Abstract
Enteric pathogens, such as Salmonella enterica and Escherichia coli O157:H7, have been shown to contaminate fresh produce. Under appropriate conditions, these bacteria will grow on and invade the plant tissue. We have developed Arabidopsis thaliana (thale cress) as a model system with the intention of studying plant responses to human pathogens. Under sterile conditions and at 100% humidity, S. enterica serovar Newport and E. coli O157:H7 grew to 109 CFU g−1 on A. thaliana roots and to 2 × 107 CFU g−1 on shoots. Furthermore, root inoculation led to contamination of the entire plant, indicating that the pathogens are capable of moving on or within the plant in the absence of competition. Inoculation with green fluorescent protein-labeled S. enterica and E. coli O157:H7 showed invasion of the roots at lateral root junctions. Movement was eliminated and invasion decreased when nonmotile mutants of S. enterica were used. Survival of S. enterica serovar Newport and E. coli O157:H7 on soil-grown plants declined as the plants matured, but both pathogens were detectable for at least 21 days. Survival of the pathogen was reduced in unautoclaved soil and amended soil, suggesting competition from indigenous epiphytes from the soil. Enterobacter asburiae was isolated from soil-grown A. thaliana and shown to be effective at suppressing epiphytic growth of both pathogens under gnotobiotic conditions. Seed and chaff harvested from contaminated plants were occasionally contaminated. The rate of recovery of S. enterica and E. coli O157:H7 from seed varied from undetectable to 19% of the seed pools tested, depending on the method of inoculation. Seed contamination by these pathogens was undetectable in the presence of the competitor, Enterobacter asburiae. Sampling of 74 pools of chaff indicated a strong correlation between contamination of the chaff and seed (P = 0.025). This suggested that contamination of the seed occurred directly from contaminated chaff or by invasion of the flower or silique. However, contaminated seeds were not sanitized by extensive washing and chlorine treatment, indicating that some of the bacteria reside in a protected niche on the seed surface or under the seed coat.
Salmonella enterica and enterohemorrhagic Escherichia coli cause severe diarrhea in humans. One particularly prevalent enterohemorrhagic type, E. coli O157:H7, can cause life-threatening hemorrhagic colitis and, in very severe cases, hemolytic uremic syndrome (47). Several outbreaks have been linked to fresh fruits and vegetables (1, 53, 59). The source of contamination likely occurs postharvest from a variety of animal sources, including humans (58) Alternatively, preharvest contamination could occur via manure application, contaminated irrigation water, animals, or use of contaminated seed. Whether the contamination originates pre- or postharvest, it has been well documented that S. enterica and E. coli O157:H7 survive and grow on fresh produce (8, 11, 21, 62), especially on sprouts, where the contamination can exceed 107 CFU per g of fresh weight (12, 57).
Disinfection of plant surfaces with a variety of agents has been tested, and reductions of several orders of magnitude are possible (57). However, in the case of E. coli O157:H7, the minimal infective dose for humans is so low (ca. 10 bacteria) that disinfection is often not practical without destroying the produce (42) or inhibiting germination (44). Furthermore, since penetration of E. coli O157:H7 into plant tissue has been demonstrated, disinfection of the tissue may not be effective (31, 51, 55, 62). Alternative intervention measures have targeted prevention of the initial infestation. For effective measures to be developed, a much better understanding of the mechanisms of pathogen colonization of plants will be needed.
Effective, long-term colonization of plants requires the ability to compete for limited moisture and nutrients. This is especially true in the phyllosphere of soil-grown plants, where the environment is harsh and nutrients are limited (6, 34). Furthermore, the environment on the surface of the leaf and root is not uniform, either spatially or temporally (32, 35). Hence, the ability to move towards or on the host plant is important for colonization by many plant-associated bacteria. Nonmotile mutants of the plant pathogens Pseudomonas syringae, Erwinia amylovora, Ralstonia solanacearum, and Agrobacterium tumefaciens show decreased virulence, primarily due to their inability to enter the plant (5, 28, 56). Nonpathogenic bacteria also benefit from motility. Movement along the leaf surface towards colonization of stomates could be visualized with green fluorescent protein (GFP)-labeled P. aeruginosa (45). Loss of flagellar motility decreases the fitness of leaf- and root-colonizing bacteria (14, 22, 60). Although motility promotes colonization outside the host, it appears that plant endophytic bacteria become nonmotile and nonchemotactic upon entering the plant (27, 33).
Efforts are under way in several laboratories to understand the molecular mechanisms of survival and growth of human pathogens on plants from the bacterial perspective (7, 15, 31, 48, 54, 61). However, the plant biology involved in the interaction with human pathogens has not been investigated. We anticipate that some of the similarities between the interactions of pathogens and their hosts (10) will also be evident in the seemingly novel interactions between human pathogens and plants. It is reasonable to propose that interactions occur frequently between human-pathogenic bacteria and plants because enteric pathogens survive outside of their host organism. Furthermore, human pathogens and some plants may have evolved mechanisms to recognize each other. Remarkably, it has been shown that Arabidopsis thaliana (thale cress) makes a protein (FLS2, flagellin sensing) that can recognize a conserved region of bacterial flagellin (18). This flagellin region is conserved in many different types of bacteria, including enteric pathogens such as E. coli O157:H7 and Salmonella enterica (M. B. Cooley, unpublished data).
An understanding of the specific interactions between human pathogens and plants will take considerable effort. This is especially true for the plant side of the interaction because many crop plants have very large genomes and are poorly characterized genetically. Hence, the aim of this research was to develop a model system for the growth of S. enterica and E. coli O157:H7 on A. thaliana. The advanced genetics of A. thaliana should greatly facilitate the discovery of the plant factors involved in these interactions. We expect that several factors important in these interactions will also play a crucial role in interactions with crop plants. In this paper, we demonstrate growth of S. enterica and E. coli O157:H7 on both the roots and shoots of A. thaliana in a gnotobiotic system similar to that used to produce sprouts and hydroponic crops. In this system we show that inoculation of the pathogen onto a portion of the root leads eventually to contamination of the whole plant and invasion of the roots. Additionally, we show that S. enterica and E. coli O157:H7 will survive and persist on soil-grown A. thaliana and that pathogens can be recovered from the flowers and seeds.
MATERIALS AND METHODS
Strains and plasmids.
All bacterial strains used in this paper are listed in Table 1. Plasmid pWM1029, containing a constitutively expressed gfp gene, was constructed by first fusing the t0 terminator from pPROBE-gfp[tagless] (40) to a consensus Campylobacter jejuni promoter and gfp in pWM1007 with a PCR overlap extension method (29). The resulting plasmid, pWM1015, was digested with AatII and FspI to remove the ColE1 origin of replication. The AatII site was converted to BamHI with an adapter, and then the broad-host-range origin of replication from pBBR1MCS2 was inserted as a BglII-SspI fragment (36).
TABLE 1.
Bacterial strains used in this studya
| Strain | Source or comments | Source and/or reference(s) |
|---|---|---|
| Salmonella enterica serovar Newport (RM1655) | From alfalfa seed | 96E01153C-TX from Greg Inami, California State Health Laboratory Berkeley |
| Escherichia coli O157:H7 Odwalla (RM1484) | From apple juice | Steve Weagant, Food and Drug Administration; strain # SEA13B88 |
| Salmonella enterica serovar Typhimurium SJW1103 (RM2967) | Wild type | R. M. Macnab (Yale University) (37, 63) |
| Salmonella enterica serovar Typhimurium SJW1368 (RM2965) | Nonmotile, ΔflhDC | R. M. Macnab (Yale University) (37, 63) |
| Salmonella enterica serovar Typhimurium SJW1809 (RM2966) | Nonmotile, flaN | R. M. Macnab (Yale University) (37, 63) |
| Enterobacter asburiae (RM3638) | From A. thaliana | This study; typed by fatty acid analysis (Microbial ID) |
All strains were transformed with pWM1029 by electroporation (13).
Growth, inoculation, and sampling of gnotobiotically grown plants.
