Abstract
The thermostability of maltogenic amylase from Thermus sp. strain IM6501 (ThMA) was improved greatly by random mutagenesis using DNA shuffling. Four rounds of DNA shuffling and subsequent recombination of the mutations produced the highly thermostable mutant enzyme ThMA-DM, which had a total of seven individual mutations. The seven amino acid substitutions in ThMA-DM were identified as R26Q, S169N, I333V, M375T, A398V, Q411L, and P453L. The optimal reaction temperature of the recombinant enzyme was 75°C, which was 15°C higher than that of wild-type ThMA, and the melting temperature, as determined by differential scanning calorimetry, was increased by 10.9°C. The half-life of ThMA-DM was 172 min at 80°C, a temperature at which wild-type ThMA was completely inactivated in less than 1 min. Six mutations that were generated during the evolutionary process did not significantly affect the specific activity of the enzyme, while the M375T mutation decreased activity to 23% of the wild-type level. The molecular interactions of the seven mutant residues that contributed to the increased thermostability of the mutant enzyme with other adjacent residues were examined by comparing the modeled tertiary structure of ThMA-DM with those of wild-type ThMA and related enzymes. The A398V and Q411L substitutions appeared to stabilize the enzyme by enhancing the interdomain hydrophobic interactions. The R26Q and P453L substitutions led potentially to the formation of genuine hydrogen bonds. M375T, which was located near the active site of ThMA, probably caused a conformational or dynamic change that enhanced thermostability but reduced the specific activity of the enzyme.
Thermostability is one of the most important properties of industrial enzymes, especially those that are used in the starch industry, such as maltogenic amylases (MAases). Investigations into the physical basis for the stability of proteins at high temperatures have identified several common features of these proteins (22, 31, 42). Crystallographic evidence suggests that hydrogen bonding and hydrophobic or ionic interactions between amino acids have profound effects on protein thermostability. Experimental evidence that supports the involvement of these interactions in thermophilic proteins has also been reported (9, 40, 44). Although none of the mechanisms proposed to date are applicable to all known thermostable enzymes, they provide insights into how the question of protein thermostability may be addressed.
A series of attempts to enhance enzyme thermostability by using site-directed mutagenesis, which were based on proposed thermostabilization mechanisms and tertiary-structure information, have been made (10, 13, 34, 36, 38). Although this method has been highly useful in understanding the structure-function relationships of enzymes, rational mutagenesis requires not only detailed information on the tertiary structures and catalytic properties of the enzymes but also the ability to predict the proper site of substitution and intuition concerning the optimal amino acid to be substituted. In cases where information on the tertiary structure is lacking, proteins can still be manipulated to have enhanced properties by random mutation of the entire gene. Among the random mutagenesis methods, DNA shuffling is a very powerful technique for obtaining mutations via random in vitro recombination during PCR (8, 39). The half-life of subtilisin S41 at 60°C was increased 1,200-fold as the result of eight successive rounds of mutagenesis and recombination, and the melting temperature of the mutant, as measured by circular dichroism, increased by 25°C relative to that of the wild-type protein (43). The half-life of β-galactosidase from Paenibacillus polymyxa at 55°C was increased 20-fold by using error-prone PCR and DNA shuffling (2).
The MAase enzyme (EC 3.2.1.133) constitutes a subfamily of amylolytic enzymes together with cyclomaltodextrinase (EC 3.2.1.54), neopullulanase (EC 3.2.1.135), and Thermoactinomyces vulgaris amylase II (33). The members of this subfamily share the characteristics of being able to hydrolyze multiple substrates, which include starch, pullulan, and cyclodextrins, and to simultaneously transfer the hydrolyzed sugar moiety to another sugar molecule, which makes them useful for the preparation of branched oligosaccharide mixtures (25) and novel carbohydrates (20, 32). Crystallographic analysis of enzymes in this subfamily revealed that they have the (α/β)8 barrel and C domain that are common to amylolytic enzymes, as well as an extra 124-residue N domain, which is involved in domain-swapped homodimer formation (15, 16, 18, 24). The optimal temperatures for MAases and related enzymes have been reported as 40 to 60°C (6, 7, 17, 19, 23, 41). The development of thermostable MAases for industrial applications has been facilitated by molecular cloning of the MAase gene from thermophiles. Currently, the most thermostable MAase known is the MAase from Thermus sp. strain IM6501 (ThMA), which has an optimal temperature of 60°C (19). Recently, two related enzymes from hyperthermophiles were reported: a cyclodextrinase from a Thermococcus sp. with an optimal temperature of 95°C (14) and a cyclodextrinase-like glucosidase from Thermotoga maritima (26). The latter enzyme exhibited its highest activity at 85°C and showed a preference for cyclodextrins over starch. However, the activity of this enzyme was very low, and its catalytic properties were somewhat different from those of a typical MAase. The transfer activities of these two enzymes were not reported.
In this study, we generated thermostable MAases from ThMA by random mutagenesis and investigated the factors that were associated with the thermostability of this enzyme. After four rounds of random mutagenesis and screening, 10 mutation sites were identified in ThMA, 6 of which contributed to enhanced thermostability and 1 of which increased the catalytic activity of the enzyme. The changes in the thermostability and catalytic properties of the selected mutants during the evolutionary process were characterized in detail. We developed a hypothesis for the enhanced thermostability contributed by each mutation site based on predictions of the tertiary structures of the mutant enzymes, which in turn were modeled on the known wild-type and related enzyme structures.
MATERIALS AND METHODS
Bacterial strains and reagents.
Escherichia coli MC1061 [F− araD139 recA13 Δ(araABC-leu)7696 galU galK ΔlacX74 rpsL thi hsdR2 mcrB] was used as the host for gene manipulation and for expression of wild-type ThMA and its derivatives. The E. coli transformants were cultured at 37°C in Luria-Bertani (LB) broth (1% [wt/vol] Bacto Tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) that contained either 100 μg of ampicillin/ml or 20 μg of kanamycin/ml. Mutants with elevated thermostability were screened on LB agar plates that contained 1% (wt/vol) soluble starch and 20 μg of kanamycin/ml. All the chemicals used in this study were of reagent grade.
Random mutagenesis and mutant library construction.
