Abstract
The chromate reductase purified from Pseudomonas ambigua was found to be homologous with several nitroreductases. Escherichia coli DH5α and Vibrio harveyi KCTC 2720 nitroreductases were chosen for the present study, and their chromate-reducing activities were determined. A fusion between glutathione S-transferase (GST) and E. coli DH5α NfsA (GST-EcNfsA), a fusion between GST and E. coli DH5α NfsB (GST-EcNfsB), and a fusion between GST and V. harveyi KCTC 2720 NfsA (GST-VhNfsA) were prepared for their overproduction and easy purification. GST-EcNfsA, GST-EcNFsB, and GST-VhNFsA efficiently reduced nitrofurazone and 2,4,6-trinitrotoluene (TNT) as their nitro substrates. The Km values for GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA for chromate reduction were 11.8, 23.5, and 5.4 μM, respectively. The Vmax values for GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA were 3.8, 3.9, and 10.7 nmol/min/mg of protein, respectively. GST-VhNfsA was the most effective of the three chromate reductases, as determined by each Vmax/Km value. The optimal temperatures of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA for chromate reduction were 55, 30, and 30°C, respectively. Thus, it is confirmed that nitroreductase can also act as a chromate reductase. Nitroreductases may be used in chromate remediation. GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA have a molecular mass of 50 kDa and exist as a monomer in solution. Thin-layer chromatography showed that GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA contain FMN as a cofactor. GST-VhNfsA reduced Cr(VI) to Cr(III). Cr(III) was much less toxic to E. coli than Cr(VI).
Environmental pollution by chromium may be severe. Chromium contamination is known to be prevalent at U.S. Department of Energy sites (29). The electroplating and leather-tanning industries also contribute to environmental contamination with Cr(VI) (23). Chromate compounds containing Cr(VI) are used widely in the cooling towers of heavy industry and atomic power plants, since Cr(VI) prevents corrosion and the growth of organisms (2). Cr(VI) is soluble, toxic, and carcinogenic, whereas Cr(III) is less soluble and less toxic (12). Thus, it is desirable to change Cr(VI) into Cr(III). This approach is taken in the bioremediation of Cr(VI) pollution. It shows promise for solving pollution problems and has advantages over various other physical and chemical methods.
Chromate-reducing activities can be found in the cell extracts of many bacteria (4, 5, 8, 10, 13, 18, 25, 30, 33-35). Chromate reductase can reduce the toxicity of Cr(VI) by reducing it to Cr(III) and lowering its solubility (5, 9). The chromate reductase from Pseudomonas ambigua has been purified and characterized (33). Chromate-reducing activities have been associated with DT-diaphorase (7) and aldehyde oxidase (1) in the cell cytoplasm. Cytochrome P450 located in the cell membrane is also known to have chromate-reducing activity (21). It seems that various reductases in the cell can function in chromate reduction. P. ambigua chromate reductase (33) has high homology with Escherichia coli NfsA (59%) and Vibrio harveyi NfsA (58%) nitroreductase.
There are two types of nitroreductase. Type I is oxygen insensitive (3, 6, 15, 17, 20, 37, 38, 39), and type II is oxygen sensitive (21, 26). In type I, there are two nitroflavin reductase superfamiles: NfsA and NfsB (3, 6, 14, 17, 20, 37, 38, 39). NfsA includes the major nitroreductase in E. coli (3, 37) and Frp in V. harveyi (14, 17). NfsB includes the minor nitroreductase in E. coli (3, 20, 38) and Frase I in Vibrio (Photobacterium) fischeri (6, 39).
The E. coli and V. harveyi nitroreductases were chosen for the present study. It will be shown that these nitroreductases are also efficient in chromate reduction.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5α was grown on Luria-Bertani (LB) broth for 12 h at 37°C. Freeze-dried V. harveyi KCTC 2720 cells were resuspended in sterile distilled water. The cell suspension was directly used in PCR. V. harveyi KCTC 2720 was from the Korean Collection for Type Cultures (KCTC), Seoul, Korea.
Cloning of nitroreductase gene.