Sterile A. thaliana ecotype Col-0 was grown on hydroponic medium to mimic commercial growth of sprouts and to monitor the growth of S. enterica serovar Newport and E. coli O157:H7. Seeds were surface sterilized by a three-step treatment (0.1% Triton X-100 for 30 min, 70% ethanol for 2 min, and 10% bleach for 5 min). The seeds were then washed five times in sterile water and sown on 0.5× Murashige and Skoog basal salts (MS; Sigma-Aldrich) with 1.2% Bacto agar in Falcon Integrid plates (VWR Scientific). The plates were incubated in a growth chamber at a nearly vertical position (80o) to encourage the roots to grow along the surface of the agar. Germination conditions were 22°C during the day, 20°C at night, a 16-h photoperiod, 5,000 lux of illumination, and approximately 100% relative humidity.
Prior to inoculation, S. enterica serovar Newport, E. coli O157:H7, and Enterobacter asburiae were grown for 24 h in M9 glucose medium with 50 μg of kanamycin (Sigma) ml−1 at 23°C and diluted to give 104 or 106 CFU ml−1 in 0.5 mM potassium phosphate, pH 7. The initial bacterial concentration was determined by plating appropriate dilutions in triplicate on Shigella and Salmonella (SS) agar. The roots were inoculated at 1 to 2 cm of length (approximately 5 days postgermination) or 3 to 4 cm of length (6 to 7 days postgermination) by pipetting the suspension directly onto the roots or shoots while the plate was held in a vertical position. Excess liquid was allowed to run off the plants and removed from the bottom of the plate. This inoculation method resulted in about 2 to 5 μl remaining on each root or 5 to 10 μl on each shoot.
For each time point, six roots or shoots were sampled with sterile scissors and forceps and then sonicated with a model FS30 sonication bath (Fisher Scientific) for 75 s in phosphate-buffered saline (PBS) before plating the supernatant on SS agar or Chromagar (DRG International). Enterobacter asburiae is distinguishable from S. enterica serovar Newport and E. coli O157:H7 as a steel blue colony on Chromagar.
Pulsed-field gel electrophoresis of selected isolates was performed according to the standardized protocol reported by the Centers for Disease Control and Prevention (2) to verify that the recovered bacteria were the same as the inoculated strains. Colonies were also examined for fluorescence with a Leica MZ-FLIII stereo epifluorescence microscope (Leica Microsystems, Exton, Pa.).
Migration of S. enterica and E. coli O157:H7 on plants.
The lower portion of the roots of sterile plants (6 to 7 days after germination) was inoculated 2.4 cm from the crown (the boundary between the shoot and the root) as described above. Care was taken to ensure that the shoot and the upper 2.4 cm of root were not contaminated inadvertently during inoculation. To avoid inadvertent movement of bacteria above the inoculated region, the plates were continuously held in a vertical or nearly vertical position after inoculation. Movement of the GFP-labeled bacteria was observed by viewing the roots with a Leica MZ-FLIII stereo epifluorescence microscope with the GFP2 filter set. The movement of the bacteria on 10 plants was recorded, and separate plates were used for each time point. In this way, only plates that had always been held in a vertical position were used. A duplicate set of plates were used to measure the growth of the bacteria. Six lower inoculated and six upper uninoculated regions were excised with sterile scissors and forceps for each time point and processed to determine bacterial concentration as described above.
Inoculation of seeds.
Fifty milligrams of nonsterile seed was suspended in water with 108 CFU of S. enterica serovar Newport or E. coli O157:H7 ml−1. Prior to inoculation, the bacteria were cultured in M9 glucose medium with 50 μg of kanamycin ml−1 at 23°C for 24 h. The seed suspension was tumbled for 24 h at 23°C. The seeds were allowed to settle and washed four times with sterile water. After the last wash, the supernatant was plated on SS agar to determine the basal level of bacteria. An aliquot of seeds were resuspended in PBS and sonicated for 75 s, and the supernatant was plated on SS agar. The amount of bacteria bound to the seeds is reported as the difference between the number of bacteria in the supernatant before and after sonication. Subsequently, the sonicated seeds were dried and weighed. The remaining seeds were suspended in 0.1% agar and pipetted directly onto soil.
Growth, inoculation, and sampling of soil-grown plants.
Gnotobiotically grown seedlings contaminated with S. enterica serovar Newport or E. coli O157:H7 were transplanted to autoclaved Sunshine mix 1 (SunGro Horticulture, Bellevue, Wash.) supplemented with Plantex 20-20-20 fertilizer (McCalif Horticulture, Ceres, Calif.) at 0.3 g per kg of soil. Alternatively, the seedlings were transplanted to unautoclaved Sunshine mix 1 or Sunshine mix 1 supplemented 1:1 (wt/wt) with Yolo fine sandy loam (Vegetable Crops Department, University of California, Davis) with Plantex 20-20-20 fertilizer. The seedlings were carefully lifted off the surface of the agar with sterile forceps without touching the roots and laid on the soil. The roots were then overlaid with soil, and the pots were covered with plastic wrap. The transplants were situated far enough apart to prevent cross-contamination during the transplantation process. The transplants were gradually released to 75% humidity over 3 days to minimize the shock.
In experiments involving inoculation of the soil, S. enterica serovar Newport or E. coli O157:H7 from a 24-h culture in M9 glucose medium at 23°C with 50 μg of kanamycin ml−1 was washed once with water and mixed into the soil to give 108 CFU g−1 before transplanting the seedlings. The seedlings (5 days postgermination) were transplanted from identical, uninoculated soils. The growth regimen for all soil-grown plants was a 16-h photoperiod, 5,000 lux intensity at 22°C in the day and 20°C at night in a Conviron MTR30 growth chamber (Controlled Environments, Inc.).
Samples of leaf and flower tissue from the apical region of the plants (36 samples of approximately 20 mg [fresh weight] each) were excised with sterile scissors and forceps and enriched for 24 h in 10 ml of lactose broth (BD Microbiology, Irvine, Calif.) at 37°C. Then 20 μl of the enrichment culture was streaked onto SS agar, Chromagar, or tryptic soy agar (Fisher Scientific).
Seed and chaff harvest, enrichment, and plating.
Seeds harvested from groups of four mature, dry plants were pooled. The number of seeds in each pool varied from 7,000 to 20,000. With A. thaliana, the developing seeds are encased in a pod-like structure called a silique. Chaff, which consists mostly of silique tissue, was cleaned from the seeds twice with a 40-mesh sieve and then saved independently. Consistent with the method of Inami and Moler (30), 1,000 seeds from each seed pool from E. coli O157:H7- or S. enterica serovar Newport-contaminated plants were first germinated on 0.5× MS with 0.8% Bacto agar prior to testing for contamination. The germinated seedlings were cultured in lactose broth for 24 h at 37°C to selectively enrich E. coli O157:H7 or S. enterica serovar Newport. Twenty microliters of the culture was streaked on SS agar and Chromagar.
Contamination in chaff samples was determined by grinding the samples to a fine powder with a mortar and pestle. Then 100 mg of ground chaff was suspended in 10 ml of lactose broth and incubated with shaking at 37°C for 24 h. The cultures were plated on SS agar and Chromagar. The correlation between seed and chaff contamination was determined by the chi-square test (49).
To test for tightly bound or internalized contamination, seeds from soil-grown plants inoculated in the gnotobiotic system and previously identified as contaminated were washed by first suspending the seeds in PBS and then vortexing at the maximum setting for 1 min. The seeds were washed five times in PBS, with careful attention to removing any chaff. An aliquot of 1,000 seeds were then plated on 0.5× MS with 0.8% agar, germinated, and tested for contamination as described above. The remaining seeds were resuspended in PBS, sonicated for 90 s with a model FS30 sonication bath (Fisher Scientific), and washed as described above.
An aliquot of 1,000 seeds were plated on 0.5× MS with 0.8% agar, germinated, and tested for contamination as described above. The remaining seeds were resuspended in 200 ppm of free chlorine (Cl2) in 10 mM sodium phosphate, pH 6.8, mixed by hand for 10 min at 23°C, and washed five times in sterile water. Free Cl2 concentration was determined by the amperometric method (9). The washed seeds were plated on 0.5× MS with 0.8% agar, germinated, and tested for contamination as described above. To test the effectiveness of washing, the supernatants from the fifth wash from all three washing cycles were enriched in lactose broth for 24 h and plated on SS agar. Cultures were then streaked on SS agar or Chromagar. Representative colonies from seeds or chaff were analyzed by pulsed-field gel electrophoresis as described above.