DNA shuffling for random mutagenesis was carried out according to the method of Zhao and Arnold (45). The 1.8-kb XbaI and HindIII fragments that carried the structural genes of the wild-type or mutant ThMA were isolated and digested with DNase I in reaction buffer (50 mM Tris-HCl [pH 7.5]-10 mM MnCl2) at 15°C for 5 min. DNA fragments of <300 bp were purified from a 2% (wt/vol) agarose gel and subjected to PCR for self-assembly by using Taq polymerase (Takara Bio Inc., Shiga, Japan). The thermocycling reaction consisted of an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, with a final extension step at 72°C for 7 min. To isolate the products of full-length self-assembly, 2 μl of the self-assembled DNA mixture was amplified using 1.0 mM (each) ThMAXba, the 5′-flanking primer (5′-CCTAGTCTAGAAAAGAAGCCATCCACCACCGCTCA-3′), and ThMASFLC, the 3′-flanking primer (5′-GCCGATATGATAAGCTTGTCCGTAACCGCCCG-3′), which contain an XbaI and a HindIII site (underlined), respectively. PCR was conducted as described above. The shuffled products were purified by using a High Pure PCR Product Purification Kit (Roche Diagnostics GmbH, Mannheim, Germany) and were digested with XbaI and HindIII. The digested DNA fragments were then ligated into p6×HTKXb119 at the corresponding sites and transformed into E. coli. The p6×HTKXb119 expression vector carried the Bacillus licheniformis MAase promoter (17), the hexahistidine tag for easy purification of recombinant proteins, the kanamycin resistance gene from pET29-b(+) (Novagen Inc., Madison, Wis.), and multiple cloning sites. The p6×HTKXb119 vector was used for expression of the N-terminally six-His tagged wild-type and mutant ThMAs in this study.
Screening of mutants with enhanced thermostability.
To screen mutants with enhanced thermostability, transformants (1,000 to 1,500 per DNA shuffling round) were inoculated onto LB agar plates that contained 20 μg of kanamycin/ml and were incubated at 37°C for 10 h. The cells were then transferred to Hybond-N nylon membranes (Amersham Pharmacia Biotech, Uppsala, Sweden), lysed, and fixed according to the method described by Song and Rhee (37). The membranes were incubated under the appropriate conditions for each DNA shuffling round (screening conditions for each round are described in Results). The heat-treated membranes were placed on fresh LB plates that contained 1% (wt/vol) soluble starch (Showa Manufacturing Co., Fukuoka, Japan) and incubated at 60°C for 12 h. Finally, mutants that showed larger clear zones than wild-type ThMA when stained with an iodine solution (I2, 0.203 g; KI, 5.2 g; H2O added to 100 ml) were selected. The mutations were verified by DNA sequencing using the BigDye Terminator Cycle Sequencing kit and the ABI 377 Prism DNA sequencer (Perkin-Elmer, Boston, Mass.).
Enzyme purification and activity assays.
Wild-type ThMA and derived mutant proteins were purified from E. coli strains that harbored the corresponding genes on p6×HTKXb119 by affinity chromatography of the lysates on a nickel-nitrilotriacetic acid column (Qiagen Inc., Valencia, Calif.), as described previously (19). The purified enzymes were concentrated by ultrafiltration (Millipore Co., Bedford, Mass.) after dialysis against 50 mM Tris-HCl (pH 7.0) and were used for further investigation. The hydrolytic activities of the wild-type and mutant proteins were assayed at their optimal temperatures by using 0.5% (wt/vol) β-cyclodextrin (β-CD) in 50 mM sodium acetate (pH 6.0). The 3,5-dinitrosalicylic acid method was used to measure the reducing sugar from β-CD (Sigma Chemical Co., St. Louis, Mo.) by using maltose as the standard (28). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of maltose per min. Protein concentrations were calculated by using the extinction coefficient at 280 nm (ɛ280), which was determined according to the method of Pace et al. (30). For both the wild-type and mutant enzymes, ɛ280 was 128,200 M−1cm−1.
Analysis of the thermostability of mutant enzymes.
The thermostability of each mutant enzyme was analyzed by determining the half-life of thermal inactivation and the melting temperature (Tm). The half-lives of the purified enzymes, which were dissolved in 50 mM sodium acetate buffer (pH 6.0), were determined by incubation of the samples (0.4 mg/ml) in water baths at different temperatures (75, 78, 80, and 85°C), from which aliquots were taken at various time points and placed immediately in an icewater bath. The residual β-CD-hydrolyzing activities of the aliquots were measured at the appropriate optimal temperatures. The first-order rate constant, kd, of irreversible thermal denaturation was obtained from the slope of the plots of ln (residual activity) versus time, and the half-lives were calculated as ln2/kd. To determine the Tm of the mutant enzymes, differential scanning calorimetry (DSC) was performed with a DSC 120 (Seiko Instrument Inc., Chiba, Japan), which was equipped with a liquid nitrogen intracooler system. The wild-type and mutant ThMAs were concentrated to 50 mg/ml in 50 mM sodium acetate buffer (pH 6.0) by using a Microcon filter (Millipore Co.) and weighed directly in a DSC aluminum pan. After sealing, the pan was heated from 30 to 130°C at a heating rate of 1°C/min. A pan that contained 50 mM sodium acetate buffer (pH 6.0) was used as the reference.
Kinetic studies of wild-type and mutant ThMAs.
The kinetics of the wild-type and mutant ThMAs for β-CD were measured by mixing appropriate concentrations of the enzymes (0.1 ml) with various concentrations of the substrate (0.9 ml) in 50 mM sodium acetate buffer (pH 6.0). The hydrolysis reactions of the wild-type and mutant proteins were performed at 60°C. Aliquots of the reaction mixtures were taken every 30 s and combined immediately with an equal volume of 0.1 M NaOH to stop the reaction. The amount of reducing sugar formed during the reaction was determined by the copper-bicinchoninate method (11), and the kinetic parameters were determined by fitting a hyperbolic Michaelis-Menten curve with the SigmaPlot program (version 5.0; SPSS Inc., Chicago, Ill.). The substrate concentrations used in the determination of kinetic parameters were 86.6 to 434 μM.
Saturation mutagenesis.
ThMA residue Met375 was replaced with a random amino acid by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) in order to investigate the effect of amino acid changes on thermostability. Primers M375X-N (5′-CAGTTTGACGCCGTCNNNAACTACCCGTTGGCG-3′) and M375X-C (5′-CGCCAACGGGTAGTTNNNGACGGCGTCAAACTG-3′) were used. The mutated genes were transformed into E. coli, and active clones were screened by the iodine test after treatment of the colonies with d-cycloserine, as described previously (17). Integration of the mutation was confirmed by sequence analysis, as described above.
Modeling of the mutant ThMA.