To clone the nfsA gene of E. coli DH5α, two primers (EcNfsA1 [5′-GTAGGATCCACGCCAACCATTGAAC-3′] containing a BamHI site, and EcNfsA2 [5′-ACTGAATTCTTAGCGCGTCGCCCAAC-3′] containing a EcoRI site) were used. The two primer sequences were deduced from E. coli AB1157 (36). To clone the nfsB gene from E. coli DH5α, two primers (EcNfsB1 [5′-GTAGGATCCGATATCATTTCTGTCGC-3′] containing a BamHI site and EcNfsB2 [5′-ACTGAATTCTTACACTTCGGTTAAGGTG-3′] containing a EcoRI site) were used. The two primer sequences were deduced from E. coli C600 (38). To clone the nfsA gene from V. harveyi KCTC 2720, two primers (VhNfsA1 [5′-GTAGGATCCAACAATACGATTGAAAC-3′] containing a BamHI site and VhNfsA2 [5′-ACTGAATTCTTAGCGTTTTGCTAGCC-3′] containing a EcoRI site) were used. The two primer sequences were from V. harveyi ATCC 33843 (17). PCRs were carried out in a 50-μl reaction volume. A 1-μl volume of suspended cells was added to 49 μl of PCR mixture (50 mM KCl; 10 mM Tris-HCl [pH 9.0]; 0.1% Triton X-100; 1.5 mM MgCl2; 0.25 mM [each] dATP, dGTP, dCTP, and dTTP [Sigma, St. Louis, Mo.]; 10 pmol (0.2 × 10−3 mM] of each primer, and 2.5 U of Taq DNA polymerase [Promega Co., Madison, Wis.]). PCR amplification was performed in a Perkin-Elmer thermal cycler with the following steps: an initial denaturation step at 95°C for 7 min; 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final extension step of 72°C for 5 min. Amplified DNA fragments were electrophoresed in a 1.5% agarose gel and were eluted by using a GeneClean II kit (Bio 101, Inc., La Jolla, Calif.). For easy purification of nitroreductase, the three nitroreductase genes were fused into the glutathione S-transferase (GST) gene (32). Each PCR product was cut with BamHI and EcoRI (Promega Corp., Madison, Wis.), electrophoresed in a 1.5% agarose gel, and eluted with the GeneClean II kit. It was ligated by T4 DNA ligase (TaKaRa Co., Kyoto, Japan) into plasmid pGEX-4T-1 (Pharmacia Biotech) containing GST that was also cut with BamHI and EcoRI. E. coli BL21 cells were transformed with the ligation mixture. The strain containing the fusion genes on the plasmids was screened by ampicillin resistance and plasmid digestion with the two enzymes. GST-EcNfsA is a fusion between GST at the N terminus and upstream from E. coli DH5α NfsA, and GST-EcNfsB is a fusion between GST and E. coli DH5α NfsB. GST-VhNfsA is a fusion between GST and V. harveyi KCTC2720 NfsA. All three nitroreductase fusion genes on the plasmid (pGST-EcNfsA, pGST-EcNfsB, and pGST-VhNfsA) were confirmed by one primer in the nitroreductase gene (EcNfsA1, EcNfsB1, and VhNfsA1) and the other on the plasmid (CR2). The sequence of CR2 is 5′-GGGAGCTGCATGTGTCAGAG-3′.
DNA sequencing and sequence analysis.
All three nitroreductase genes were obtained from pGST-EcNfsA, pGST-EcNfsB, and pGST-VhNfsA and were then subcloned into pUC18. Their nucleotide sequences were determined by using ThermoSequenase (Amersham Life Science, Arlington Heights, Ill.) and the dideoxy termination method. The accession numbers for GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA deposited in GenBank are AF394662, AF394661, and AF395832, respectively.
Purification of fused nitroreductase.
E. coli BL21 harboring the plasmids containing fused nitroreductase genes was cultured at 37°C for 14 h and diluted 100-fold in fresh LB broth. It was cultured again to reach its exponential phase. In this phase, protein expression was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3.5 h. The cells were harvested by centrifugation at 13,000 × g for 5 min. To purify the fused nitroreductase, affinity chromatography was used. Cell extracts were obtained by sonication. The extracts and glutathione-agarose beads (Sigma) were mixed. The fused nitroreductase bound to the glutathione-agarose beads was eluted with 10 mM reduced glutathione (pH 8.0). The protein concentration was determined either by measuring the optical density values at A280 or by the Bradford method with a Serva dye and with bovine serum albumin as a standard (28).
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a 12.5% gel by the method of Laemmli (16). Protein bands were visualized with Coomassie brilliant blue R-250 (Sigma).
Gel filtration.
