Abstract
A new in situ DNA amplification technique for microscopic detection of bacteria carrying a specific gene is described. Loop-mediated isothermal amplification (LAMP) was used to detect stxA2 in Escherichia coli O157:H7 cells. The mild permeabilization conditions and low isothermal temperature used in the in situ LAMP method caused less cell damage than in situ PCR. It allowed use of fluorescent antibody labeling in the bacterial mixture after the DNA amplification for identification of E. coli O157:H7 cells with an stxA2 gene. Higher-contrast images were obtained with this method than with in situ PCR.
Understanding of the structures and functions of microbial communities in natural environments often requires enumeration of specific microorganisms within that ecosystem. In natural ecosystems, nutritional requirements for bacterial growth are often lacking; most microorganisms cannot be grown in cultures with conventional techniques and many exhibit low levels of metabolic activity, making their detection with methods such as fluorescent in situ hybridization (FISH) problematic (3, 24). In the field of environmental microbiology, many culture-independent techniques have come into use over the last decade (23, 26).
Among such culture-independent techniques, FISH and fluorescent antibodies have been widely used because they can provide information about absolute abundance, morphology, cell size, and physiological activity of specific bacteria in their natural environment (1, 6, 9, 10, 18, 30). However, it has still been difficult to detect single-copy genes present in the cell by FISH (3, 13). Methods for phylogenetical identification of bacteria carrying specific genes have considerable potential in microbial ecology.
In situ amplification of a specific sequence can facilitate the detection of a single-copy functional gene inside a bacterial cell (7, 11). Various versions of prokaryotic in situ PCR and reverse transcription-PCR have been developed to detect specific genes at the single-cell level (4, 5, 8, 22, 29). For previous reports, the HNPP-Fast Red TR system (10, 32) was used for high-sensitivity detection of in situ PCR products (12, 27). However, in situ PCR has several disadvantages: (i) high background which can result from leakage of amplified products and cell destruction due to permeabilization treatments and/or thermal cycling, which makes it difficult to use fluorescent antibodies for simultaneous cell identification, (ii) false-negative results caused by impermeability of the cell wall and prevention of PCR amplification by DNA polymerase inhibitors, and (iii) false-positive results due to diffusion of the labeled amplicon and its adhesion to cells giving negative results and/or nonspecific incorporation of labeled nucleotide by nick translation activity.
Recently, Notomi et al. have developed a new DNA amplification method called loop-mediated isothermal amplification (LAMP) of DNA (16). The characteristics of this method are high specificity, high sensitivity, and the use of isothermal reactions. The possible advantages of in situ LAMP include (i) the use of an isothermal and low reaction temperature (63°C), causing less cell damage and making it possible to use fluorescent antibodies for simultaneous cell identification, (ii) the generation by LAMP of large tandem repeats of the target sequence, preventing amplicons from leakage outside the cell, and (iii) the use of low-molecular-weight DNA polymerase (2), which enters the cell easily, for in situ LAMP.
In this report, the LAMP reaction was adapted to reduce background while amplifying a specific gene inside the cell. Fluorescent antibody labeling was also applied to identify specific cells and examine the reliability of in situ LAMP. Simultaneous visualization of a functional gene and surface antigen was carried out by both in situ LAMP and in situ PCR for comparison.