The three-dimensional structure of mutant ThMA-DM was modeled by using the SWISS-MODEL (version 3.51) program at the ExPASy server (35). The structures of wild-type ThMA and of two related enzymes were used as modeling templates. The RCSB Protein Data Bank (4) entries for these proteins are 1SMA, 1JI2, and 1EA9. Visualization and analysis of the modeled structure were carried out by using the Swiss-PdbViewer (version 3.51) (35). The figures were created with MOLSCRIPT (version 2.1) (21).
RESULTS
Screening of ThMA mutants for increased thermostability.
ThMA, which is the most thermostable wild-type MAase known to date, has a half-life of 5.3 min at 75°C (19). To produce ThMA mutants with increased thermostability, mutant libraries were created by DNA shuffling using random <300-bp fragments from the ThMA gene. The screening strategy used in this study was based on the retention of starch-hydrolyzing activity during incubation at elevated temperatures. Upon heat treatment at 80°C or higher, the parental ThMA enzyme did not form a detectable clear zone on the starch plate when stained with iodine. Variants that showed increased halo formation were presumed to have higher thermostability than the parental enzyme. The thermostability of each halo-positive mutant was verified further by measuring the half-life of thermal inactivation of the purified enzyme. The DNA sequences of the mutants selected during the DNA shuffling process were determined in order to verify the association between the mutation introduced and the increase in thermal resistance.
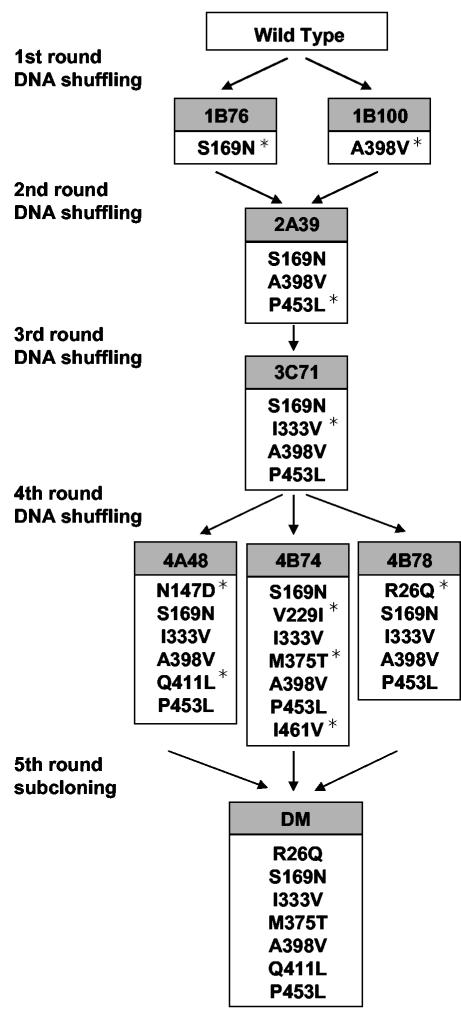
Approximately 1,500 clones were screened for increased thermostability after heat treatment at 80°C for 60 min. The positive clones 1B76 and 1B100, which were obtained in the first round of shuffling, had 1.25- and 1.55-fold-longer half-lives, respectively, than the wild-type enzyme at 75°C (Table 1). DNA sequence analysis revealed that 1B76 and 1B100 contained single amino acid substitutions (S169N and A398V, respectively) (Fig. 1). These two clones were used as parental clones in the second round of shuffling. More than 1,000 clones from the second-generation library were screened after heat treatment at 85°C for 60 min. Clone 2A39 exhibited the highest thermostability and had a half-life at 75°C that was about fivefold longer than that of the wild-type enzyme (Table 1). The second round of DNA shuffling introduced the additional mutation P453L into 2A39, along with the 1B76 and 1B100 mutations (Fig. 1). The third round of shuffling, which used 2A39 as the parental clone, produced 3C71 after heat treatment at 90°C for 30 min (Table 1). Clone 3C71 had accumulated an additional mutation, I333V (Fig. 1). The fourth round of shuffling used 3C71 as the parent, and 3 mutants were isolated from the 1,500 clones that were subjected to heat treatment at 95°C for 15 min. The selected mutants, 4A48, 4B74, and 4B78, showed half-lives of 18.3, 77.1, and 8.2 min, respectively, at 78°C, whereas the parental clone 3C71 had a half-life of only 3.6 min at this temperature (Table 1). The results of DNA sequence analysis showed that N147D and Q411L in 4A48, V229I, M375T, and I461V in 4B74, and R26Q in 4B78 were new additions to the existing mutations (Fig. 1).
TABLE 1.
Thermostability of wild-type ThMA and the evolved ThMA mutantsa
| Generation | Variantb | Tmc (°C) |
t1/2 (min)d at:
|
|||
|---|---|---|---|---|---|---|
| 75 °C | 78 °C | 80 °C | 85 °C | |||
| 0 | WT | 76.2 ± 0.2 | 5.3 ± 0.6 | — | ||
| 1 | 1B76 | 76.2 ± 0.2 | 6.6 ± 0.3 | — | ||
| 1B100 | 77.2 ± 0.5 | 8.2 ± 0.4 | — | |||
| 2 | 2A39 | 78.7 ± 0.3 | 24.6 ± 1.3 | — | ||
| 3 | 3C71 | 79.1 ± 0.1 | 60.2 ± 3.7 | 3.6 ± 0.2 | — | |
| 4 | 4A48 | 81.2 ± 0.1 | 149.7 ± 5.6 | 18.3 ± 0.9 | 5.2 ± 0.2 | — |
| 4B74 | 83.1 ± 0.1 | 263.8 ± 9.9 | 77.1 ± 6.5 | 42.4 ± 2.4 | — | |
| 4B78 | 79.4 ± 0.3 | 95.1 ± 7.4 | 8.2 ± 0.6 | 1.0 ± 0.1 | — | |
| 5 | DM | 86.9 ± 0.1 | ND | ND | 172.0 ± 5.9 | 10.7 ± 0.5 |
Values in this table are averages based on values from triplicate experiments.
WT, wild type.
Measured by DSC.
t1/2, half-life; —, less than 1 min; ND, not determined.
FIG. 1.
Lineage of ThMA variants. Amino acid substitutions accumulated by five generations of mutants are shown. Newly introduced mutations in each generation are marked with asterisks.