Gel filtration was carried out in a 2.5-by-50-cm column of Sephadex G-75 (Sigma). The Sephadex G-75 column was equilibrated in 20 mM Tris-HCl (pH 7.0) buffer at a flow rate of 0.6 ml/min. The void volume (Vo) was determined by using blue dextran (Sigma). The standard proteins (Sigma) used for the estimation of molecular mass included bovine albumin (66 kDa), egg albumin (45 kDa), rabbit glyceraldehyde-3-phosphate dehydrogenase (36 kDa), bovine carbonic anhydrase, (29 kDa), and bovine trypsinogen (24 kDa). The elution profiles were monitored by UV absorption at 280 nm. SDS-PAGE was used to characterize the sizes and types of subunits in the purified chromate reductases.
Spectral measurements.
The absorption spectrum of nitroreductase was measured with spectrophotometer (UVICON930) in a range between 250 and 500 nm.
Identification of the nitroreductase-bound flavin.
To characterize the cofactor in the nitroreductases, the purified nitroreductases were boiled and precipitated to extract the cofactor. After centrifugation, the supernatant (20 μl) containing the cofactor was spotted and chromatographed in the dark on silica gel 60 F254 (Merck) with FAD and FMN standards by using a solvent system (distilled water-ethanol-acetic acid [20:6:1]). The position of each spot was visualized by using 365-nm UV.
Determination of nitroreductase activity.
Nitrofurazone nitroreductase activity was assayed in a reaction mixture containing 10 μM Tris-HCl (pH 7.5), 10 μM nitrofurazone (Sigma), 0.1 mM NADH (Sigma), and GST-EcNfsA, GST-EcNfsB, or GST-VhNfsA enzyme. Nitroreductase activity was determined by a decrease in absorbance at 400 nm (36). The transformation of TNT was indicated by a decrease in the absorbance of TNT at 447 nm (22). The assay mixture contained 50 mM Tris-HCl (pH 7.0), 0.1 mM TNT, 1 mM NADH, and either GST-EcNfsA, GST-EcNfsB, or GST-VhNfsA enzyme.
Determination of chromate reductase activity.
A standard curve for the reaction between chromate and its binding dye, 1,5-diphenylcarbazide (Sigma), was used to determine the chromate concentration (13). The diphenylcarbazide forms a pink complex with Cr(VI) but not with Cr(III). Reduction of Cr(VI) can produce Cr(V) and Cr(IV). However, Cr(V) and Cr(IV) are not stable in aqueous states at neutral pH (25). Thus, it is valid to measure Cr(VI) conversion to Cr(III) by using the dye method.
The chromate reductase assay was performed in 10 mM Tris-HCl (pH 7.0) buffer containing 1 mM NADH, 1 mM K2CrO4 (Sigma), and the enzyme at 30°C for 1 h. Then, 10 μl of 0.1 M H2SO4 and 15 μl of 0.5% (vol/vol) 1,5-diphenyl carbazide were added to the tube, and the residual Cr(VI) content was determined at 540 nm. The optimal temperature of chromate reductase activity was determined at various temperatures ranging from 4 to 80°C.
Determination of chromate reductase kinetics.
Various concentrations of K2CrO4 were added to the basic enzyme reaction mixture. Km and Vmax values were determined by using a Lineweaver-Burk plot.
RESULTS
Cloning of nitroreductase genes.
The chromate reductase from P. ambigua (33) was compared to the E. coli and P. ambigua nitroreductases. Surprisingly, a high degree of homology between P. ambigua chromate reductase and E. coli nitroreductase NfsA (59%) and between P. ambigua reductase and V. harveyi nitroreductase NfsA (58%) was found, as determined by using the BLAST function of the National Center for Biotechnology Information. It was of interest to determine whether the E. coli NfsA and NfsB and the V. harveyi NfsA nitroreductases had chromate reductase activities similar to that of the P. ambigua chromate reductase.
The PCR products of E. coli DH5α nfsA and nfsB and of V. harveyi KCTC 2720 nfsA nitroreductase genes consisted of 738, 669, and 738 bp, respectively, just as predicted (data not shown).
The three nitroreductase genes were sequenced. No differences were found between the E. coli DH5α and AB1157 nfsA genes and the E. coli DH5α and C600 nfsB genes (GenBank numbers AF394662 and AF394661, respectively). However, eight nucleotide sequences in the nfsA nitroreductase genes were different between the two V. harveyi strains (GenBank no. AF395832). Five amino acid residues in the nitroreductase in V. harveyi ATCC 33843 were replaced by different residues in V. harveyi KCTC 2720 (data not shown).
Some biochemical characteristics of fused nitroreductase.