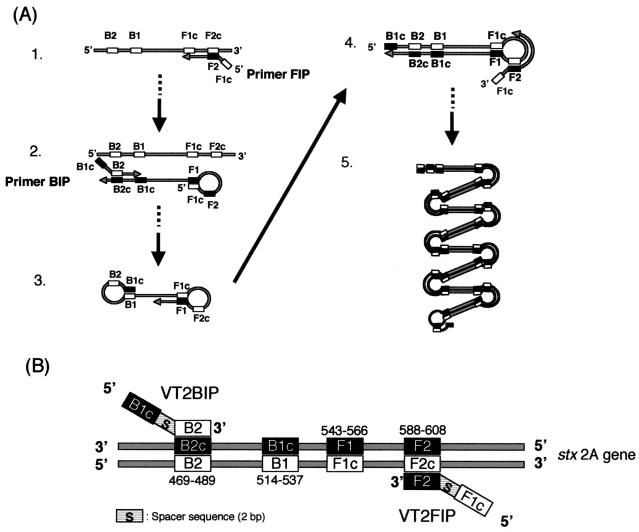
The mechanism of LAMP is summarized in Fig. 1A (16). Primer FIP hybridizes to F2c in the target DNA and initiates complementary strand synthesis, leading to the production of dumbbell form DNA (structure 2), which quickly converts a stem-loop DNA by self-primed DNA synthesis (structure 3). This stem-loop DNA then serves as the starting material for LAMP cycling. FIP hybridizes to the loop in the stem-loop DNA and primer strand displacement DNA synthesis (structure 4), generating as an intermediate one-gapped stem-loop DNA with an additional inverted copy of the target sequence in the stem and loop formed at the opposite end via the primer BIP sequence. The final products (structure 5) are mixtures of stem-loop DNAs with various stem lengths and cauliflower-like structures with multiple loops formed by annealing between alternately inverted repeats of the target sequence in the same strand. The positions of the LAMP primers for this study are summarized in Fig. 1B. Primers VT2BIP (5′-TGCTCTGGATGCATCTCTGGTCATATCTGGTTTCATCATATCTGGCG-3′) and VT2FIP (5′-GCGTTTTGTCACTGTCACAGCAGATAGCCTGACGAAATTCTCTCTGT-3′) were used for in situ LAMP. Each primer for the stxA2 gene contains two distinct sequences corresponding to the sense and antisense sequences of target DNA. Outer primers were used in the original LAMP method (16), but only inner primers (VT2BIP and VT2FIP) were used in this study because no difference was observed at the end of the reaction with and without the outer primers.
FIG. 1.
(A) Summary of steps in the LAMP reaction. (B) Systematic representation of the location of primers used in this study. The strand represents the positions of primers used for in situ LAMP. The numbers listed indicate the numbers of bases for the beginning of the stxA2 gene in E. coli O157:H7 (GenBank accession no. AP000422).
Escherichia coli K-12 W3110 and E. coli O157:H7 Okayama O27 were used. E. coli O157:H7 Okayama O27 has one stx1 gene and two stx2 genes (31), while E. coli K-12 has no stx gene. These two strains were grown in cultures at 37°C in aerobic Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl [pH 7.0]).
Exponentially growing E. coli K-12 and E. coli O157:H7 cells were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.2]), and suspended in freshly filtered paraformaldehyde (4% in PBS) for 16 h at 4°C. After fixation, cells were washed twice in PBS and suspended in 50% ethanol with sterile distilled and deionized water. Fixed cells were washed twice with PBS, and a 10-μl aliquot (about 106 to 107 cells) was spotted inside a hole (6 mm in diameter) of ML-250 polyester seal (Nichiban, Tokyo, Japan), which was attached to gelatin-coated glass slides (Matsunami Glass Ltd., Osaka, Japan), and allowed to vacuum dry. After the polyester seal was peeled off of the glass slides and dehydration in an ethanol series (50, 80, and 100% ethanol for 1 min each) was completed, samples were incubated with lysozyme solution (0.5 mg of lysozyme [Nacalai Tesque Inc., Kyoto, Japan] ml−1, 100 mM Tris-HCl [pH 8.2], 50 mM EDTA) for 10 min at room temperature, rinsed with sterile distilled and deionized water, and dehydrated again as described above.
Permeabilization was furthered by treatment for 5 min at room temperature with proteinase K (Roche Diagnostics) at a final concentration of 0.1 μg ml−1. After permeabilization, RNA was removed from cells by DNase-free RNase A (Sigma) treatment at a final concentration of 0.5 mg ml−1 for 20 min at room temperature. Finally, samples were rinsed with sterile distilled and deionized water and dehydrated in an ethanol series. Treated bacterial cells on the glass slides were sealed with a total of 50 μl of the LAMP buffer containing VT2FIP and VT2BIP primers (0.8 μM each), dATP, dGTP, and dTTP (0.02 mM each), 0.002 mM Cy3-labeled dCTP (Amersham Pharmacia), 1 M betaine, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100, and 16 U of Bst DNA large-fragment polymerase (New England Biolabs) under an AmpliCover Disk (Perkin-Elmer Applied Biosystems), following the manufacturers' instructions. The LAMP reaction was carried out by incubation at 63°C for 2 h with a thermal cycler (GenAmp In Situ PCR System 1000; Perkin-Elmer Applied Biosystems). After the amplification, the glass slides were rinsed twice with sterile distilled and deionized water at room temperature for 15 min. Then, samples were counterstained with 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) ml−1 for 10 min.