To select the mutations responsible for thermostability among those accumulated in 4A48 or 4B74, eight chimeric genes were constructed by general recombination using restriction enzymes. The thermostabilities of the purified chimeric enzymes were investigated by determining their half-lives at 78°C and their Tms by using DSC (Table 2). Three chimeric mutants that were generated from 4B74 (ThMA-RII, -RIV, and -RVI) and one that was generated from 4A48 (ThMA-RVIII) showed higher thermostabilities than the other chimeric mutants. In the group of 4B74 derivatives, ThMA-RII, which had only one mutation (M375T), exhibited the highest thermostability. The 4A48 derivative ThMA-RVIII, with the Q411L mutation, had a longer half-life at 78°C than the parental clone. The V229I, I461V, or N147D mutation alone (ThMA-RI, -RIII, or -RVII) caused a significant decrease in the half-life at 78°C and a reduction in the Tm, which indicates that these changes are detrimental to thermostability. Consequently, the results suggest that the M375T and Q411L mutations enhance the thermostability of ThMA.
TABLE 2.
Specific activities and thermostabilities of the chimeric mutants for selection of positive mutations in 4B74 and 4A48a
| Variant | Mutations | Sp actb (102 U/mg) | Tm (°C) | t1/2c at 78 °C (min) |
|---|---|---|---|---|
| 3C71 | S169N, A398V, P453L | 2.1 (±0.1) | 79.1 ± 0.1 | 3.6 ± 0.2 |
| 4B74 | S169N, A398V, P453L, V229I, M375T, I471V | 0.3 (±0.0) | 83.1 ± 0.1 | 77.1 ± 6.5 |
| RI | S169N, A398V, P453I, V229I | 2.1 (±0.1) | 77.6 ± 0.3 | — |
| RII | S169N, A398V, P453L, M375T | 0.3 (±0.0) | 83.9 ± 0.1 | 78.4 ± 6.5 |
| RIII | S169N, A398V, P453L, I471V | 2.2 (±0.1) | 77.7 ± 0.1 | — |
| RIV | S169N, A398V, P453L, V229I, M375T | 0.3 (±0.0) | 83.3 ± 0.3 | 62.6 ± 0.7 |
| RV | S169N, A398V, P453L, V229I, I471V | 2.1 (±0.1) | 77.6 ± 0.2 | — |
| RVI | S169N, A398V, P453L, M375T, I471V | 0.3 (±0.0) | 83.4 ± 0.1 | 52.4 ± 0.4 |
| 4A48 | S169N, A398V, P453L, N147D, Q411L | 2.0 (±0.1) | 81.2 ± 0.1 | 18.3 ± 0.9 |
| RVII | S169N, A398V, P453L, N147D | 2.0 (±0.1) | 78.3 ± 0.1 | — |
| RVIII | S169N, A398V, P453L, Q411L | 2.2 (±0.1) | 81.5 ± 0.3 | 23.8 ± 5.1 |
Values in this table are averages based on values from triplicate experiments.
Activities were determined in 50 mM sodium acetate buffer (pH 6.0) at 70 °C.
t1/2, half-life; —, less than 1 min.
Based on these findings, a derivative of ThMA (ThMA-DM) that contained the seven mutations R26Q, S169N, I333V, M357T, A398V, Q411L, and P453L was constructed via several subcloning steps. ThMA-DM showed the highest thermostability among all the mutants screened, with half-lives of 172 and 10.7 min at 80 and 85°C, respectively (Table 1).
Tms of the evolved ThMA mutants.
The Tms of ThMA-DM and the mutants that were obtained in each round of DNA shuffling were determined by DSC. With the exception of 1B76, all the evolved enzymes had Tms that were 0.3 to 4°C higher than that of the parent enzyme (Table 1). Among the mutants obtained throughout the evolution process, 4B74 exhibited the greatest increase in thermostability, with Tms that were 4.0 and 6.9°C higher than those of its parent clone (3C71) and wild-type ThMA, respectively. Finally, the Tm of ThMA-DM was 86.9°C, which was 10.7°C higher than that of wild-type ThMA.
Catalytic properties of the evolved ThMA mutants.
All the evolved mutants showed higher optimal temperatures than wild-type ThMA (65 to 75°C). The optimal temperature of ThMA-DM was 75°C, which is the highest value reported to date for MAases. Although the introduction of either S169N or A398V raised the specific activities of the evolved mutants to levels 1.3- to 1.4-fold higher than that of wild-type ThMA at 60°C, the introduction of additional mutations up to the third generation did not improve this level of activity (Table 3). One of the clones obtained in the fourth generation, 4A48, also maintained higher activity than the wild type. However, the specific activities of 4B74 and AB78 dropped to only 20 and 67% of the wild-type activity, respectively. The kinetic parameters of 4B74 suggested that the reduction in 4B74 activity was mostly affected by a decrease in turnover number (Table 3). Interestingly, the Km was also decreased for the 4B74 mutant (90 μM), although most of mutations did not cause any significant difference in Km (160 to 190 μM). The 4B74 mutant showed a Km that was 56% that of the wild-type and parental (3C71) enzymes. Chimeric enzymes based on 4B74 and the wild-type enzyme revealed that the reduction in specific activity was caused by the M375T mutation, which by itself contributed to the thermostabilization of the enzyme (Table 2). Therefore, the kinetic parameters of 4B74 suggest that the M375T mutation may lead to change around the ThMA active site and result in an increase in substrate affinity as well as a reduction in turnover number. ThMA-DM also showed 13% of wild-type activity, presumably due to the M375T mutation. Although the kcat of ThMA-DM was similar to that of 4B74, the catalytic efficiency (kcat/Km) of ThMA-DM was higher than that of 4B74 due to the decrease in Km.
TABLE 3.
Specific activities and kinetic parameters of wild-type ThMA and the evolved ThMA variantsa
| Generation | Variantb | Sp actc (102 U/mg) | kcatc (102 s−1) | Kmc (102 μM) | kcat/Km (s−1 μM−1) |
|---|---|---|---|---|---|
| 0 | WT | 1.5 (±0.0) | 1.6 (±0.0) | 1.6 (±0.1) | 1.0 |
| 1 | 1B76 | 2.1 (±0.1) | 2.0 (±0.1) | 1.7 (±0.2) | 1.2 |
| 1B100 | 2.0 (±0.0) | 1.9 (±0.1) | 1.7 (±0.3) | 1.1 | |
| 2 | 2A39 | 1.7 (±0.0) | 1.7 (±0.1) | 1.9 (±0.2) | 0.9 |
| 3 | 3C71 | 1.7 (±0.1) | 1.2 (±0.1) | 1.6 (±0.3) | 0.8 |
| 4 | 4A48 | 1.7 (±0.1) | 1.3 (±0.1) | 1.6 (±0.2) | 0.8 |
| 4B74 | 0.3 (±0.1) | 0.2 (±0.0) | 0.9 (±0.1) | 0.2 | |
| 4B78 | 1.0 (±0.1) | 1.1 (±0.1) | 1.6 (±0.2) | 0.7 | |
| 5 | DM | 0.2 (±0.0) | 0.2 (±0.0) | 0.5 (±0.0) | 0.4 |
Values in this table are averages based on values from triplicate experiments.