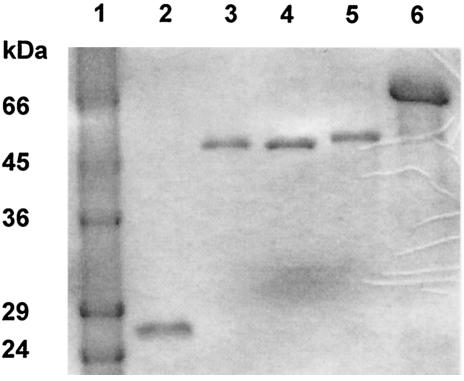
Using the affinity between glutathione and its S-transferase, the three fused nitroreductases were purified to homogeneity and used in biochemical studies (Fig. 1). The molecular masses of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA were each determined to be 50 kDa on an SDS-PAGE gel (Fig. 1). The absorbance of GST-EcNfsA (GST-EcNfsB and GST-VhNfsA) was maximal at 370 and 450 nm (data not shown). This suggests that GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA are typical flavoproteins containing FAD or FMN. Thin-layer chromatography analysis showed that GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA contain FMN rather than FAD (data not shown). GST used as a control did not contain FMN at all (data not shown).
FIG. 1.
Purification of nitroreductases fused to GST. E. coli DH5α (pGST-EcNfsA), E. coli DH5α (pGST-EcNfsB), and E. coli DH5α (pGST-VhNfsA) were cultured to exponential phase in LB broth and then induced by the addition of 0.5 mM IPTG for 3.5 h. Induced cells were broken by sonication and separated into supernatant and pellet fractions by centrifugation. By using affinity chromatography of glutathione and GST, GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA (lanes 3, 4, and 5, respectively) were purified. Lanes 2 and 6 show GST (26 kDa) and BSA (66 kDa), respectively. Lane 1 has molecular-mass standards.
Nitroreductase activity.
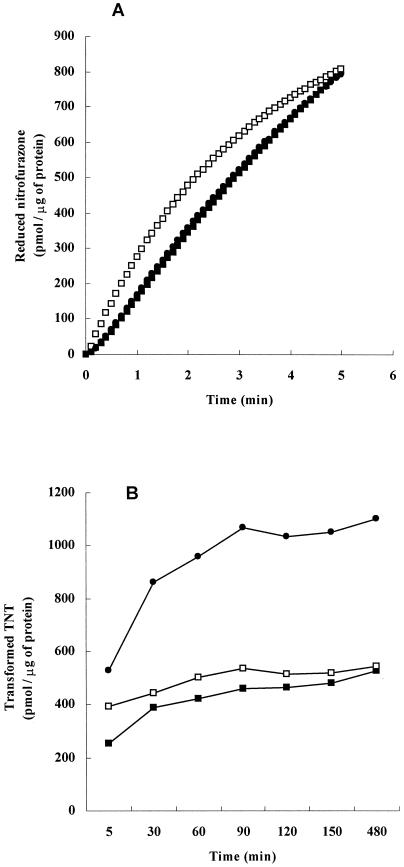
We determined whether a fusion of GST and nitroreductase still contains nitroreductase activity. Substrates for E. coli nitroreductase are menadione (vitamin K), nitrofurazone, TNT, etc. (7, 11, 19, 22, 36). We chose nitrofurazone and TNT as the substrates for GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA. We postulated that electrons move from NADH to substrate nitrofurazone or TNT in the presence of nitroreductase. We found that GST-EcNfsB was the most efficient in nitrofurazone-reducing activity and that GST-VhNfsA was as efficient as GST-EcNfsA (Fig. 2A). GST-VhNfsA was most efficient in TNT reduction, GST-EcNfsB is next most efficient, and GST-EcNfsA is least efficient (Fig. 2B). These results demonstrated clearly that all three—GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA—retained nitroreductase activities.
FIG. 2.
(A) Nitrofurazone-reducing activity. The reduction of nitrofurazone was determined by measuring the decrease in the absorbance of nitrofurazone at 400 nm (molar extinction coefficient, 12,960 M−1 cm−1). A basic enzyme reaction mixture (1.5 ml) containing 10 mM Tris-HCl buffer (pH 7.0), 0.1 mM NADH, 10 μM nitrofurazone, and 10 μg of either GST-EcNfsA (▪), GST-EcNfsB (□), or GST-VhNfsA (•) was incubated at 30°C. The reaction was started by the addition of NADH. One unit was defined as the amount of the enzyme that reduced 1 pmol of nitrofurazone/μg of protein. (B) TNT-reducing activity. The transformation of TNT was determined by measuring the decrease in the absorbance of TNT at 447 nm. The standard assay mixture (1 ml) containing 50 mM Tris-HCl (pH 7.0), 0.1 mM TNT, 1 mM NADH, and either 172 μg of GST-EcNfsA (▪), 166 μg of GST-EcNfsB (□), or 82 μg of GST-VhNfsA (•) was incubated at 30°C. Theenzymatic reaction was initiated by adding NADH. The reaction was quenched by adding 160 μl of 1 M NaOH, resulting in a pH of 12.2. Quantitative measurements were made 5 min after the addition of NADH to the TNT solution. One unit was defined as the amount of the enzyme that reduced 1 pmol of TNT/μg of protein.