Confocal microscopy.
GFP-labeled bacteria were viewed on plant roots or leaves with a TCS NT scanning confocal microscope (Leica Microsystems) with either HC PL APO 10×/0.4 or PL APO 40×/1.25 to 0.75 oil immersion objectives. An argon laser was used to excite GFP (excitation wavelength [λexc] = 488 nm). GFP fluorescence was detected with a BP525/50 filter and assigned the color green. A krypton laser (λexc = 568 nm) was used to excite chloroplast and vasculature autofluorescence. A helium-neon laser (λεωχ = 633 nm) was used to excite epidermal tissue stained with TO-PRO-3 (Molecular Probes, Eugene, Oreg.). TO-PRO-3 stain usually did not penetrate the root deeper than the epidermis. Chloroplast and vascular autofluorescence and TO-PRO-3 fluorescence were detected with the LP590 filter and assigned the color red. Dual-color images were obtained with the TCS NT software (version 2; Leica Microsystems).
RESULTS
Growth and survival of S. enterica serovar Newport and E. coli O157:H7 on A. thaliana.
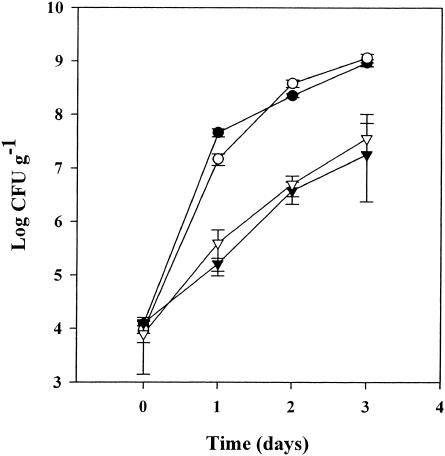
S. enterica serovar Newport and E. coli O157:H7 were monitored for growth on A. thaliana roots and shoots. Different inoculation and growth conditions were used to mimic the conditions that S. enterica serovar Newport and E. coli O157:H7 might experience during commercial production of sprouts or hydroponics and on field-grown plants. Commercial sprout facilities commonly germinate the seeds in water. However, growth of both pathogens on A. thaliana sprouts in sterile water was minimal, and results were inconsistent (data not shown). The addition of a hydroponic solution produced vigorously growing A. thaliana sprouts and induced the growth of the pathogens (Fig. 1). Inoculation and growth of the bacteria were performed with the roots exposed on the surface of agar plates. This gnotobiotic technique allowed sampling and in situ observation of GFP-labeled bacteria on the roots with minimal disturbance. Both S. enterica serovar Newport and E. coli O157:H7 grew to 109 CFU g−1 in 3 days. On leaf tissue, growth of both pathogens was 30- to 50-fold less than growth on roots, probably due to a reduction of available nutrients on the foliage.
FIG. 1.
Growth of S. enterica serovar Newport (○, ▿) and E. coli O157:H7 (•, ▾) on A. thaliana shoots (▾, ▿) and roots (•, ○). Inoculation and growth of the bacteria were performed with the roots exposed on the surface of agar plates. Bacterial concentrations are averages of six samples. Error bars show standard errors.
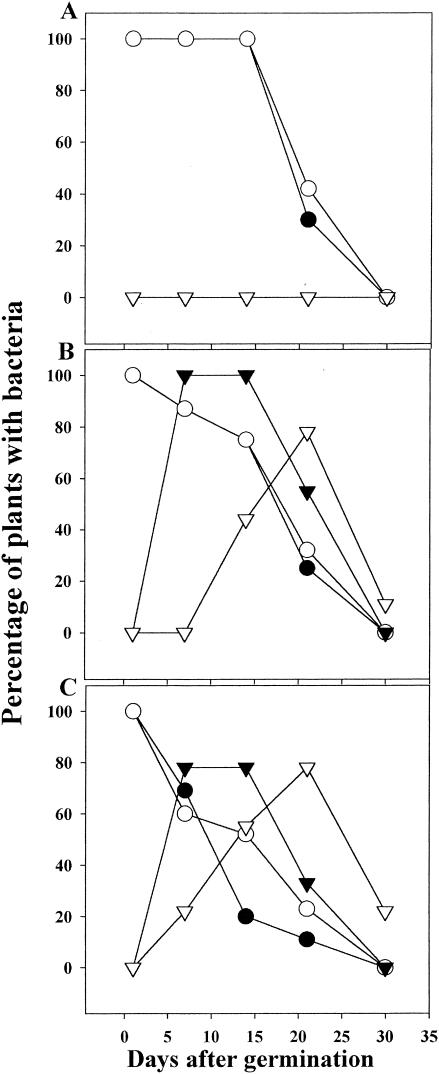
Growth of S. enterica serovar Newport and E. coli O157:H7 on soil-grown plants was decreased compared to growth in the gnotobiotic system described above. The bacteria are exposed to desiccation, UV light, and competition from other microbes in this environment. In fact, efficient recovery of S. enterica serovar Newport and E. coli O157:H7 from the foliage of soil-grown plants required incubation of the plant samples in enrichment broth. Therefore, quantification of the number of bacteria present on the plant samples prior to enrichment was not possible. The inoculum was applied to seeds and sown separately on autoclaved potting mix, unautoclaved potting mix, and a mixture of potting soil plus an agricultural soil, Yolo fine sandy loam. A sample of seeds were analyzed to determine the extent of bound pathogen. S. enterica serovar Newport- and E. coli O157:H7-inoculated seeds were contaminated at 24,000 and 334 CFU per seed, respectively. Seedlings in the three soil types were sampled 1 day after germination (day 1) and periodically thereafter, always from apical tissue, either a leaf or a flower (Fig. 2). At day 1, 100% of the samples were contaminated. Thereafter, the incidence of contamination decreased, with S. enterica serovar Newport and E. coli O157:H7 becoming undetectable at 30 days.
FIG. 2.
Incidence of recovery of S. enterica serovar Newport (○) and E. coli O157:H7 (•) from plants grown in autoclaved (A), unautoclaved (B), and amended (C) soils and coincident isolation of Enterobacter asburiae from S. enterica serovar Newport-infected (▿) and E. coli O157:H7-infected (▾) plants. Each data point represents samples from 36 plants.
The rate of decline was similar for S. enterica serovar Newport and E. coli O157:H7. Nevertheless, survival of both pathogens was different on different soils. Autoclaved soil allowed the longest survival of the pathogens (Fig. 2A). One hundred percent of the plant samples were still contaminated when sampled after 14 days. In contrast, S. enterica serovar Newport and E. coli O157:H7 survived less well on plants grown on unautoclaved potting soil and the mixture of potting soil and Yolo fine sandy loam (Fig. 2B and C). Similar results were obtained when seedlings were transplanted into the three soils previously inoculated at 108 CFU g−1 with either pathogen. Specifically, mixed soils and unautoclaved soils supported the epiphytic survival of S. enterica serovar Newport and E. coli O157:H7 less well than autoclaved soils, and neither pathogen was detectable on plants grown on any of the three soils at day 30 (data not shown). Also, in a related experiment, seedlings inoculated in the gnotobiotic system above were transplanted to autoclaved soil 10 days later, and survival of S. enterica serovar Newport and E. coli O157:H7 was monitored. S. enterica serovar Newport and E. coli O157:H7 were detectable on these plants at 30 days in 87% and 75% of the apical flowers, respectively.