WT, wild type.
Activities and kinetic parameters were determined in 50 mM sodium acetate buffer (pH 6.0) at 60 °C.
Predicting thermostabilization mechanisms.
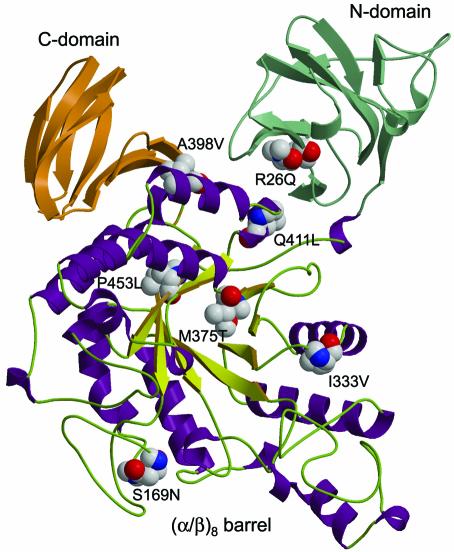
We produced homology-based models of ThMA-DM by using the SWISS-MODEL program, in an effort to understand the effect of each mutation on the evolution of ThMA toward increased thermostability (35). The models were constructed with reference to the known structure of wild-type ThMA (1SMA) (18) as well as to those of cyclomaltodextrinase I-5 from alkalophilic Bacillus sp. strain I-5 (1EA9) (24) and T. vulgaris α-amylase II (1JI2) (16), whose amino acid sequences are 56 and 46% identical, respectively, to that of ThMA. Figure 2 shows a structural model of the ThMA-DM monomer, which was based on ThMA and displays the seven mutations that were introduced into the enzyme.
FIG. 2.
Overall structural model of a ThMA-DM monomer, showing secondary-structure elements and the positions of seven stabilizing mutations. Green, N domain; orange, C domain. In the (α/β)8 barrel, chartreuse arrows stand for β-strands, violet spirals represent α-helices, and loops are depicted as light green. This model is displayed using MOLSCRIPT.
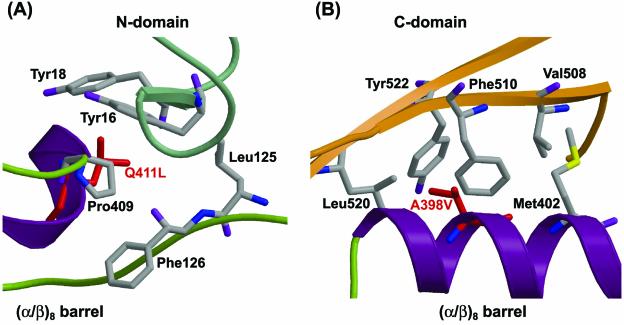
The modeled structure of ThMA-DM shows that A398V is located at the interface between the (α/β)8 barrel and the C domain and that Q411L is located at the interface between the (α/β)8 barrel and the N domain (Fig. 2). Q411L is surrounded by a hydrophobic core consisting of Tyr16, Tyr18, Leu125, Phe126, and Pro409 (Fig. 3A). Replacement of the hydrophilic Gln411 with a hydrophobic leucine in ThMA-DM might stabilize the hydrophobic interaction with the hydrophobic core, which is organized by the amino acids in the (α/β)8 barrel and the N domain of ThMA. Ala398 constitutes the hydrophobic core with Met402, Val508, Tyr522, and Leu520, and faces toward Phe510, located in the center. This hydrophobic interaction between the hydrophobic core and Phe510 also represents an interdomain interaction between the (α/β)8 barrel and the C domain. Therefore, replacement with a more hydrophobic amino acid, i.e., valine (A398V), might enhance the hydrophobic interaction (Fig. 3B). Consequently, the hydrophobic interactions involving A398V and Q411L might participate in stabilizing the interaction between the (α/β)8 barrel and the N and C domains, and the improved interdomain interactions might stabilize the overall structure of the ThMA mutant. Our results confirm that tightening of hydrophobic interactions is one of the most powerful strategies for protein stabilization.
FIG. 3.
Predicted hydrophobic interactions in ThMA-DM with the Q411L (A) and A398V (B) mutations. This is a representation of secondary-structure elements as depicted in Fig. 2. Red indicates mutated residues. The amino acids participating in the hydrophobic core are also represented.
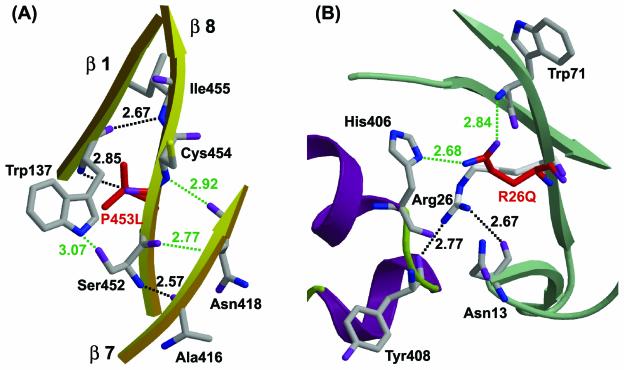
The three-dimensional structure of the wild-type enzyme shows that the first, seventh, and eighth β-strands of ThMA interact via two H bonds between Trp137 (β-strand 1) and Pro453 (β-strand 8) or Ile455 (β-strand 8) and via one H bond between Ser452 (β-strand 8) and Ala416 (β-strand 7) (Fig. 4A). The modeled structure of ThMA-DM shows distortion of the backbone around P453L and three possible hydrogen bonds. The carboxyl and hydroxyl groups of Ser452 (β-strand 8) probably form hydrogen bonds with the amino group of Asn418 (β-strand 7) and the indole group of Trp137 (β-strand 1), respectively. In addition, the amino group of Cys454 (β-strand 8) may be able to form hydrogen bonds with the carboxyl group of Asn418 (β-strand 7). Therefore, thermostabilization by P453L is due to an increase in H bonding between adjacent β-strands, which stabilizes the (α/β)8 barrel around the P453L mutation.
FIG. 4.
Predicted hydrogen bonds in ThMA-DM with the P453L (A) and R26Q (B) mutations. This is a representation of secondary-structure elements as depicted in Fig. 2. Red indicates mutated residues. Green dotted lines, newly generated H bonds; black dotted lines, H bonds in the wild-type enzyme. Distances are given along the dotted lines.