The molecular masses of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA were each also determined to be 50 kDa by gel filtration (data not shown). They were 50 kDa on SDS-PAGE (Fig. 1). Thus, purified GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA are monomers in solution. In contrast, P. ambigua chromate reductase is a dimer (33).
Chromate reductase activity.
We determined whether the fused nitroreductase had chromate reductase activity like P. ambigua chromate reductase.
Glutathione (0.2 mM) is known to have chromate-reducing activity (33). However, 0.025 mM glutathione, which was present in the assay buffer, did not demonstrate any chromate-reducing activity. The GST used in the present study did not retain any chromate-reducing activity (data not shown).
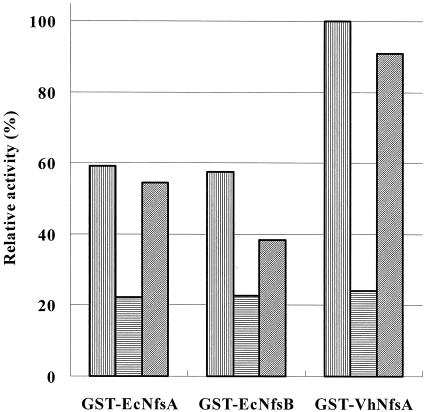
NADH alone showed chromate-reducing activity (Fig. 3). However, when chromate reductase was added to the reaction mixture, the chromate-reducing activities of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA increased by 2.6, 2.6, and 4.2 fold, respectively (Fig. 3). Thus, GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA are real chromate reductases in the presence of coenzyme NADH. When the NADH concentration in the reaction mixture was increased from 0.05 to 1 mM, the chromate reductase activities of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA increased (Fig. 3). This confirms again that GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA require NADH for their activities. P. ambigua chromate reductase is also NADH dependent (33). NADPH was almost as efficient as NADH in chromate reduction by GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA (data not shown).
FIG. 3.
NADH-dependent chromate reductase activity. Three kinds of reaction mixtures were prepared: 10 mM Tris-HCl (pH 7.0), 0.1 mM NADH, 20 μM K2CrO4, and either GST-EcNfsA, GST-EcNfsB, or GST-VhNfsA (▥); 10 mM Tris-HCl (pH 7.0), 0.1 mM NADH, 20 μM K2CrO4 (no enzyme) (▤); and 10 mM Tris-HCl (pH 7.0), 0.05 mM NADH, 20 μM K2CrO4, and either GST-EcNfsA, GST-EcNfsB, or GST-VhNfsA (▨). They were incubated at 30°C for 1 h, and the chromate reductase activity was determined by the dye method described in the text. One unit of chromate reductase activity was defined as the amount of the enzyme which decreased 1 nmol of Cr(VI) per min at 30°C per mg of protein. The chromate reductase activity of GST-VhNfsA in the reaction buffer (▥) was regarded as 100%.
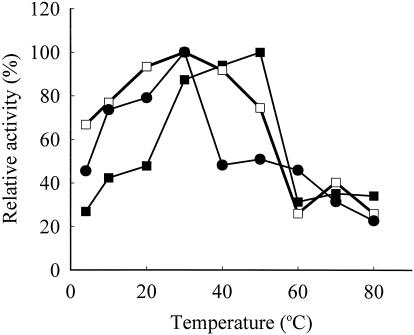
Activity of GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA was optimal at 55, 30, and 30°C, respectively (Fig. 4). GST-EcNfsB is characteristic of being still active at 4°C, retaining 65% of its maximum activity. GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA all lost considerable activities at >60°C (Fig. 4).
FIG. 4.
Effect of temperature on chromate reductase activity. The basic enzyme reaction mixtures containing GST-EcNfsA (▪), GST-EcNfsB (□), or GST-VhNfsA (•) were incubated at 4 to 80°C, and their chromate-reducing activities were determined. The chromate reductase activity of GST-EcNfsA at 55°C, GST-EcNfsB at 30°C, or GST-VhNfsA at 30°C was regarded as the 100% value.