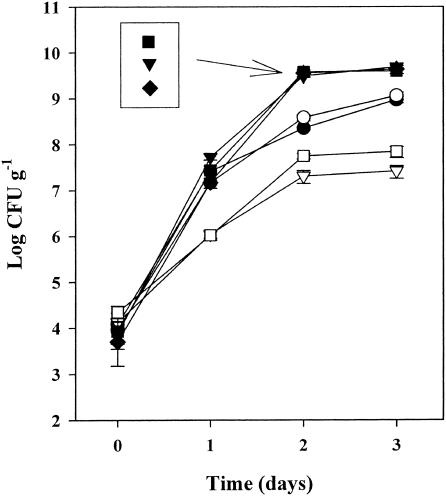
The differences in pathogen survival on plants growing in these three soils suggested that competing bacteria might have been present initially in the soils and subsequently colonized the plants. Seven different bacteria were isolated from the enrichment culture from soil-grown plants. One coliform strain, Enterobacter asburiae, was selected for further investigation because it was isolated from plants 10 times more frequently than all of the other strains combined and its appearance correlated with the selection against S. enterica serovar Newport and E. coli O157:H7 (Fig. 2B and C). The appearance of Enterobacter asburiae in plant samples grown from E. coli O157:H7-infected seeds occurred earlier than with samples grown from S. enterica-infected seeds. Recovery of Enterobacter asburiae from the plant samples also declined with the age of the plants and, similar to the pathogens, was undetectable in many samples at day 30. Also, when Enterobacter asburiae was coinoculated with S. enterica serovar Newport or E. coli O157:H7 onto roots grown in sterile conditions, growth of S. enterica serovar Newport and E. coli O157:H7 was suppressed greater than 10-fold (Fig. 3).
FIG. 3.
Growth of Enterobacter asburiae (▾), S. enterica serovar Newport (•), and E. coli O157:H7 (○) separately on roots of sterilely grown plants or coinoculated as S. enterica serovar Newport (□) or E. coli O157:H7 (▿) with Enterobacter asburiae (▪ ♦, respectively). Bacterial concentrations are averages of six samples. Error bars show standard errors.
Competition against E. coli O157:H7 was approximately 0.5 log greater than that against S. enterica serovar Newport. The growth of Enterobacter asburiae reached a fivefold higher level than that of either S. enterica serovar Newport or E. coli O157:H7 and was unchanged by the presence of the pathogens. Additionally, when these plants were transplanted to autoclaved soil 10 days after inoculation, neither pathogen was detectable in 36 plant samples tested, and all the samples contained Enterobacter asburiae (data not shown).
Migration of bacteria on the plant surface.
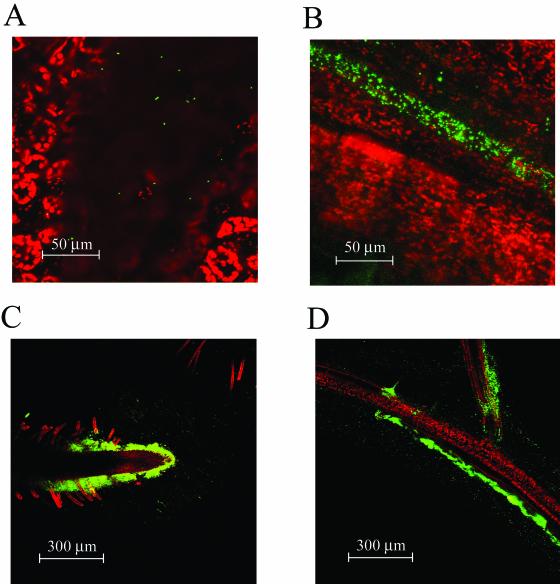
Colonization of soil-grown plants for extended periods of time, especially on the terminal flowers, may indicate that S. enterica serovar Newport and E. coli O157:H7 migrate along the surface of the plant, similar to plant epiphytes. To observe this, we used GFP-labeled S. enterica serovar Newport and E. coli O157:H7 and examined colonization of sterilely grown plants under the microscope. Colonization was not uniform, and on the leaves it was concentrated in shallow depressions or near the veins (Fig. 4A and B). On the roots, bacteria were often observed in many locations but were concentrated initially at the root tips and the branch points of lateral roots, possibly because more nutrients were available there (Fig. 4C and D). At later incubation times, the roots were often colonized uniformly with S. enterica serovar Newport and E. coli O157:H7.
FIG. 4.
Colonization of A. thaliana leaves by GFP-labeled E. coli O157:H7 localized in small depressions (A) and over veins (B). Chloroplast autofluorescence is colored red. Initial colonization of roots occurs at root tips (C) and the branch points of lateral roots (D). Red indicates either vascular autofluorescence or TO-PRO-3 staining of epidermis and root hairs (see Materials and Methods).
This progression toward colonization of the whole root indicated that the bacteria were spreading by either active motility or simple diffusion. Salmonella enterica serovar Typhimurium mutants defective in flagellin synthesis (S. enterica serovar Typhimurium SJW1368) or motility functions (S. enterica serovar Typhimurium SJW1809) were used to determine if flagellar function is necessary for movement of the pathogen on sterilely grown plants. The lower portion of the roots was inoculated with GFP-labeled mutants and wild-type pathogens 2.4 cm below the crown (the boundary between the root and the shoot). In this situation, the bacteria move upwards from the inoculated region to the uninoculated region of the root and eventually the shoot. Occasionally, a few plants failed to become inoculated and remained uncontaminated throughout the experiment (data not shown). Therefore, cross-contamination through swarming or aerosols does not occur readily between adjacent plants.
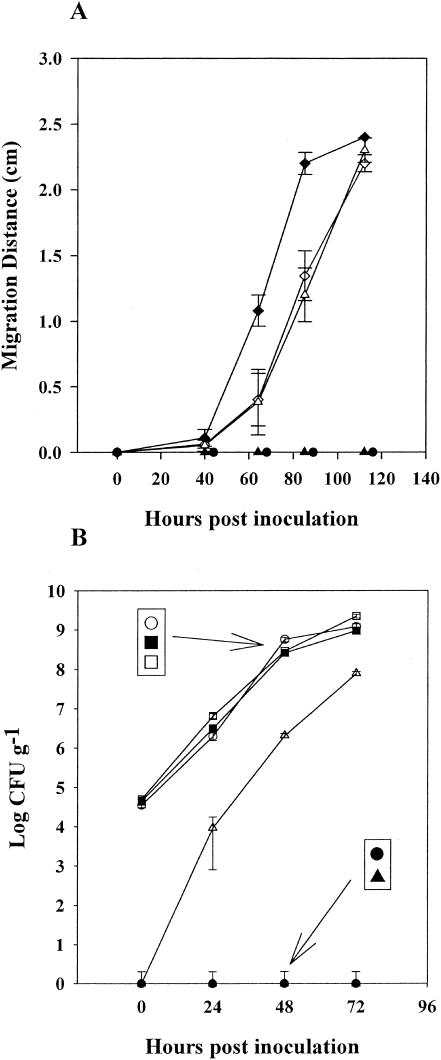
Mutant strains S. enterica serovar Typhimurium SJW1368 and S. enterica serovar Typhimurium SJW1809 failed to move upwards beyond this region (Fig. 5A). However, both mutants grew as effectively as the S. enterica serovar Typhimurium wild-type strain (SJW1103) in the lower portion of the root (Fig. 5B). At 85 h postinoculation, 60% of the E. coli O157:H7-inoculated plants showed bacteria at the crown. The migration rates of S. enterica serovar Newport and SJW1103 were slower than that of E. coli O157:H7. At 112 h postinoculation, 60% of the S. enterica-inoculated plants showed bacteria at the crown. Analysis of the slopes of these graphs indicated that both wild-type Salmonella serovars were migrating at approximately 0.9 cm per day and that E. coli O157:H7 was migrating significantly faster, at 1.4 cm per day.
FIG. 5.
Growth of bacteria and migration along the root following inoculation 2.4 cm below the crown. (A) Migration of GFP-labeled E. coli O157:H7 (♦), S. enterica serovar Typhimurium SJW1103 (▵), S. enterica serovar Newport (⋄), S. enterica serovar Typhimurium SJW1368 (•), and S. enterica serovar Typhimurium SJW1809 (▴) above the region of inoculation was observed by stereo epifluorescence microscope. Migration distance is the extent of bacterial movement during the elapsed time and is the average of 10 measurements on different plants. (B) Growth of GFP-labeled S. enterica serovar Typhimurium SJW1103, S. enterica serovar Typhimurium SJW1368 and S. enterica serovar Typhimurium SJW1809 in the inoculated region of the root (○, ▪, and □, respectively) and above the inoculated region (▵, •, and ▴, respectively). Bacterial concentrations are averages of six samples. Error bars show standard errors.