The glass slides were observed under an epifluorescence microscope (E-400; Nikon, Tokyo, Japan) with the Nikon filter sets UV-2A (30-350, DM400, and BA420), B-2A (Ex450/490, DM505, and BA520), and HQ-Cy3 (G535/50, FT565, and BP610/75) for UV, blue, and green excitation, respectively. Images were taken by a cooled charge-coupled device camera (Sensys 1401; Photometrics, Tucson, Ariz.) and stored as digital files. Exposure times for UV, blue, and green excitation were 0.2, 1, and 2 s, respectively.
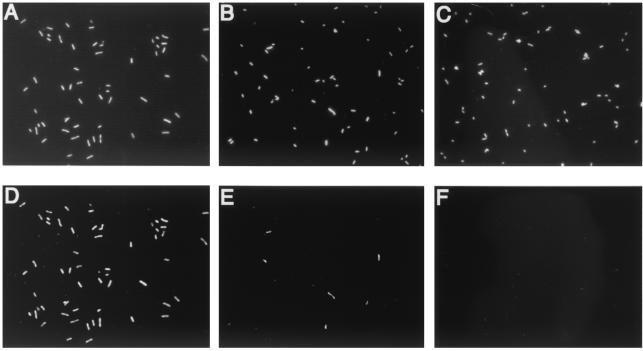
As shown in Fig. 2, the bacterial cells having the stxA2 gene (E. coli O157:H7 Okayama O27) were detected under green excitation without using image analysis. Enzymatic signal amplification in direct in situ PCR has been described previously (11, 12, 27), and the drawbacks of this method include high levels of background and the necessity of image analysis for precise detection. A labeled nucleotide incorporated in DNA synthesis by Taq polymerase to DNA nicks invariably causes a false-positive signal (17). The DNA polymerase for in situ LAMP is Bst DNA large-fragment polymerase, which does not have nick translation activity. The specificity of the reaction was further confirmed by using E. coli O157:H7 ATCC 43888 without stxA2 and E. coli O157:H7 Okayama O27 with other primers that do not have target sites in Okayama O27 (data not shown).
FIG. 2.
In situ LAMP of the stxA2 gene in fixed cells of E. coli O157:H7 Okayama O27 (A and D) and E. coli K-12 W3110 (C and F) and a fixed-cell mixture of E. coli O157:H7 Okayama O27 and E. coli K-12 W3110 (B and E). (A, B, and C) Under UV excitation (exposure time, 0.2 s), all DAPI-stained bacterial cells were visualized. (D, E, and F) Under green excitation (exposure, 2 s), only cells having stxA2-amplified products emitted red fluorescence of Cy3-labeled dCTP.
Permeabilization employed here for in situ LAMP was milder than that employed for in situ PCR because the largest molecule required for LAMP is the Bst DNA polymerase (67 kDa), which passes more easily into target cells than the 94-kDa Taq DNA polymerase used for PCR (2, 14).
The specificity of in situ LAMP was confirmed, and then this method was applied to serological identification of bacteria carrying specific genes. After in situ amplification by LAMP, the sample glass slides were rinsed with sterile distilled and deionized water at room temperature for 15 min. They were then stained at room temperature for 30 min with 2 μg of FITC-labeled anti-E. coli O157:H7 antibody (goat, polyclonal; Kirkegaard and Perry Laboratories Inc., Gaithersburg, Md.) ml−1 (12) in PBS including 30 mg of bovine serum albumin ml−1. Finally, samples were counterstained with 1 μg of DAPI ml−1 for 10 min and observed as described above.
In situ PCR was also carried out as described by Tani et al. (27) except using different primers. Primers VT2B3 and VT2F3c for the stxA2 gene (12) were used for indirect in situ PCR. The Cy3-labeled oligonucleotide probe VT2c (5′-ATGCATCTCTGGTCATTGTAT-3′) was used for detection by in situ hybridization of in situ PCR-amplified gene products, which are located between VT2B3 and VT2F3c.