All three nitroreductases showed chromate reductase activity (Table 1). Vmax/Km value of each nitroreductase can be used as a measure of the efficiency of the chromate reductase activity. GST-VhNfsA was most efficient, and GST-EcNfsA was twice as efficient as GST-EcNfsB (Table 1). The chromate reductase activity of GST-VhNfsA (12 times as efficient as GST-EcNfsB) matched that of P. ambigua chromate reductase (13 times as efficient as GST-EcNfsB) (Table 1) (33). This confirms that the nitroreductases are effective in chromate reduction.
TABLE 1.
Comparison of Cr(VI) reductase kinetics
| Chromate reductase | Km (mM) | Vmax (nmol/min/mg of protein) | Vmax/Km |
|---|---|---|---|
| GST-EcNfsA | 11.8 × 10−3 | 3.8 | 322 |
| GST-EcNfsB | 23.5 × 10−3 | 3.9 | 163 |
| GST-VhNfsA | 5.4 × 10−3 | 10.7 | 2,000 |
| P. ambigua | 13.0 × 10−3 | 27.0 | 2,100 |
Cr(III) production from Cr(VI) (chromate) by GST-VhNfsA was confirmed by the appearance of an absorption peak at 567 nm, as determined by Puzon et al. (27; data not shown). A typical Cr(III) greenish color was also observed in chromate reduction by GST-VhNfsA. Surprisingly, Cr(III) produced by GST-VhNfsA enzyme was soluble, which is consistent with data of Puzon et al. (27). CrCl3 (5 mM) in pH 7 solution was insoluble (data not shown). Although Cr(III) produced by GST-VhNfsA was soluble, Cr(III) toxicity was reduced considerably, since it did not inhibit growth of E. coli (data not shown).
NADH consumption.
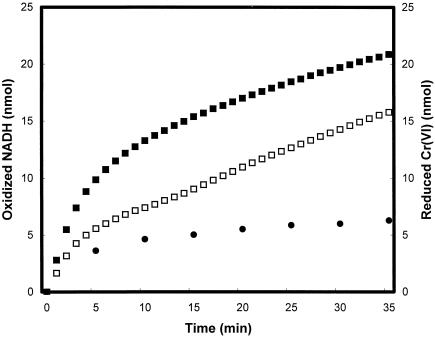
In the presence of enzyme GST-VhNfsA, NADH is oxidized even without chromate (Fig. 5). When chromate was added into the reaction buffer containing NADH and enzyme GST-VhNfsA, NADH consumption rate increased (Fig. 5), suggesting that electrons move from NADH onto Cr(VI). The ratio between the amount of NADH consumed for chromate reduction and that of reduced chromate was approximately 3 (Fig. 5).
FIG. 5.
NADH consumption. The reaction mixture contained 164 μg of enzyme GST-VhNfsA with 30 μM chromate (▪) or without chromate (□) in 1 ml of 10 mM Tris-HCl (pH 7.0), and NADH (final concentration 100 μM) was then added. The absorbance change was determined at 340 nm after every 1-min interval. The amount of reduced chromate in the reaction buffer containing 30 μM chromate, 100 μM NADH, and GST-VhNfsA was determined by the dye method described in the text at 5-min intervals (•).
DISCUSSION
We selected E. coli and V. harveyi nitroreductases and determined their chromate reductase activities, since P. ambigua chromate reductase was homologous with them. We did find that the nitroreductases are effective in chromate reduction. To our knowledge, we are the first to recognize that nitroreductase can also act as a chromate reductase.
The chromate-reducing activity of GST-VhNfsA was 12 times as efficient as that of GST-EcNfsB, and that of GST-EcNfsA is twice as efficient as that of GST-EcNfsB (Table 1), suggesting that the nfsA(GST-VhNfsA, GST-EcNfsA) family is more efficient than nfsB(GST-EcNfsB) as a chromate reductase.
GST-EcNfsA, GST-EcNfsB, and GST-VhNfsA showed optimum activities at physiological temperatures (20 to 40°C) (Fig. 4). Most of the enzymes were inactive at 4°C. However, GST-EcNfsB still showed 60% of its maximum activity at 4°C (Fig. 4). This property might be exploited when the enzyme is used under in situ bioremediation conditions even in winter.