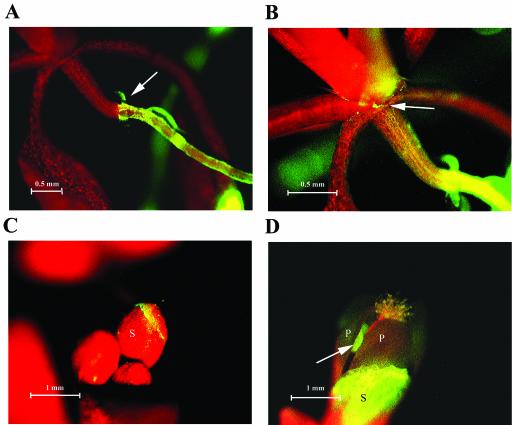
Movement of the bacteria up the plant appeared to slow greatly when they reached the crown of sterilely grown plants (Fig. 6A). Furthermore, unlike migration on roots, no migrating front of bacteria could be detected microscopically due to a much lower concentration of bacteria. Hence, the rate of migration along the shoot was not determined. Nevertheless, bacteria were visible in the meristematic region of sterilely grown plants 9 days after reaching the crown (a distance of 5 to 7 mm) (Fig. 6B). In several sterilely grown plants where the meristematic region appeared contaminated, floral buds were also contaminated. The bacteria were concentrated at the edges of the sepals and at later incubation times could also be seen on the flowers of sterilely grown plants (Fig. 6C and D).
FIG. 6.
Migration of GFP-labeled E. coli O157:H7 from the crown to the flowers. (A) Bacteria have moved up the root from the lower right to the crown (indicated by the arrow). (B) Longer exposure of the meristematic region shown in panel A, showing colonization at the base of the petioles (indicated by the arrow). (C) Colonization of three floral buds. (D) Colonization of the sepal of an opening flower. Note: green autofluorescence of the anther (indicated by the arrow) was also observed in flowers of uninoculated plants. The red in panels A to D is chloroplast autofluorescence. S, sepal; P, petal.
Recovery of S. enterica serovar Newport and E. coli O157:H7 from seeds.
Since bacteria were detectable on flowers, seeds from these plants were tested for contamination. Seeds were harvested from dry plants 45 to 60 days after germination. Seeds were collected from plants inoculated by three methods and grown in three soils, and the incidence of recovery of S. enterica serovar Newport and E. coli O157:H7 was determined (Table 2). Recovery of pathogen from seeds were highest where the pathogens were inoculated on seedlings grown in a gnotobiotic system and then transplanted to soil (Table 2). Escherichia coli O157:H7 and S. enterica serovar Newport were recovered from 34 (26.7%) and 18 (12.5%) seed pools, respectively, from plant grown on autoclaved soil. The rates of recovery of contaminated seeds from unautoclaved potting and amended potting soils were not significantly different from those on autoclaved soil with plants inoculated as seedlings in the gnotobiotic system. In contrast, plants coinoculated with a pathogen and Enterobacter asburiae and grown in autoclaved potting soil failed to produce contaminated seeds. Also, seeds harvested from plants grown from inoculated seeds were not contaminated. However, seeds from plants grown on inoculated soil were found to be contaminated, but only one seed pool from each soil type. Selected colonies isolated from seeds and analyzed by pulsed-field gel electrophoresis were shown to have the same restriction pattern as the initial inoculum strains of S. enterica serovar Newport and E. coli O157:H7 (data not shown).
TABLE 2.
Incidence of seed contamination using different inoculation methods and soilsa
| Sample | No. of contaminated seed pools (no. tested)
|
|||||
|---|---|---|---|---|---|---|
| Autoclaved soil
|
Unautoclaved soil
|
Amended soil
|
||||
| S. enterica | E. coli O157:H7 | S. enterica | E. coli O157:H7 | S. enterica | E. coli O157:H7 | |
| Inoculated seedlings | 18 (143) | 34 (127) | 3 (9) | 3 (9) | 1 (9) | 4 (9) |
| Coinoculated seedlings | 0 (18) | 0 (18) | NDb | ND | ND | ND |
| Inoculated seed | 0 (9) | 0 (9) | 0 (9) | 0 (9) | 0 (9) | 0 (9) |
| Inoculated soil | 1 (9) | 0 (9) | 0 (9) | 1 (9) | 1 (9) | 0 (9) |
Incidence of contamination in 1,000-seed samples from each seed pool. The number in parentheses is the number of seed pools tested. Seedlings were growing sterilely (gnotobiotic) when inoculated and transferred to soil after 10 days or coinoculated with E. asburiae and one of the indicated pathogens.
ND, not determined.
Recovery of S. enterica serovar Newport and E. coli O157:H7 from chaff.
Since cleaned seeds always contained small amounts of chaff, it was possible that the seed itself was not the source of contamination, but rather the contamination was due entirely to small amounts of contaminated chaff. Consequently, we monitored the extent of contamination in 74 chaff and seed pools from soil-grown plants originally inoculated in the gnotobiotic system (29 S. enterica serovar Newport- and 45 E. coli O157:H7-infected pools). Contamination was discovered in 10 chaff pools where the corresponding seed pools were not contaminated (Table 3). Likewise, four seed pools were contaminated without contamination occurring in the corresponding chaff pools, and nine pools were infected in both the seed and chaff pools. Chi-square analysis indicated that pools with contaminated chaff had a higher probability of also producing contaminated seeds (P = 0.025).
TABLE 3.
Incidence of seed contamination before and after decontamination and concurrent chaff contaminationa
| Seed and chaff contamination
|
Seed sanitation
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of pools tested | Incidence of contamination (no. of pools)
|
Probability | No. of pools tested | Incidence of contamination after treatmentb (no. of pools)
|
|||||
| Chaff | Seed | Both | Vortexing | Sonication | Chlorine | ||||
| S. enterica serovar Newport | 29 | 4 | 1 | 3 | 0.017 | 10 | 10 | 8 | 5 |
| E. coli O157:H7 | 45 | 6 | 3 | 6 | 0.033 | 8 | 8 | 8 | 3 |
| Total | 74 | 10 | 4 | 9 | 0.025c | 18 | 18 | 16 | 8 |
A total of 1,000 seeds, from each pool were tested (see Materials and Methods). The number of pools where contamination was found in only chaff or seed or in both chaff and seed is shown as incidence of contamination.
Number of pools where contamination was found in samples of 1,000 seeds after vortexing, sonication, and chlorine treatment.
Average chi-square probability for S. enterica serovar Newport and E. coli O157:H7-contaminated pools.
Seed contamination caused by contaminated chaff would probably be bound loosely to the seeds. Therefore, we attempted to vigorously clean the seeds to determine if the bacteria isolated from the seed pools were more directly associated with seeds rather than with chaff. Each of 18 S. enterica serovar Newport- and E. coli O157:H7-contaminated seed pools were sanitized progressively in three steps: vortexing, sonication, and Cl2 treatment. All 18 seed pools remained contaminated after vortexing, and only two pools were negative for the pathogen after sonication (Table 3). Furthermore, 8 of 18 pools (44%) remained contaminated even after a 5-min treatment with Cl2, whereas no bacteria could be detected in the posttreatment washes.
Invasion of primary roots.