In the case of R26Q, the two hydrogen bonds between Arg26 and Asn13 or Tyr408 disappeared, but the amide and carbonyl groups at the side chain of R26Q formed new hydrogen bonds with the amino group of Trp71 and the imidazole group of His406, respectively, based on the modeled structure (Fig. 4B). No other newly generated interactions were found. The H bond between R26Q and His406 in ThMA-DM stabilized the interaction between the N domain and the (α/β)8 barrel, similarly to the H bond between Arg26 and Tyr408 in the wild-type enzyme. However, the type of H bond changed from a main-chain/side chain hydrogen bond (Arg26 and Tyr408 in the wild type) to side chain/side chain hydrogen bonds (R26Q and His406) in ThMA-DM. The stabilization afforded by R26Q suggests that H-bond formation by R26Q in ThMA-DM may be more stable than that provided by Arg26 in the wild-type enzyme.
The M375T mutation was located in the middle of the barrel at the C-terminal of β-strand 6, in close proximity to the catalytic center of the enzyme. Although a few side chain distortions were found, it is not clear why the M375T mutation simultaneously causes dramatic increases in thermostability and decreases in catalytic activity. The modeled structure did not show any residues that interacted directly with threonine at position 375 in ThMA-DM. One possible explanation is that the absence of the bulky side chain of methionine in the catalytic dyad may perturb the environment of these residues or result in interactions that increase thermostability but produce a less favorable catalytic geometry. To test this hypothesis, we attempted saturation mutagenesis of the Met375 of ThMA-3C71. Six active clones that had a Phe, Tyr, Lys, Val, Ser, or Thr substitution at Met375 were obtained. Investigations into the thermostabilities and activities of these enzymes and those of 3C71 revealed that mutation from Met to Val, Ser, or Thr improved thermostability but dramatically decreased enzymatic activity, as shown for 4B74 (Table 4). On the other hand, mutation to amino acids with bulky side chains, such as Phe, Tyr, or Lys, resulted in less thermostability but higher enzymatic activity than that observed with mutation to small amino acids. The M375T mutation, which was selected during the evolution process, gave the highest level of thermostabilization. This result indicates that bulky amino acids at position 375 in ThMA assist the overall folding of the active site of ThMA and ensure catalytic activity. Replacement of this residue with a relatively smaller one produces conformational changes around the active site of ThMA and results in enhanced thermostability but decreased catalytic activity.
TABLE 4.
Activities and thermostabilities of the mutants constructed by saturation mutagenesis at M375 in ThMA-3C71a
| Variant | Mutation at M375 | Sp actb (102 U/mg) | Tm (°C) |
|---|---|---|---|
| 3C71 | 2.1 (±0.1) | 79.1 ± 0.1 | |
| 3C71MK | K | 1.8 (±0.0) | 74.7 ± 0.3 |
| 3C71MYc | Y | 1.1 (±0.1) | 78.6 ± 0.1 |
| 3C71MF | F | 1.5 (±0.1) | 76.7 ± 0.2 |
| 3C71MV | V | 0.1 (±0.0) | 80.8 ± 0.1 |
| 3C71MS | S | 0.2 (±0.0) | 81.8 ± 0.1 |
| 3C71MT | T | 0.3 (±0.0) | 83.9 ± 0.1 |
Values in this table are averages based on values from triplicate experiments.
Determined in 50 mM sodium acetate buffer (pH 6.0) at 70 °C.
The ɛ280 of 3C71MY was 126,710 M−1 cm−1.
DISCUSSION
In this study, we attempted to develop a highly thermostable derivative of ThMA by random mutagenesis using DNA shuffling. This approach allowed us to select the most thermostable ThMA mutant, ThMA-DM, which contained seven point mutations (Fig. 1; Table 1). During four subsequent rounds of DNA shuffling, only seven different mutations that led to increased thermostability were identified in the ThMA mutant; these seven mutations accumulated in the final mutant (ThMA-DM), which had the highest level of thermostability. However, the dramatic increase in thermostability of ThMA-DM was achieved at the expense of decreases in kcat and catalytic efficiency (kcat/Km) due to the M375T mutation (Table 2). Interestingly, many substitutions of catalytic residues of AmpC β-lactamase decrease the enzymatic activity 103- to 105-fold as they increase in the stability of the enzyme significantly, by as much as 4.7 kcal/mol (3). This result suggests that enzymes can gain significant stability from replacement of active-site residues. The observation that the removing the bulky side chain of residue 375 leads to increased thermostability and decreased enzymatic activity can be interpreted by the stability-function tradeoff, although this residue itself is not a catalytic residue in this enzyme (Table 4).
The tightening of hydrophobic interactions in a protein is one of the most powerful strategies for protein stabilization (2, 10, 29). Replacement of amino acids with more-hydrophobic residues in a hydrophobic environment optimizes protein packing. Thermostabilization by replacement of the hydrophilic residues that are surrounded by the hydrophobic core with hydrophobic residues was reported for Thermus thermophilus isopropylmalate dehydrogenase (IPMDH) (27). Leu80 in T. thermophilus IPMDH is surrounded by hydrophobic residues, but the corresponding residue in the mesophilic IPMDH is Asn. The replacement of Asn with Leu in the mesophilic IPMDH produced a 2°C increase in Tm. In our study, the A398V and Q411L mutations showed the same effect. The hydrophobic interactions involving the A398V and Q411L mutations stabilized the overall structure of ThMA by promoting interdomain interactions between the (α/β)8 barrel and the N or C domain (Fig. 3).
Hydrogen bonding is another major mechanism for protein thermostabilization. The crystal structure of T. maritima ferredoxin showed that an increased number of H bonds enhanced stabilization (27). Furthermore, the thermal resistance of Flavobacterium meningosepticum glycerol kinase was increased by the introduction via site-directed mutagenesis of a hydrogen bond (36). Based on the modeled structure of ThMA-DM, the increased H bonds play an important role in stabilizing the protein. The P453L mutation generated three new H bonds, which stabilized the structure among β-strands in the (α/β)8 barrel (Fig. 4A). Some reports have suggested that the type of H bond is a more critical parameter for thermostabilization than the number of H bonds. The charged neutral hydrogen bond and side chain/side chain hydrogen bonds are more important than other types of H bonds in stabilizing proteins (22, 27, 42). In our study, the R26Q mutation, which increased the half-life 1.6-fold at 75°C, did not increase the number of H bonds but changed the type of H bond to a side chain/side chain hydrogen bond (Fig. 4B). Therefore, we expect that the change in H-bond type brought about by R26Q leads to the stabilization of ThMA-DM.