P. putida chromate reductase has a high Vmax value of 1.72 μmol/min/mg of protein (24). However, its Km value of 374 μM (24) is 70 times higher than that of GST-VhNfsA. Overall, the chromate reductase efficiency of P. putida is similar to that of GST-VhNfsA. GST-VhNfsA activity was optimal at 30°C, whereas P. putida chromate reductase efficiency is optimal at 80°C. In actual bioremediation, GST-VhNfsA activity is expected to be more useful than that of P. putida chromate reductase. Puzon et al. (27) reported on a bacterial flavin reductase, Fre, that reduces chromate. It exhibits a very high Vmax value. Since these authors did not report its Km value, GST-VhNfsA cannot be compared to it. Fre did not contain a bound flavin and needed free FMN for its chromate reduction (27), whereas GST-VhNfsA contained a bound FMN.
Cr(III) is assumed to precipitate as Cr(OH)3 or Cr2O3 in neutral solution (27). Chromate remediation has been based on the fact that Cr(III) is less soluble and less toxic. However, we found that the Cr(III) produced by GST-VhNfsA was soluble. Puzon et al. (27) reported that Cr(III)-NAD+ complex produced by Fre enzyme was also soluble. The Cr(III) produced by GST-VhNfsA might have the similar structure. The fact that the Cr(III) produced by enzyme is mobile was disappointing at first. However, the Cr(III) produced by GST-VhNfsA was much less toxic to E. coli, and this is a favorable aspect in chromate remediation.
Some reductase in the cell might have diverse functions, including a chromate-reducing activity. It was demonstrated here that some E. coli and V. harveyi nitroreductases are also efficient in reducing chromate. The reductase or operons for copper, arsenite, and mercury are known (31). However, the operon for chromate is not yet known.
REFERENCES
- 1.Banks, R. B., and R. T. Cooke, Jr. 1986. Chromate reduction by rabbit liver aldehyde oxidase. Biochem. Biophys. Res. Commun. 137:8-14. [DOI] [PubMed] [Google Scholar]
- 2.Bhide, J. V., P. K. Dhakephallker, and K. M. Paknikar. 1996. Microbiological process for the removal of Cr(VI) from chromate-bearing cooling tower effluent. Biotechnol. Lett. 18:667-672. [Google Scholar]
- 3.Bryant, D. W., D. R. McCalla, M. Leeksma, and P. Laneuville. 1981. Type I nitroreductases of Escherichia coli. Can. J. Microbiol. 27:81-86. [DOI] [PubMed] [Google Scholar]
- 4.Campos, J., M. Martinez-Pacheco, and C. Cervantes. 1995. Hexavalent-chromium reduction by a chromate-resistant Bacillus sp. strain. Antonie Leeuwenhoek 68:203-208. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes, C., and S. Silver. 1992. Plasmid chromate resistance and chromate reduction. Plasmid 27:65-71. [DOI] [PubMed] [Google Scholar]
- 6.Daune, W., and J. W. Hastings. 1975. Flavin mononucleotide reductase of luminous bacteria. Mol. Cell. Biochem. 6:53-64. [DOI] [PubMed] [Google Scholar]
- 7.De Flora, S., A. Morelli, C. Basso, M. Romano, D. Serra, and A. De Flora. 1985. Prominent role of DT-diaphorase as a cellular mechanism reducing chromium(VI) and reverting its mutagenicity. Cancer Res. 45:3188-3196. [PubMed] [Google Scholar]
- 8.DeLeo, P. C., and H. L. Ehrlich. 1994. Reduction of hexavalent chromium by Pseudomonas fluorescens Lb300 in batch and continuous cultures. Appl. Microbiol. Biotechnol. 40:756-759. [Google Scholar]
- 9.Gadd, G. M., and C. White. 1993. Microbial treatment of metal pollution: a working biotechnology. Trends Biotechnol. 11:353-359. [DOI] [PubMed] [Google Scholar]
- 10.Gopalan, R., and H. Veeramani. 1994. Studies on microbial chromate reduction by Pseudomonas sp. in aerobic continuous suspended growth cultures. Biotechnol. Bioeng. 43:471-476. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, M., H. Ohzeki, H. Shimada, and T. Unemoto. 1996. NADH-specific quinone reductase is induced by 2-methylene-4-butylactone in Escherichia coli. Biochim. Biophys. Acta 1273:165-170. [DOI] [PubMed]
- 12.Imai, A., and E. F. Gloyna. 1990. Effect and oxidation state of chromium on the behavior of chromium in the activated sludge process. Water Chem. 24:1143-1150. [Google Scholar]
- 13.Ishibashi, Y., C. Cervantes, and S. Silver. 1990. Chromium reduction in Pseudomonas putida. Appl. Environ. Microbiol. 56:2268-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jablonski, E., and M. DeLuca. 1977. Purification and properties of the NADH and NADPH specific FMN oxidoreductases from Beneckea harveyi. Biochemistry 16:2932-2936. [DOI] [PubMed] [Google Scholar]