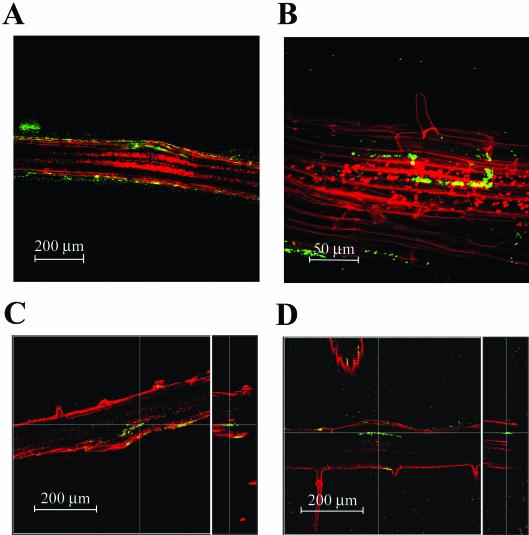
Plant pathogens often invade the roots through breaks in the epidermis upon development of lateral roots (19). Growth and inoculation of plants in the gnotobiotic system allowed us to see if S. enterica serovar Newport and E. coli O157:H7 were able to invade the plant tissue. GFP-labeled S. enterica serovar Newport and E. coli O157:H7 could be detected within the root tissue of sterilely grown plants and appeared exclusively within the primary root near the base of the lateral roots (Fig. 7). The earliest examples of invasion were seen on newly forming lateral roots where the epidermis had been broken recently by expansion of the underlying cell layers (Fig. 7A). The bacteria could be seen between and under the epidermal cells. At lateral root junctions when the lateral root was fully developed, colonization was often deeper within the primary root (Fig. 7B to D). The larger numbers of bacteria in the older roots indicate that bacteria not only invade the root tissue but also appear to proliferate within the root tissue. Bacterial invasion rarely occurred as deeply as the outer layers of the vasculature, and bacteria were never observed to be moving systemically within the plant. Invasion occurred in about 2 to 5% of the lateral roots, or about 1 occurrence per 3 cm of root. This frequency could reflect both the timing of lateral root development and the growth or spread of the bacteria. The earliest invasion detected was 18 h postinoculation, and new invasions could be observed wherever new lateral roots were produced (>10 days after inoculation) (data not shown).
FIG. 7.
Invasion of GFP-labeled bacteria into the primary root at lateral root junctions. (A) Invasion of S. enterica serovar Newport at a newly developed lateral root that has just broken the epidermis of the primary root. Fluorescent bacteria are seen just below and adjacent to the epidermal cells. (B) S. enterica serovar Newport within the primary root appears to occupy the intercellular regions (apoplast). The bacteria delineate cells within the primary root. (C) E. coli O157:H7 within the primary root at a mature lateral root junction. The right panel is a computer-generated vertical slice at the line in the left panel, showing that the bacteria are within the root, not on the surface. The lateral root is out of the focal plane. (D) E. coli O157:H7 invasion of a mature lateral root, similar to panel C but showing greater spread of the bacteria along the primary root. The red in panels A to D is either vascular autofluorescence or TO-PRO-3 staining of epidermis and root hairs (see Materials and Methods).
GFP-labeled mutant S. enterica serovar Typhimurium strains defective in flagellin synthesis (S. enterica serovar Typhimurium SJW1368) or motility functions (S. enterica serovar Typhimurium SJW1809) grew as quickly as the wild-type strain on the surface of the root (Fig. 5B), but microscopic investigation did not reveal any invasion at lateral root junctions when the inoculum was 104 CFU ml−1. The lack of invasion could be due to an inability of the bacteria to position themselves near the developing lateral root at the time of emergence. To test this possibility, we reasoned that increasing the inoculum 100-fold, to 106 CFU ml−1, could overcome the limitation of lack of motility, and invasion would occur. Inoculation at this higher concentration resulted in invasion of root tissue with both S. enterica serovar Typhimurium mutants (SWJ1368 and SWJ1809). However, the number of lateral roots where invasion was evident was very small (less than 0.1% of the lateral roots) compared to the number invaded by the wild-type strain S. enterica serovar Typhimurium SWJ1103 inoculated at 104 CFU ml−1 (data not shown).
DISCUSSION
We have shown that S. enterica serovar Newport and E. coli O157:H7 grow epiphytically on A. thaliana in a gnotobiotic system. Growth on roots was significantly higher than that reported for bean and alfalfa sprouts in similar studies (12, 57). However, in those studies, bean and alfalfa seeds were germinated in water. In contrast, demonstration of pathogen growth on A. thaliana in water was not possible. The reason might be due to the very small size of A. thaliana seeds, which may limit the duration that the seedling remains viable in water.
GFP-labeled bacteria were observed deep within the primary root but not in the vasculature. In contrast, by using immunofluorescence microscopy and scanning electron microscopy, Itoh et al. showed invasion of E. coli O157:H7 in radish tissue on sprouts grown from contaminated seeds (31). The bacteria were found in and on the xylem elements of the hypocotyl, supporting a conclusion that E. coli O157:H7 can move through the vasculature. However, bacteria may have been introduced during sample preparation. Recently, Solomon et al. also concluded that E. coli O157:H7 was moving within the plant, presumably via the vasculature, when the bacteria were shown to be present within surface-sterilized lettuce leaves after root inoculation (51). However, movement of E. coli O157:H7 on the surface of the plant and subsequent invasion of the leaf tissue were not excluded in these experiments. Whether E. coli O157:H7 is capable of movement through the vasculature is unknown.
Movement along the exterior of the root to the crown occurred relatively quickly (approximately 2.4 cm in 2 days), with E. coli O157:H7 moving at nearly twice the rate of S. enterica. These rates occurred on plants incubated at 23°C. In comparison, the bacteria alone on tryptone soft agar plates at 37°C swarmed at 5.4 and 2.7 cm per day for S. enterica serovar Typhimurium and E. coli O157:H7, respectively (52, 63). The rate of movement of S. enterica and E. coli O157:H7 in soil and on soil-grown plants is likely to be slower. However, the rate of movement of Pseudomonas spp. in soil (0.5 to 1 cm per day) was comparable to that of S. enterica and E. coli O157:H7 (3, 50). Movement from the crown to the meristematic region appeared to be substantially slower, and the bacteria were observed in only a few select niches, at the base of the petioles, on floral buds, and eventually on open flowers. Bacteria could be cultured from leaves even though we were not able to observe them microscopically. Movement may be a selective advantage for S. enterica and E. coli O157:H7, since it allows the bacteria to respond to changes in pH, moisture, or nutrients as epiphytes do (6). Motility may also help S. enterica and E. coli O157:H7 find a protected niche (e.g., inside the root) for extended colonization of the plant. Movement on sterilely grown plants did not occur with either of the two motility mutants of S. enterica serovar Typhimurium, indicating that motility requires functional flagella.
The motility mutants were also severely attenuated with respect to invasion, as has been demonstrated with plant-pathogenic bacteria (43, 56). Experiments with 100-fold-higher inoculum levels of the mutants resulted in invasion at approximately 1% of the wild-type level. These results suggest that motility is necessary only to position the bacteria to an appropriate spot where invasion can occur, such as where the lateral root emerges through the epidermis. Increasing the inoculum would increase the probability that bacteria are in good positions for invasion. One of the mutants, S. enterica serovar Typhimurium SWJ1809, produces flagella but is nonmotile due to a mutation in flaN, which encodes a protein that controls rotation of the flagellum. Thus, the fact that this mutant was also poorly invasive indicates that the flagellum must be active to support invasion. Whether an active flagellum is beneficial for S. enterica and E. coli O157:H7 colonization of soil-grown plants is under investigation.
S. enterica and E. coli O157:H7 have been shown to persist for extended periods in soil and on plant surfaces (4, 7, 15, 16, 20, 25, 57, 61). Furthermore, enteric bacteria frequently come in contact with plants once they are shed from their animal host, and survival on plants would help ensure that they are ingested again. Seemingly, there is a benefit to survival of enteric bacteria in the plant environment long enough and in sufficient numbers to ensure infection of a new host.
Survival of S. enterica serovar Newport and E. coli O157:H7 was monitored on soil-grown A. thaliana. In the most extreme situation, A. thaliana was inoculated on agar plates and then transplanted to soil. Growth on the plates produced a strong inoculum of both S. enterica serovar Newport and E. coli O157:H7. Furthermore, this contamination persisted after transplantation to soil and throughout the life of the plants. The bacteria could be recovered from seeds and chaff harvested from the plants after the plants had died (about 60 days after germination). In contrast, persistence was greatly reduced when seeds were inoculated and sowed directly on soil, probably due to the much lower inoculum. Although 100% of the plants grown on autoclaved soil were still contaminated after 14 days, both S. enterica serovar Newport and E. coli O157:H7 were undetectable at 30 days. The percentage of contaminated plants decreased further when the seeds were sown on unautoclaved soil and decreased again in amended soil. These results are probably due to the presence of indigenous competitors present in unautoclaved potting soil or the Yolo fine sandy loam. In the latter case, the persistence of S. enterica serovar Newport and E. coli O157:H7 was decreased to 50% of that observed on plants grown in autoclaved soil.