A FISH step was used for detection of amplicons inside cells, because detecting an unlabeled amplicon of in situ PCR by FISH is more specific for the gene target and yields a lower level of background (28). Following in situ PCR, the samples were sealed with hybridization solution (900 mM NaCl, 5 mM EDTA, 20 mM Tris-HCl [pH 7.5], 0.01% sodium dodecyl sulfate [SDS], 50% formamide) under an AmpliCover Disk. Samples were heated at 94°C for 10 min, chilled on ice, and then incubated at 42°C for 16 h in a moisture chamber with the hybridization solution described above containing Cy3-labeled VT2c probe (5 ng μl−1 final concentration). After the incubation, glass slides were washed at 45°C for 30 min with washing buffer 1 (300 mM NaCl, 5 mM EDTA, 20 mM Tris-HCl [pH 8.2], 0.01% SDS), washed at 45°C for 30 min with washing buffer 2 (300 mM NaCl, 5 mM EDTA, 20 mM Tris-HCl [pH 8.2], 0.01% SDS, 1 μg of DAPI ml−1, 10 μg of fluorescein isothiocyanate [FITC]-labeled O157 antibody ml−1, 1 mg of bovine serum albumin ml−1), and observed as described above.
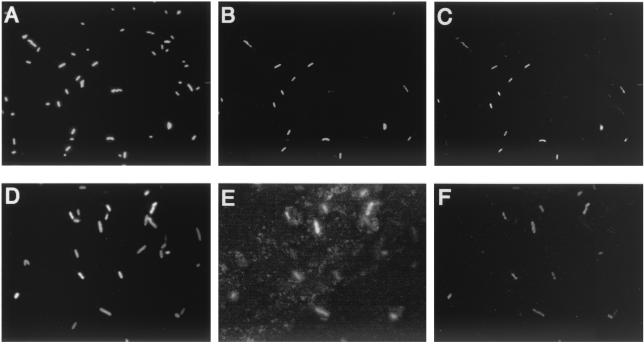
Triple staining with Cy3-dCTP for detection of the stxA2 gene, FITC-labeled antibody for detection of O157 antigens, and DAPI for nucleic acid was performed to permit simultaneous serological identification of cells and detection of a specific toxin gene. Figure 3 shows the results of in situ LAMP with an antibody-staining method for bacterial mixtures of E. coli K-12 W3110 and E. coli O157:H7 Okayama O27 (Fig. 3A, B, and C). Under UV excitation (Fig. 3A), all bacterial cells showed blue fluorescence of DAPI. Under blue excitation (Fig. 3B), E. coli O157:H7 Okayama O27 cells showed green fluorescence of FITC-labeled antibody. Under green excitation (Fig. 3C), only the stxA2-possessing cells (E. coli O157:H7 Okayama O27) showed bright orange-red fluorescence of Cy3-labeled dCTP incorporated into the LAMP product, allowing easy distinction between the cells giving positive results and those giving negative results. The fluorescently labeled antibody-positive cells were identical to the LAMP-positive cells (Fig. 3B and C). The LAMP reaction is isothermal and occurs at a lower temperature (63°C) than PCR (94°C at maximum). This milder temperature and the elimination of thermal cycling may make it possible to use antibody after amplification, so this method might detect the stxA2 gene and identify the cells at the single-cell level.
FIG. 3.
(A, B, and C) In situ LAMP of stxA2 gene in fixed-cell mixture of E. coli K-12 W3110 and E. coli O157:H7 Okayama O27. (D, E, and F) In situ PCR amplification of stxA2 gene in a fixed-cell mixture of E. coli K-12 W3110 and E. coli O157:H7 Okayama O27. Under UV excitation (exposure time, 0.2 s), all DAPI-stained bacterial cells were visualized (A and D). Under blue excitation (exposure time, 1 s), E. coli O157:H7 cells emitted green fluorescence due to the presence of FITC-labeled anti-E. coli O157:H7 antibody (B and E). Under green excitation (exposure, 2 s), cells having stxA2-amplified products emitted red fluorescence of Cy3-labeled dCTP (C) or Cy3-labeled probe (F).
Figure 3 also shows the results of direct in situ PCR-FISH with the fluorescent antibody method used for the bacterial mixtures of E. coli K-12 W3110 and E. coli O157:H7 Okayama O27 (Fig. 3D, E, and F). Under UV excitation (Fig. 3D), all bacterial cells showed blue fluorescence of DAPI. Under blue excitation (Fig. 3E), E. coli O157:H7 cells showed green fluorescence of FITC-labeled antibody. Under green excitation (Fig. 3F), only the stxA2-possessing cells (E. coli O157:H7 Okayama O27) showed bright orange-red fluorescence of Cy3-labeled VT2c probe targeting the stxA2 amplicon, allowing easy discrimination between the cells giving positive results and those giving negative results. By in situ PCR, however, the antibody combination resulted in high levels of background, perhaps caused by antigen falling off the cell surface (Fig. 3E) due to the high temperatures required for PCR cycles. Thus, fluorescent antibody labeling was not able to be combined with in situ PCR to confirm whether results were falsely negative or falsely positive. In addition, all excitation images (UV, blue, and green) showed altered cell morphology consistent with collapse compared to those of cells subjected to in situ LAMP. The in situ PCR cells appeared blurred due to the destruction of cell structures.