- 15.Kitts, C. L., C. E. Green, R. A. Otley, M. A. Alvarez, and P. J. Unkefer. 2000. Type I nitroreductases in soil enterobacteria reduce TNT (2,4,6-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can. J. Microbiol. 46:278-282. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lei, B., M. Liu, S. Huang, and S. C. Tu. 1994. Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing and overexpression of the gene and purification and characterization of the cloned enzyme. J. Bacteriol. 176:3552-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llovera, S., R. Bonet, M. D. Simon-Pujol, and F. Congregado. 1993. Chromate reduction by resting cells of Agrobacterium radiobacter EPS-916. Appl. Environ. Microbiol. 59:3516-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCalla, D. R., A. Reuvers, and C. Kaiser. 1970. Mode of action of nitrofurazone. J. Bacteriol. 104:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael, N. P., J. K. Brehm, G. M. Anlezark, and N. P. Minton. 1994. Physical characterization of the Escherichia coli B gene encoding nitroreductase and its overexpression in Escherichia coli K-12. FEMS Microbiol. Lett. 124:195-202. [DOI] [PubMed] [Google Scholar]
- 21.Mikalsen, A., J. Alexander, H. Wallin, M. Ingelman-Sundberg, and R. A. Andersen. 1991. Reductive metabolism and protein binding of chromium(VI) by P450 protein enzymes. Carcinogenesis 12:825-831. [DOI] [PubMed] [Google Scholar]
- 22.Oh, B. T., G. Sarath, P. J. Shea, R. A. Drijber, and S. D. Comfort. 2000. Rapid spectrophotometric determination of 2,4,6-trinitrotoluene in a Pseudomonas enzyme assay. J. Microbiol. Methods 42:149-158. [DOI] [PubMed] [Google Scholar]
- 23.Oh, Y. S., and S. C. Choi. 1997. A comparison between bacterial Cr(VI) resistance and Cr(VI) reduction among environmental isolates. Korean J. Microbiol. 33:125-130. [Google Scholar]
- 24.Park, C. H., M. Keyhan, B. Wielinga, S. Fendorf, and A. Matin. 2000. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 66:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peitzsch, N., G. Eberz, and D. H. Nies. 1998. Alcaligenes eutrophus as a bacterial chromate sensor. Appl. Environ. Microbiol. 64:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson, F. J., R. P. Mason, J. Hovsepian, and J. L. Holtzman. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009-4014. [PubMed] [Google Scholar]
- 27.Puzon, G. J., J. N. Petersen, A. G. Roberts, D. M. Kramer, and L. Xun. 2002. A bacterial flavin reductase system reduces chromate to a soluble chromium(III)-NAD+ complex. Biochem. Biophys. Res. Commun. 294:76-81. [DOI] [PubMed] [Google Scholar]
- 28.Read, S. M., and D. H. Northcote. 1981. Minimization of variation in the reponse to different proteins of the Coomassie blue G dye-binding assay for protein. Anal. Biochem. 116:53-64. [DOI] [PubMed] [Google Scholar]
- 29.Riley, R. G., J. M. Zachara, and F. J. Wobber. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. Report DOE/ER-0547T. U.S. Department of Energy, Washington, D.C.
- 30.Shen, H., and Y. T. Wang. 1994. Modeling hexavalent chromium reduction in Escherichia coli 33456. Biotechnol. Bioeng. 43:293-300. [DOI] [PubMed] [Google Scholar]
- 31.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 32.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, Y. Tai, and M. Okazaki. 1992. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J. Bacteriol. 174:5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, P. C., T. Mori, K. Komori, M. Sasatsu, K. Toda, and H. Ohtake. 1989. Isolation and characterization of an Enterobacter cloacae strain that reduces hexavalent chromium under anaerobic conditions. Appl. Environ. Microbiol. 55:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, P. C., T. Mori, K. Toda, and H. Ohtake. 1990. Membrane-associated chromate reductase activity from Enterobacter cloacae. J. Bacteriol. 172:1670-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zenno, S., H. Koike, A. N. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroredctase from Escherichia coli, similar in biochemical properties to Frase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736-744. [DOI] [PubMed] [Google Scholar]
- 39.Zenno, S., K. Saigo, H. Kanoh, and S. Inouye. 1994. Identification of gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J. Bacteriol. 176:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]