Persistence was affected more significantly if a strain cultured from the phyllosphere, Enterobacter asburiae, was coinoculated with S. enterica serovar Newport and E. coli O157:H7 (initially on sterilely grown plants prior to transfer to soil). Remarkably, S. enterica serovar Newport and E. coli O157 were undetectable 1 day after transfer of the plants to autoclaved soil, whereas Enterobacter asburiae infected all the plants. The possibility that Enterobacter asburiae is a competitor of S. enterica serovar Newport and E. coli O157 on A. thaliana is indicated by the observation that its appearance on the plant was correlated with the highest competition against S. enterica serovar Newport and E. coli O157:H7. Enterobacter asburiae has been reported to be an endophyte and a frequent inhabitant of the rhizosphere of cotton, bean, and cucumber (23, 24, 39) and to move systemically in the plant (46). Evidence that Enterobacter asburiae inhabits the phyllosphere has not been reported. Also, it is interesting that Enterobacter asburiae is a coliform bacterium, since E. coli O157:H7 has been shown to compete with indigenous and manure-applied coliforms in soil and on plant roots (17). The mechanisms by which Enterobacter asburiae competes with S. enterica serovar Newport and E. coli O157:H7 on A. thaliana are the subject of further investigation in our laboratory.
Contaminated samples from soil-grown plants analyzed for pathogen contamination were always from the apical regions of the plant and were commonly flowers. It is obvious that contamination of flowers could lead directly to seed contamination. Nevertheless, there was a correlation between chaff and seed contamination, indicating that seed contamination may be due solely to cross-contamination by chaff. However, a direct correlation between chaff and seed contamination could also occur if siliques are contaminated when seed contamination occurs by invasion of the flower or silique. Invasion of both ovules and seeds has been shown to occur with many different epiphytic and rhizosphere bacteria (41). Bacteria may enter the seeds via a variety of routes, such as the vasculature, pollen germ tube, and the dorsal suture of the silique or hilum of the mature seed (26, 38, 64). We observed binding of S. enterica on the hilum of mung bean seeds inoculated in the laboratory (J. Barak, personal communication). Furthermore, it has been shown recently that root inoculation of lettuce with E. coli O157:H7 leads to internalization of bacteria within the leaves (51). This result suggests that E. coli O157:H7 may move throughout the plant. Importantly, if invasion of the plant leads to seed contamination, it could occur before maturation and hardening of the seed coat. In our experiments, seed contamination could not be eliminated in nearly half of the seed pools by a combination of washing, sonication, and chlorine treatment. Thus, it appears that the bacteria adhere tightly to the outside of the seeds or are located under the seed coat.
The incidence of seed contamination was sensitive to the inoculation method (inoculum strength). Inoculation of the seeds prior to sowing on soil and coinoculation with a competitor resulted in plants that failed to yield contaminated seeds in the next generation. In contrast, inoculation of sterilely grown seedlings or soil led to contaminated seeds. We expected a correlation between seed contamination and the persistence of S. enterica serovar Newport and E. coli O157:H7 on the foliage, yet persistence was limited to 21 days on plants grown on inoculated soil, and the pathogens were discovered on the seeds. However, contamination on the plants may not be uniform, and plant samples were always selected from the apical portion of the plant. Seeds that come from subapical flowers may be more likely to harbor the pathogens because these seeds mature before the seeds developing at terminal flowers. Also unexpectedly, the incidence of seed contamination was not sensitive to the type of soil. Contaminated seeds were recovered from plants grown in each of the three soils and at approximately the same efficiency. However, the inoculum was very high in the soil and sterile seedling inoculation methods and could have overwhelmed any competition by indigenous bacteria. The contamination of seeds that we observed in our experiments is of interest. However, the controlled environments in our model preclude us from making any strong conclusions regarding potential seed contamination in the field when soil or plant tissue becomes contaminated by irrigation or fertilization.
Knowledge of how a host plant supports or limits the growth and invasion of bacteria could facilitate the development of new control measures. While the gnotobiotic method does not mimic the complexity of the natural environment, it does mimic commercial sprout production and hydroponic farming. Further investigations with A. thaliana may identify factors important for binding, growth, and persistence of S. enterica serovar Newport and E. coli O157:H7 on and in sprouts. Furthermore, use of the gnotobiotic method is an important and nearly indispensable tool towards discovery of the host genes involved in these interactions. The advanced knowledge of A. thaliana genetics and the methods of studying global gene expression (e.g., microarrays) have led to discovery of host genes responsive to S. enterica serovar Newport and E. coli O157:H7 (M. B. Cooley, unpublished data). Some of these genes may also be important in crop plants grown in the natural environment and thus provide clues helpful in discovering novel methods to minimize contamination of produce.
Acknowledgments
We thank Lee-Shin Tsai (WRRC, ARS, USDA, Albany, Calif.) for help in analysis of free Cl2 and Maria Brandl (WRRC, ARS, USDA, Albany, Calif.) for help with confocal microscopy.
This research was funded by U.S. Department of Agriculture Agricultural Research Service CRIS project 5325-42000-022.
REFERENCES
- 1.Anonymous. 1997. Outbreaks of Escherichia coli O157:H7 infection and cryptosporidiosis associated with drinking unpasteurized apple cider—Connecticut and New York, October 1996. Morbid. Mortal. Wkly. Rep. 46:4-8. [PubMed] [Google Scholar]
- 2.Anonymous. 1996. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: a manual. National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Ga.
- 3.Arora, D. K., A. B. Filonow, and J. L. Lockwood. 1983. Bacterial chemotaxis to fungal propagules in vitro and in soil. Can. J. Microbiol. 29:1104-1109. [Google Scholar]
- 4.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayot, R. G., and S. M. Ries. 1986. Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology 76:441-445. [Google Scholar]
- 6.Beattie, G. A., and S. E. Lindow. 1999. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89:353-359. [DOI] [PubMed] [Google Scholar]
- 7.Beuchat, L. R. 1999. Survival of enterohemorrhagic Escherichia coli O157:H7 in bovine feces applied to lettuce and the effectiveness of chlorinated water as a disinfectant. J. Food. Prot. 62:845-849. [DOI] [PubMed] [Google Scholar]
- 8.Brandl, M. T., and R. E. Mandrell. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaway, J. O. 1989. Amperometric titration method, p. 4-54. In L. S. Clesceri, A. E. Greenberg, and R. R. Trussell (ed.), Standard methods for the examination of water and wastewater, 17th ed. American Public Health Association, Washington, D.C.
