Abstract
Gene transfer systems for Gordonia polyisoprenivorans strains VH2 and Y2K based on electroporation and conjugation, respectively, were established. Several parameters were optimized, resulting in transformation efficiencies of >4 × 105 CFU/μg of plasmid DNA. In contrast to most previously described electroporation protocols, the highest efficiencies were obtained by applying a heat shock after the intrinsic electroporation. Under these conditions, transfer and autonomous replication of plasmid pNC9503 was also demonstrated to proceed in G. alkanivorans DSM44187, G. nitida DSM44499T, G. rubropertincta DSM43197T, G. rubropertincta DSM46038, and G. terrae DSM43249T. Conjugational plasmid DNA transfer to G. polyisoprenivorans resulted in transfer frequencies of up to 5 × 10−6 of the recipient cells. Recombinant strains capable of polyhydroxyalkanoate synthesis from alkanes were constructed.
Since reclassification of the gram-positives Rhodococcus aichiensis and Nocardia amarae to the genus Gordonia (13), this taxon is now a well-defined genus among the Corynebacterium, Mycobacterium, and Nocardia (CMN) group of actinomycetes. Species of Gordonia have attracted much interest in recent years due to their unusual and diverse capabilities to catalyze biotransformations and biodegradation of poorly approachable substances (2, 7, 9, 16). Although the number of reports of newly identified species of this genus steadily increases, no suitable genetic transfer systems have yet been described. Molecular analysis of rubber degradation by G. polyisoprenivorans and of other interesting pathways of Gordonia species is hampered by the lack of suitable and efficient gene transfer systems. Therefore, the present study identified plasmids, which can be transferred to G. polyisoprenivorans and other species of this genus by conjugational transfer or electroporation and which are stably maintained.
Identification of vector systems for G. polyisoprenivorans.
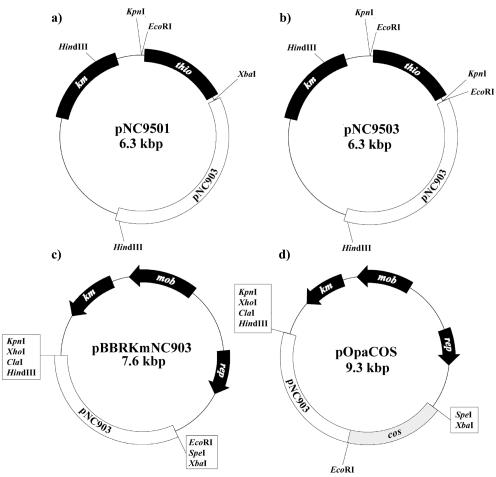
The 6.3-kbp plasmid pNC9503 (Fig. 1b) was recently described as an E. coli/Rhodococcus shuttle vector (12). It possesses a unique restriction site for XbaI and comprises the kanamycin resistance gene from Tn903 (24) for selection in E. coli and Rhodococcus/Gordonia. In addition, pNC9503 carries a thiostrepton resistance gene from Streptomyces azureus for selection in coryneform bacteria. The origin of replication (oriV) in actinomycetes is located on a fragment derived from the native Rhodococcus rhodochrous plasmid pNC903. A partial sequence revealed that it was 90% similar to the sequence of the R. rhodochrous plasmid pRC4 (10), which encodes a RepA and RepB protein. This plasmid, in turn, shares sequence similarity with the Mycobacterium fortuitum plasmid pAL5000 (17). Plasmid pNC9501 (Fig. 1a) is a derivative of pNC9503 differing from the latter only in possessing two additional unique restriction sites for KpnI and EcoRI.
FIG. 1.
Molecular organization of the E. coli/Rhodococcus (Gordonia) shuttle vectors used in the present study. pNC9501 (a) and pNC9503 (b) differ in the localization and orientation of restriction sites for EcoRI, KpnI, and XbaI. (c) Mobilizable plasmid pBBRKmNC903. (d) Mobilizable cosmid pOpaCOS. Relevant structural genes and other elements are indicated: km, kanamycin resistance gene; thio, thiostrepton resistance gene; pNC903, fragment from pNC903 comprising the ori for replication in coryneform actinomycetes; mob, required for mobilization; rep, required for replication in E. coli; cos, cos site required for lambda packaging.