The change in absolute cell numbers on glass slides was measured during each step of in situ LAMP and in situ PCR (i.e., after the first ethanol dehydration, lysozyme permeabilization, proteinase K permeabilization, RNase A treatment, and amplification of target DNA) (Table 1). After each operation, glass slides were stained with DAPI at room temperature for 10 min and mounted in nonfluorescence immersion oil for observation and cell counting. For each step, at least 1,000 cells in 20 different fields were counted. The microscopic enumeration results were obtained from three parallel samples.
TABLE 1.
Percentages of cells remaining on glass slide during each experimental stepa
| Amplification method | % (SD)b of cells remaining on slide after:
|
||||
|---|---|---|---|---|---|
| Ethanol dehydration | Lysozyme permeabilization | Proteinase K permeabilization | RNase A treatment | Gene amplification | |
| In situ LAMP | 100 (3) | 101 (4) | 102 (9) | 106 (2) | 96 (6) |
| In situ PCR | 100 (4) | 104 (7) | 96 (4) | 99 (1) | 81 (7) |
Cell number after first ethanol dehydration was defined as 100%.
Values in parentheses indicate standard deviations of triplicate samples.
Gene amplification in both in situ LAMP and in situ PCR resulted in the loss of cells from the glass slide (Table 1). Increasing temperatures may cause detachment of cells from the glass slides. In situ LAMP, however, showed smaller cell decreases than in situ PCR; that is, it may be more quantitative.
Although the original LAMP method requires an initial denaturation of target DNA (95°C and 5 min) along with Bst DNA polymerase denaturation at the end of the reaction (80°C and 10 min) (16), the in situ LAMP method used in this study eliminated the initial heating for denaturation of template DNA and final denaturation of the polymerase to avoid cell destruction. The elimination merely resulted in a slower initiation of amplification, and no difference was observed at the end of the reaction. Thus, the method is technically simple and does not necessitate the use of special equipment such as a thermal cycler.
Microbial communities in natural environments are complex, and the permeability of their cell wall structures is not uniform. Permeabilization conditions for in situ gene amplification need to be optimized so that all reagents can enter the cell without diffusing the amplified products outside. Many different permeabilization procedures have been used for in situ hybridization with horseradish peroxidase-labeled probes (19, 20, 25), tyramide signal amplification (15), and multiply-labeled polyribonucleotide probes (21). By concentration of samples onto membrane filters and subsequent embedding of filters in agarose, efficient permeabilization was achieved without decreases in bacterial cell numbers under severe conditions (19). Embedding samples in gel before in situ LAMP enables enumeration of bacterial cells carrying specific genes in a natural environment without diffusing amplified products outside the cell or causing species-selective cell loss and destruction in mixed microbial communities.
Acknowledgments
This work was fully supported by The Development of Monitoring Methods for Microorganisms in Environment Project supported by the New Energy and Industrial Technology Development Organization, Tokyo, Japan.