- 10.Cao, H., R. L. Baldini, and L. G. Rahme. 2001. Common mechanisms for pathogens of plants and animals. Annu. Rev. Phytopathol. 39:259-284. [DOI] [PubMed] [Google Scholar]
- 11.Carmichael, I., I. Harper, M. Coventry, P. Taylor, J. Wan, and M. Hickey. 1999. Bacterial colonization and biofilm development on minimally processed vegetables. J. Appl. Microbiol. 85:45S-51S. [DOI] [PubMed]
- 12.Charkowski, A. O., J. D. Barak, C. Z. Sarreal, and R. E. Mandrell. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl. Environ. Microbiol. 68:3114-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooley, M. B., M. R. D'Souza, and C. I. Kado. 1991. virC and virD operons of the Agrobacterium Ti plasmid are regulated by the ros chomosomal gene: analysis of the cloned ros gene. J. Bacteriol. 173:2608-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.deWeger, L. A., C. I. vanderVlugt, A. Wijfjies, P. A. Bakker, B. Shippers, and B. Lugtenberg. 1987. Flagella of a plant growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 169:2769-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingman, D. W. 2000. Growth of Escherichia coli O157:H7 in bruised apple (Malus domestica) tissue as influenced by cultivar, date of harvest and source. J. Environ. Microbiol. 66:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagliardi, J. V., and J. S. Karns. 2002. Persistence of Escherichia coli O157:H7 in soil and on plant roots. Environ. Microbiol. 4:89-96. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Gomez, L., and T. Boller. 2000. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5:1003-1011. [DOI] [PubMed] [Google Scholar]
- 19.Gough, C., J. Vasse, C. Galera, G. Webster, E. Cocking, and J. Denarie. 1997. Interactions between bacterial diazotrophs and non-legume dicots: Arabidopsis thaliana as a model plant. Plant Soil 194:123-130. [DOI] [PubMed] [Google Scholar]
- 20.Guo, X., J. Chen, R. E. Brackett, and L. R. Beuchat. 2001. Survival of salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67:4760-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, X. A., M. W. van Iersel, J. R. Chen, R. E. Brackett, and L. R. Beuchat. 2002. Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl. Environ. Microbiol. 68:3639-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haefele, D. M., and S. E. Lindow. 1987. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl. Environ. Microbiol. 53:2528-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallmann, J., A. Quadt-Hallmann, W. F. Mahaffee, and J. W. Kloepper. 1997. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43:895-914. [Google Scholar]
- 24.Hallmann, J., A. Quadt-Hallmann, R. Rodriguez-Kabana, and J. W. Kloepper. 1998. Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biol. Biochem. 30:925-937. [Google Scholar]
- 25.Hara-Kudo, Y., H. Konuma, M. Iwaki, F. Kasuga, Y. Sugita-Konishi, Y. Ito, and S. Kumagai. 1997. Potential hazard of radish sprouts as a vehicle of Escherichia coli O157:H7. J. Food. Prot. 60:1125-1127. [DOI] [PubMed] [Google Scholar]
- 26.Harman, G. E. 1983. Mechanisms of seeds infection and pathogenesis. Phytopathology 73:326-329. [Google Scholar]
- 27.Hatterman, D. R., and S. M. Ries. 1989. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology 79:284-289. [Google Scholar]
- 28.Hawes, M. C., and L. Y. Smith. 1989. Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil-grown pea plants. J. Bacteriol. 171:5668-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes with the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 30.Inami, G. B., and S. E. Moler. 1999. Detection and isolation of Salmonella from naturally contaminated alfalfa seeds following an outbreak investigation. J. Food. Prot. 62:662-664. [DOI] [PubMed] [Google Scholar]
- 31.Itoh, Y., Y. Sugita-Konishi, F. Kasuga, M. Iwaki, Y. Hara-Kudo, N. Saito, Y. Noguchi, H. Konuma, and S. Kumagai. 1998. Enterohemorrhagic Escherichia coli O157:H7 present in radish spouts. Appl. Environ. Microbiol. 64:1532-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaeger, C. H., S. E. Lindow, W. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamoun, S., and C. I. Kado. 1990. Phenotypic switching affecting chemotaxis, xanthan production, and virulence in Xanthomonas campestris. Appl. Environ. Microbiol. 56:3855-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinkel, L. L. 1997. Microbial population dynamics on leaves. Annu. Rev. Phytopathol. 35:327-347. [DOI] [PubMed] [Google Scholar]
- 35.Kinkel, L. L., M. Wilson, and S. E. Lindow. 1995. Effects of scale on estimates of epiphytic bacterial populations. Microb. Ecol. 29:283-297. [DOI] [PubMed] [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Kutsukake, K., T. Iino, Y. Komeda, and S. Yamaguchi. 1980. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol. Gen. Genet. 178:59-67. [DOI] [PubMed] [Google Scholar]
- 38.Mabagala, R. B. 1997. The effect of populations of Xanthomonas campestris pv. phaseoli in bean reproductive tissues on seeds infection of resistant and susceptible bean genotypes. Eur. J. Plant Pathol. 103:175-181. [Google Scholar]
- 39.McInroy, J. A., and J. W. Kloepper. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 173:337-342. [Google Scholar]
- 40.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 41.Mundt, J. O., and N. F. Hinkle. 1976. Bacteria within ovules and seeds. Appl. Environ. Microbiol. 32:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panapoulos, N., and M. Schroth. 1974. Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64:1389-1397. [Google Scholar]
- 44.Piernas, V., and J. Guiraud. 1997. Disinfection of rice seeds prior to sprouting. J. Food Sci. 62:611-615. [Google Scholar]
- 45.Plotnikova, J. M., L. G. Rahme, and F. M. Ausubel. 2000. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 124:1766-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quadt-Hallmann, A., N. Benhamou, and J. W. Kloepper. 1997. Bacterial endophytes in cotton: mechanisms of entering the plant. Can. J. Microbiol. 43:577-582. [Google Scholar]
- 47.Rasmussen, M., and T. Casey. 2001. Environmental and food safety aspects of Escherichia coli O157:H7 infection in cattle. Crit. Rev. Microbiol. 27:57-73. [DOI] [PubMed] [Google Scholar]
- 48.Salter, M., T. Ross, and T. McMeekin. 1998. Applicability of a model for non-pathogenic Escherichia coli for predicting the growth of pathogenic Escherichia coli. J. Appl. Microbiol. 85:357-364. [DOI] [PubMed] [Google Scholar]
- 49.Samuels, M., and J. Witmer. 1999. Statistics for the life sciences. Prentice Hall, Upper Saddle River, N.J.
- 50.Scher, F. M., J. W. Kloepper, and C. A. Singleton. 1985. Chemotaxis of fluorescent Pseudomonas spp. to soybean seeds exudates in vitro and in soil. Can. J. Microbiol. 31:570-574. [Google Scholar]
- 51.Solomon, E. B., S. Yaron, and K. R. Matthews. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steele, B. T., N. Murphy, G. S. Arbus, and C. P. Rance. 1982. An outbreak of hemolytic uremic syndrome associated with ingestion of fresh apple juice. Clin. Lab. Observ. 101:963-965. [DOI] [PubMed] [Google Scholar]
- 54.Stewart, D., K. Reineke, J. Ulaszek, and M. Tortorello. 2001. Growth of Salmonella during sprouting of alfalfa seeds associated with salmonellosis outbreaks. J. Food. Prot. 64:618-622. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi, K., and J. F. Frank. 2000. Penetration of Escherichia coli O157:H7 into lettuce tissues as affected by inoculum size and temperature and the effect of chlorine treatment on cell viability. J. Food. Prot. 63:434-440. [DOI] [PubMed] [Google Scholar]
- 56.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taormina, P. J., L. R. Beuchat, and L. Slutsker. 1999. Infections associated with eating seeds sprouts: an international concern. Emerg. Infect. Dis. 5:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tauxe, R. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tauxe, R., H. Kruse, C. Hedberg, M. Potter, J. Madden, and K. Wachsmuth. 1997. Microbial hazards and emerging issues associated with produce, a preliminary report to the national advisory committee on microbiologic criteria for foods. J. Food. Prot. 60:1400-1408. [DOI] [PubMed] [Google Scholar]
- 60.VanderBroek, A. M., M. Kambrecht, and J. Vanderleyden. 1998. Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasiliense. Microbiology 144:2599-2606. [DOI] [PubMed] [Google Scholar]
- 61.Vernon-Campbell, J., J. Mohle-Boetani, R. Reporter, S. Abbott, J. Farrar, M. T. Brandl, R. E. Mandrell, and S. B. Werner. 2001. An outbreak of Salmonella serotype Thompson associated with fresh cilantro. J. Infect. Dis. 183:984-987. [DOI] [PubMed] [Google Scholar]
- 62.Wachtel, M. R., L. C. Whitehand, and R. E. Mandrell. 2002. Association of Escherichia coli O157:H7 with preharvest leaf lettuce upon exposure to contaminated irrigation water. J. Food. Prot. 65:18-25. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaumeyer, W. J. 1957. A monographic study of bean diseases and methods of their control. USDA Tech. Bull. 868. U.S. Department of Agriculture, Washington, D.C.