These vectors were introduced into G. polyisoprenivorans strains VH2 and Y2K applying a basic electroporation protocol previously developed for R. opacus PD630 (12). Electroporation of strain Kd2 failed. The electroporated cells were plated on media containing 25 μg of thiostrepton or 25 μg of kanamycin/ml for the selection of transformants. Resistant colonies appeared after 4 to 6 days of incubation at 30°C. Plasmid DNA was isolated from each 20 randomly chosen transformants and then analyzed with respect to their restriction patterns. All transformants harbored plasmid DNA, indicating that autonomous replication of both plasmids occurs in G. polyisoprenivorans. They were therefore suitable as E. coli-G. polyisoprenivorans shuttle vectors. However, restriction analysis revealed that ca. 50% of the plasmids recovered from the recombinant clones had undergone identical modifications resulting in truncations of the 5.1-kbp EcoRI fragment of plasmid pNC9503 by deletion of ca. 800 bp. By changing the electroporation protocol, these modifications were prevented (see below).
Because first electroporation experiments led to transformation rates of only about 103 transformants/μg of plasmid DNA, the electroporation protocols for both strains of G. polyisoprenivorans were systematically optimized. For this, one parameter of cultivation or of the electroporation conditions was altered at a time, whereas the others were kept constant. The optimum of the field strength was 10 kV/cm. Transformation efficiencies depended strongly on the cultivation conditions, the medium, and the type and concentration of cell wall-weakening additives. The most suitable basic medium to obtain electrocompetent cells was Luria-Bertani (LB) broth (18); LB broth was twofold more efficient than nutrient broth (ADSA-Micro, Barcelona, Spain) or standard I complex nutrient broth (Merck, Darmstadt, Germany). Highest transformation efficiencies were obtained if cells were used from the early growth phase when the cultures had reached optical densities of 0.5 at 600 nm. Therefore, all subsequent alterations of medium composition and cultivation conditions were done with LB medium, and the effects of sucrose, glycine, and isonicotinic acid hydrazide on transformation efficiency were investigated in a range previously described for Rhodococcus spp. (12). Glycine and sucrose in the medium enhanced the electroporation efficiency most effectively at concentrations of 0.5% (wt/vol) and 1.5% (wt/vol), respectively. Optimal concentration of isonicotinic acid hydrazide was 1.5 μg/ml; its addition increased the efficiency of electroporation about twofold. Plasmid DNA concentrations of ≤0.25 μg/ml resulted in the highest transformation rates. Temperatures and the duration of temperature shifts used for preincubation or incubation after the electroporation pulse also affected transformations. For G. polyisoprenivorans highest transformation efficiencies of up to 4 × 105 CFU/μg of plasmid DNA were obtained with cells grown at 30°C, and if they were incubated for 10 min at 0°C before and for 6 min at 46°C after the electroporation pulse. This heat shock also suppressed the 800-bp deletion of transformed plasmid DNA. The optimized electroporation protocol is as follows: DNA was purified from E. coli strains and dialyzed against distilled H2O by using microfilters (pore size of 0.025 μm; Millipore, Eschborn, Germany). For growth of G. polyisoprenivorans 50 ml of LB medium supplemented with 0.5% (wt/vol) glycine, 1.5% (wt/vol) sucrose, and 1.5 μg of isonicotinic acid hydrazide/ml in a 250-ml Erlenmeyer flask were inoculated with 1 ml of an overnight preculture in standard I complex nutrient broth medium, and the cells were grown at 30°C to an optical density of 0.5 at 600 nm. Cells were harvested, washed twice, and concentrated 20-fold in cold double-distilled H2O. Competent cells were either used directly for electroporation or stored at −70°C. Immediately before electroporation, 400 μl of competent cells were mixed with 0.001 to 10 μg of DNA and preincubated 10 min on ice. Electroporation