REFERENCES
- 1.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F. O. Glöckner, A. Wille, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aliotta, J. M., J. J. Pelletier, J. L. Ware, L. S. Moran, J. S. Benner, and H. Kong. 1996. Thermostable Bst DNA polymerase I lacks a 3′→5′ proofreading exonuclease activity. Genet. Anal. 12:185-195. [PubMed]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, F., B. Binder, and R. E. Hodson. 2000. Flow cytometric detection of specific gene expression in prokaryotic cells using in situ RT-PCR. FEMS Microbiol. Lett. 184:291-296. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., W. A. Dustman, and R. E. Hodson. 1999. Microscopic detection of toluene dioxygenase gene and its expression inside bacterial cells in seawater using in situ PCR. Hydrobiologica 401:131-138. [Google Scholar]
- 6.Eberl, L., R. Schulze, A. Ammendola, O. Geisenberger, R. Erhart, C. Sternberg, S. Molin, and R. Amann. 1997. Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol. Lett. 149:77-83. [Google Scholar]
- 7.Hodson, R. E., W. A. Dustman, R. P. Garg, and M. A. Moran. 1995. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl. Environ. Microbiol. 61:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs, D., M. L. Angles, A. E. Goodman, and B. A. Neilan. 1997. Improved methods for in situ enzymatic amplification and detection of low copy number genes in bacteria. FEMS Microbiol. Lett. 152:65-73. [DOI] [PubMed] [Google Scholar]
- 9.Kenzaka, T., N. Yamaguchi, B. Prapagdee, E. Mikami, and M. Nasu. 2001. Bacterial community composition and activity in urban rivers in Thailand and Malaysia. J. Health Sci. 47:353-361. [Google Scholar]
- 10.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085-2093. [DOI] [PubMed] [Google Scholar]
- 11.Kurokawa, K., K. Tani, and M. Nasu. 1997. Direct in situ PCR method for the detection of verotoxin-producing Escherichia coli. Jpn. J. Bacteriol. 52:513-518. [DOI] [PubMed] [Google Scholar]
- 12.Kurokawa, K., K. Tani, M. Ogawa, and M. Nasu. 1999. Abundance and distribution of bacteria carrying sltII gene in natural river water. Lett. Appl. Microbiol. 61:405-410. [DOI] [PubMed] [Google Scholar]
- 13.Lanoil, B. D., and S. J. Giovannoni. 1997. Identification of bacterial cells by chromosomal painting. Appl. Environ. Microbiol. 63:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawyer, F. C., S. Stoffel, R. K. Saiki, K. Myambo, R. Drummond, and D. H. Gelfand. 1989. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J. Biol. Chem. 264:6427-6437. [PubMed] [Google Scholar]
- 15.Lebaron, P., P. Catala, C. Fajon, F. Joux, J. Baudart, and L. Bernard. 1997. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl. Environ. Microbiol. 63:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuovo, G. J., R. J. Hohman, G. A. Nardone, and I. A. Nazarenko. 1999. In situ amplification using universal energy transfer-labeled primers. J. Histochem. Cytochem. 47:273-279. [DOI] [PubMed] [Google Scholar]
- 18.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pernthaler, P., J. Pernthaler, M. Schattenhofer, and R. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. A comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porter, J., R. Pickup, and C. Edwards. 1995. Flow cytometric detection of specific genes in genetically modified bacteria using in situ polymerase chain reaction. FEMS Microbiol. Lett. 34:463-465. [DOI] [PubMed] [Google Scholar]
- 23.Ranjard, L., F. Poly, and S. Nazaret. 2000. Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res. Microbiol. 151:167-177. [DOI] [PubMed] [Google Scholar]
- 24.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schönhuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro, H. M. 2000. Microbial analysis at the single-cell level: tasks and techniques. J. Microbiol. Methods 42:3-16. [DOI] [PubMed] [Google Scholar]
- 27.Tani, K., K. Kurokawa, and M. Nasu. 1998. Development of a direct in situ PCR method for detection of specific bacteria in natural environments. Appl. Environ. Microbiol. 64:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tani, K., M. Muneta, K. Nakamura, K. Shibuya, and M. Nasu. 2002. Monitoring of Ralstonia eutropha KT1 in groundwater in an experimental bioaugmentation field by in situ PCR. Appl. Environ. Microbiol. 68:412-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolker-Nielson, T., K. Holmstrøm, and S. Molin. 1998. Non-genetic population heterogeneity studied by in situ polymerase chain reaction. Mol. Microbiol. 27:1099-1105. [DOI] [PubMed] [Google Scholar]
- 30.Wallner, G., I. Steinmetz, D. B. Suermann, and R. Amann. 1996. Combination of rRNA-targeted hybridization probes and immuno-probes for the identification of bacteria by flow cytometry. Syst. Appl. Microbiol. 19:569-576. [Google Scholar]
- 31.Watarai, M., T. Sato, M. Kobayashi, T. Shimizu, S. Yamasaki, T. Tobe, C. Sasakawa, and Y. Takeda. 1998. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga toxin-producing Escherichia coli. Infect. Immun. 66:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi, N., S. Inaoka, K. Tani, K. Kenzaka, and M. Nasu. 1996. Detection of specific bacterial cells with 2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate and fast red TR in situ hybridization. Appl. Environ. Microbiol. 62:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]