Deamination of the Asn and Gln residues represents a major pathway for protein degradation at high temperatures (12, 42). The Asn or Gln side chain amide group attacks the n + 1 peptide nitrogen and forms the succinimide intermediate, which breaks down to yield an α-linked (Asp or Glu) or β-linked (iso-Asp or iso-Glu) residue (42). According to this mechanism, Gly, Ser, and Ala are favored in n + 1 because their small side chains do not obstruct cyclization into the succinimide intermediate. The elimination of the heat-labile residue in the Q411L mutant may enhance the thermostability of ThMA as well as hydrophobic interactions. In the case of R26Q, which produced a new heat-labile residue, the R26Q side chain participated in the formation of the H bond. Therefore, the amide side chain of R26Q was not free to act as a nucleophile, and a large side chain of the Leu27 residue, which followed R26Q, also retarded the formation of the succinimide intermediate.
However, we found no evidence in the models for thermostabilization by S169N, I333V, and M375T. The elimination of chemically labile residues, such as Met or Cys, which undergo oxidation at high temperatures, is expected to render the enzyme more resistant to irreversible thermal denaturation (1, 2, 5). However, the inactivation rate of the wild-type enzyme in the presence of 10 mM dithiothreitol was not increased over that in the absence of dithiothreitol (data not shown). Therefore, thermostabilization via M375T does not occur for this reason, and we expect that the conformational change occurs as a result of the removal of the bulky methionine side chains and the generation of various stabilizing interactions. Valine is conserved among the other MAases and related enzymes at the site corresponding to Ile333 in ThMA (33). Therefore, mutation to the consensus residue (I333V) probably contributes more to stability than the nonconsensus isoleucine residue. In an attempt to understand the effects of mutations M375T and S169N on the catalytic properties, the model for binding of β-CD to the ThMA-DM mutant was examined. The S169N mutation, which contributed to increased enzymatic activity but not thermostability (Tables 1 and 2), was located on the surface of the overall structure, with the side chain of Asn directed outwards, and β-CD did not interact directly with S169N. Likewise, the binding of a substrate to the catalytic site was not affected by the M375T mutation (data not shown).
Acknowledgments
This study was supported by the Industrial Technology Development Program of the Ministry of Commerce, Industry, and Energy and also, in part, by the NRL program of the Korean Ministry of Science and Technology. We acknowledge the financial support of the Brain Korea 21 Project in the form of scholarships to Y.-W. Kim, J.-H. Choi, and J.-W. Kim.
REFERENCES
- 1.Akanuma, S., A. Yamagishi, N. Tanaka, and T. Oshima. 1998. Serial increase in the thermal stability of 3-isopropylmalate dehydrogenase from Bacillus subtilis by experimental evolution. Protein Sci. 7:698-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrizubieta, M. J., and J. Polaina. 2000. Increased thermal resistance and modification of the catalytic properties of a β-glucosidase by random mutagenesis and in vitro recombination. J. Biol. Chem. 275:28843-28848. [DOI] [PubMed] [Google Scholar]
- 3.Beadle, B. M., and B. K. Shoichet. 2002. Structural bases of stability-function tradeoffs in enzymes. J. Mol. Biol. 321:285-296. [DOI] [PubMed] [Google Scholar]
- 4.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchert, T. V., S. F. Lassen, A. Svendsen, and H. B. Frantzen. 1995. Oxidation stable amylases for detergents, p. 175-179. In B. Petersen, B. Svensson, and S. Pedersen (ed.), Carbohydrate bioengineering. Elsevier Science, Amsterdam, The Netherlands.
- 6.Cha, H. J., H. G. Yoon, Y. W. Kim, H. S. Lee, J. W. Kim, K. S. Kweon, B. H. Oh, and K. H. Park. 1998. Molecular and enzymatic characterization of novel maltogenic amylase that hydrolyzes and transglycosylates acarbose. Eur. J. Biochem. 253:251-262. [DOI] [PubMed] [Google Scholar]
- 7.Cho, H. Y., Y. W. Kim, T. J. Kim, J. W. Kim, and K. H. Park. 1999. Molecular characterization of a dimeric intracellular maltogenic amylase of Bacillus subtilis SUH4-2. Biochim. Biophys. Acta 1478:333-340. [DOI] [PubMed] [Google Scholar]
- 8.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 9.Dalhus, B., M. Saarinen, U. H. Sauer, P. Eklund, K. Johansson, A. Karlsson, S. Ramaswamy, A. Bjork, B. Synstad, K. Naterstad, R. Sirevag, and H. Eklund. 2002. Structural basis for thermophilic protein stability: structures of thermophilic and mesophilic malate dehydrogenases. J. Mol. Biol. 318:707-721. [DOI] [PubMed] [Google Scholar]
- 10.Declerck, N., M. Machius, R. Chambert, G. Wiegand, R. Huber, and C. Gaillardin. 1997. Hyperthermostable mutants of Bacillus licheniformis α-amylase: thermodynamic studies and structural interpretation. Protein Eng. 10:541-549. [DOI] [PubMed] [Google Scholar]
- 11.Fox, J. D., and J. F. Robyt. 1991. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195:93-96. [DOI] [PubMed] [Google Scholar]
- 12.Geiger, T., and S. Clarke. 1987. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 262:785-794. [PubMed] [Google Scholar]
- 13.Grinberg, A. V., and R. Bernhardt. 2001. Contribution of a salt bridge to the thermostability of adrenodoxin determined by site-directed mutagenesis. Arch. Biochem. Biophys. 396:25-34. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, Y., T. Yamamoto, S. Fujiwara, M. Takagi, and T. Imanaka. 2001. Extracellular synthesis, specific recognition, and intracellular degradation of cyclomaltodextrins by the hyperthermophilic archaeon Thermococcus sp. strain B1001. J. Bacteriol. 183:5050-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hondoh, H., T. Kuriki, and Y. Matsuura. 2003. Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J. Mol. Biol. 326:177-188. [DOI] [PubMed] [Google Scholar]
- 16.Kamitori, S., A. Abe, A. Ohtaki, A. Kaji, T. Tonozuka, and Y. Sakano. 2002. Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 α-amylase 1 (TVAI) at 1.6 Å resolution and α-amylase 2 (TVAII) at 2.3 Å resolution. J. Mol. Biol. 318:443-453. [DOI] [PubMed] [Google Scholar]
