. 1998. Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42:9-27. [DOI] [PubMed] [Google Scholar]
- 23.Lam, D. S., E. Houang, D. S. Fan, D. Lyon, D. Seal, and E. Wong. 2002. Incidence and risk factors for microbial keratitis in Hong Kong: comparison with Europe and North America. Eye 16:608-618. [DOI] [PubMed] [Google Scholar]
- 24.Learn, D. B., E. P. Brestel, and S. Seetharama. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 55:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsay, D., and A. Von Holy. 1999. Different responses of planktonic and attached Bacillus subtilis and Pseudomonas fluorescens to sanitizer treatment. J. Food Prot. 62:368-379. [DOI] [PubMed] [Google Scholar]
- 26.Losso, J. N., S. Nakai, and E. A. Charter. 2000. Lysozyme, p. 185-210. In A. S. Naidu (ed.), Natural food antimicrobial systems. CRC Press, Boca Raton, Fla.
- 27.Maijala, R., O. Lyytikainen, T. Johansson, T. Autio, T. Aalto, L. Haavisto, and T. Honkanen-Buzalski. 2001. Exposure of Listeria monocytogenes within an epidemic caused by butter in Finland. Int. J. Food Microbiol. 70:97-109. [DOI] [PubMed] [Google Scholar]
- 28.McLean, R. J. C., J. C. Nickel, and M. E. Olson. 1995. Biofilm associated urinary tract infections, p. 261-273. In H. M. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 29.Miller, M. J., D. E. Callahan, D. McGrath, R. Manchester, and S. E. Norton. 2001. Disinfection efficacy of contact lens care solutions against ocular pathogens. CLAO J. 27:16-22. [PubMed] [Google Scholar]
- 30.Naidu, A. S. 2000. Natural food antimicrobial systems. CRC Press, Boca Raton, Fla.
- 31.Nirenberg, H. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-sektion Liseola. Mitteillungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft. Kommissionsverlag Paul Parey, Berlin-Dahlem, Germany.
- 32.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owens, J. D., D. S. Thomas, P. S. Thompson, and J. W. Timmerman. 1989. Indirect conductimetry: a novel approach to the conductimetric enumeration of microbial populations. Lett. Appl. Microbiol. 9:245-249. [Google Scholar]
- 34.Passerini, L., K. Lam, J. W. Costerton, and E. G. King. 1992. Biofilms on indwelling vascular catheters. Crit. Care Med. 20:665-673. [DOI] [PubMed] [Google Scholar]
- 35.Pitt, J. I. 1973. An appraisal of identification methods for Penicillium species: novel taxonomic criteria based on temperature and water relations. Mycologia 65:1135-1157. [PubMed] [Google Scholar]
- 36.Radford, C. F., O. J. Lehmann, and J. K. Dart. 1998. Acanthamoeba keratitis: multicentre survey in England 1992-6. Br. J. Ophthalmol. 82:1387-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radford, C. F., D. C. Minassian, and J. K. Dart. 2002. Acanthamoeba keratitis in England and Wales: incidence, outcome, and risk factors. Br. J. Ophthalmol. 86:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiter, B., and G. Härnulv. 1984. Lactoperoxidase antibacterial system: natural occurrence, biological functions and practical applications. J. Food Prot. 47:724-732. [DOI] [PubMed] [Google Scholar]
- 39.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaeffer, A. J., J. M. Jones, and S. K. Amundsen. 1980. Bactericidal effect of hydrogen peroxide on urinary tract pathogens. Appl. Environ. Microbiol. 40:337-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorland, L. H. 1999. Lactoferrin: a multifunctional glycoprotein. APMIS 107:971-981. [DOI] [PubMed] [Google Scholar]
- 42.Willcox, M. D. P., N. Harmis, B. A. Cowell, T. Williams, and B. A. Holden. 2001. Bacterial interactions with contact lenses; effects of lens material, lens wear and microbial physiology. Biomaterials 22:3235-3247. [DOI] [PubMed] [Google Scholar]
- 43.Wolfson, L. M., and S. S. Sumner. 1993. Antibacterial activity of the lactoperoxidase system—a review. J. Food Prot. 56:887-892. [DOI] [PubMed] [Google Scholar]