- 17.Kim, I. C., J. H. Cha, J. R. Kim, S. Y. Jang, B. C. Seo, T. K. Cheong, D. S. Lee, Y. D. Choi, and K. H. Park. 1992. Catalytic properties of the cloned amylase from Bacillus licheniformis. J. Biol. Chem. 267:22108-22114. [PubMed] [Google Scholar]
- 18.Kim, J. S., S. S. Cha, H. J. Kim, T. J. Kim, N. C. Ha, S. T. Oh, H. S. Cho, M. J. Cho, M. J. Kim, H. S. Lee, J. W. Kim, K. Y. Choi, K. H. Park, and B. H. Oh. 1999. Crystal structure of a maltogenic amylase provides insight into a catalytic versatility. J. Biol. Chem. 274:26279-26286. [DOI] [PubMed] [Google Scholar]
- 19.Kim, T. J., M. J. Kim, B. C. Kim, J. C. Kim, T. K. Cheong, J. W. Kim, and K. H. Park. 1999. Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl. Environ. Microbiol. 65:1644-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, Y. K., M. J. Kim, C. S. Park, and K. H. Park. 2002. Modification of sorbitol by transglycosylation using Bacillus stearothermophilus maltogenic amylase. Food Sci. Biotechnol. 11:401-406. [Google Scholar]
- 21.Kraulis, P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 22.Kumar, S., C. J. Tsai, and R. Nussinov. 2000. Factors enhancing protein thermostability. Protein Eng. 13:179-191. [DOI] [PubMed] [Google Scholar]
- 23.Kuriki, T., J. H. Park, S. Okada, and T. Imanaka. 1988. Purification and characterization of thermostable pullulanase from Bacillus stearothermophilus and molecular cloning and expression of the gene in Bacillus subtilis. Appl. Environ. Microbiol. 54:2881-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H. S., M. S. Kim, H. S. Cho, J. I. Kim, T. J. Kim, J. H. Choi, C. Park, H. S. Lee, B. H. Oh, and K. H. Park. 2002. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J. Biol. Chem. 277:21891-21897. [DOI] [PubMed] [Google Scholar]
- 25.Lee, H. S., J. H. Auh, H. G. Yoon, M. J. Kim, J. H. Park, S. S. Hong, M. H. Kang, T. J. Kim, T. W. Moon, J. W. Kim, and K. H. Park. 2002. Cooperative action of α-glucanotransferase and maltogenic amylase for an improved process of isomaltooligosaccharide (IMO) production. J. Agric. Food Chem. 50:2812-2817. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. H., Y. W. Kim, T. J. Kim, C. Park, J. W. Kim, T. W. Moon, and K. H. Park. 2002. A novel amylolytic enzyme from Thermotoga maritima with cyclodextrinase- and α-glucosidase-like activities, liberating glucose from the reducing end of the substrates. Biochem. Biophys. Res. Commun. 295:818-825. [DOI] [PubMed] [Google Scholar]
- 27.Macedo-Ribeiro, S., B. M. Martins, P. J. Pereira, G. Buse, R. Huber, and T. Soulimane. 2001. New insights into the thermostability of bacterial ferredoxins: high-resolution crystal structure of the seven-iron ferredoxin from Thermus thermophilus .J. Biol. Inorg. Chem. 6:663-674. [DOI] [PubMed] [Google Scholar]
- 28.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 29.Numata, K., Y. Hayashi-Iwasaki, J. Kawaguchi, M. Sakurai, H. Moriyama, N. Tanaka, and T. Oshima. 2001. Thermostabilization of a chimeric enzyme by residue substitutions: four amino acid residues in loop regions are responsible for the thermostability of Thermus thermophilus isopropylmalate dehydrogenase. Biochim. Biophys. Acta 1545:174-183. [DOI] [PubMed] [Google Scholar]
- 30.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panasik, N., J. E. Brenchley, and G. K. Farber. 2000. Distributions of structural features contributing to thermostability in mesophilic and thermophilic α/β barrel glycosyl hydrolases. Biochim. Biophys. Acta 1543:189-201. [DOI] [PubMed] [Google Scholar]
- 32.Park, K. H., M. J. Kim, H. S. Lee, N. S. Han, D. M. Kim, and J. F. Robyt. 1998. Transglycosylation reactions of Bacillus stearothermophilus maltogenic amylase with acarbose and various acceptors. Carbohydr. Res. 313:235-246. [DOI] [PubMed] [Google Scholar]
- 33.Park, K. H., T. J. Kim, T. K. Cheong, J. W. Kim, B. H. Oh, and B. Svensson. 2000. Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim. Biophys. Acta 1478:165-185. [DOI] [PubMed] [Google Scholar]
- 34.Pedone, E., M. Saviano, M. Rossi, and S. Bartolucci. 2001. A single point mutation (Glu85Arg) increases the stability of the thioredoxin from Escherichia coli .Protein Eng. 14:255-260. [DOI] [PubMed] [Google Scholar]
- 35.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 36.Sakasegawa, S., H. Takehara, I. Yoshioka, H. Misaki, H. Sakuraba, and T. Ohshima. 2002. Stabilization of Flavobacterium meningosepticum glycerol kinase by introduction of a hydrogen bond. Biosci. Biotechnol. Biochem. 66:1374-1377. [DOI] [PubMed] [Google Scholar]
- 37.Song, J. K., and J. S. Rhee. 2000. Simultaneous enhancement of thermostability and catalytic activity of phospholipase A1 by evolutionary molecular engineering. Appl. Environ. Microbiol. 66:890-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriprapundh, D., C. Vieille, and J. G. Zeikus. 2000. Molecular determinants of xylose isomerase thermal stability and activity: analysis of thermozymes by site-directed mutagenesis. Protein Eng. 13:259-265. [DOI] [PubMed] [Google Scholar]
- 39.Stemmer, W. P. C. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389-391. [DOI] [PubMed] [Google Scholar]
- 40.Sziligyi, A., and P. Zavodszky. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8:493-504. [DOI] [PubMed] [Google Scholar]
- 41.Tonozuka, T., M. Ohtsuka, S. Mogi, H. Sakai, T. Ohta, and Y. Sakano. 1993. A neopullulanase-type α-amylase gene from Thermoactinomyces vulgaris R-47. Biosci. Biotechnol. Biochem. 57:395-401. [DOI] [PubMed] [Google Scholar]
- 42.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wintrode, P. L., K. Miyazaki, and F. H. Arnold. 2001. Patterns of adaptation in a laboratory evolved thermophilic enzyme. Biochim. Biophys. Acta 1549:1-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X., W. Meining, M. Fischer, A. Bacher, and R. Ladenstein. 2001. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 Å resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306:1099-1114. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]