with a model 2510 electroporator was performed in electrocuvettes (Eppendorf-Netheler-Hinz, Hamburg, Germany) with gaps of 2 mm and at the following settings: 10 kV/cm, 600 Ω, and 25 μF. Time constants of 4 to 5 ms were reached. Pulsed cells were immediately diluted with 600 μl of LB, incubated for 6 min at 46°C, regenerated at 30°C for 4 h, and plated on appropriate selective media, and transformants were identified after 4 to 6 days of incubation. In controls, no spontaneous kanamycin-resistant colonies occurred. The survival rate without heat shock was 68% (VH2) and 63% (Y2K) after electroporation and dropped to 44% (VH2) and 36% (Y2K) if heat shock was applied. This protocol was also applied to 16 different strains belonging to 12 different species of the genus Gordonia (Table 1). The transformation efficiencies for the other Gordonia strains were significantly lower than for G. polyisoprenivorans VH2 and Y2K and ranged between 102 and 104 CFU/μg of plasmid DNA. Autonomous replication of plasmid pNC9503 was shown to occur in G. alkanivorans DSM44187, G. nitida DSM44499T, G. rubropertincta DSM43197T, G. rubropertincta DSM46038, and G. terrae DSM43249T.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Gordonia spp. | ||
| G. alkanivorans MoAcy2 | Alkane-degrading wild type | DSM44187 |
| G. alkanivorans HKI 0136 | Alkane-degrading wild type | DSM44369T |
| G. amarae 9c | DSM43391 | |
| G. amarae Se6 | DSM43392T | |
| G. amicalis IEGM | Benzothiophene-desulfurizing wild type | DSM44461T |
| G. desulfuricans 213E | Benzothiophene-desulfurizing wild type | DSM44462T |
| G. hirsuta K718a | DSM44140T | |
| G. hydrophobica 1610/1b | DSM44015T | |
| G. nitida LE31 | 3-Ethylpyridine- and 3-methylpyridine-degrading wild type | DSM44499T |
| G. polyisoprenivorans Kd2 | Rubber-degrading wild type | DSM44302T |
| G. polyisoprenivorans VH2 | Rubber-degrading wild type | DSM44266 |
| G. polyisoprenivorans Y2K | Rubber-degrading wild type | 2 |
| G. rhizosphera 141 | DSM44383T | |
| G. rubropertincta N4 | DSM43197T | |
| G. rubropertincta 60017 | DSM46038 | |
| G. terrae NCTC 10669 | DSM43249T | |
| G. terrae T6 | DSM43342 | |
| G. westfalica Kb1 | Rubber-degrading wild type | DSM44215T |
| E. coli | ||
| S17-1 | thi-1 proA hsdR17 (rK− mK+) recA1, tra genes of RP4 integrated into the chromosome | 22 |
| XL1-Blue | recA1 endA gyrA96 thi hsdR17 (rK− mK+) supE44 relA1 λ−lac[F proAB lacIq ZΔ M15 Tn10(Tcr)] | 4 |
| Plasmids | ||
| pNC9501 | E. coli/Rhodococcus (Gordonia) shuttle vector; km, thio, pNC903 ori | H. Saeki, Japan Energy Corporation (12) |
| pNC9503 | E. coli/Rhodococcus (Gordonia) shuttle vector; km, thio, pNC903 ori | H. Saeki, Japan Energy Corporation (12) |
| pBBRKmNC903 | E. coli/Rhodococcus (Gordonia) shuttle vector, mobilizable; km, pNC903 ori | This study |
| pOpaCOS | E. coli/Rhodococcus (Gordonia) shuttle vector, mobilizable; cos, km, pNC903 ori | This study |
| pAK71 | phaC1 from P. aeruginosa; thio, pNC903 ori | 12 |
| pHC79 | ap, tc, cos | 11 |
| pBBR1MCS-2 | Gram-negative broad-host-range vector, mobilizable; km | 14 |
Construction of mobilizable vectors for conjugational transfer.
Because efficiencies of plasmid DNA transfer by electroporation decrease with increasing plasmid sizes (23), transfer of vectors by conjugation using E. coli S17-1 as a donor for G. polyisoprenivorans was also investigated. Two mobilizable vectors were constructed. (i) oriV, comprising a 2.4-kbp EcoRI/HindIII fragment of plasmid pNC9503, which mediates stable replication in G. polyisoprenivorans, was cloned into EcoRI/HindIII-digested DNA of the gram-negative broad-host-range vector pBBR1MCS-2 (14) (GenBank accession no. U23751), yielding plasmid pBBRKmNC903 (Fig. 1c). (ii) A 1.7-kbp BglII fragment containing the cos sites enabling lambda packaging of large DNA molecules for creating genomic libraries was derived from vector pHC79 (11) (GenBank accession no. L08873). It was treated with mung bean nuclease and subsequently cloned into SmaI-digested pBBR1MCS-2 DNA, yielding pBBR1MCS-2cos (data not shown). Afterward, the 2.4-kbp EcoRI/HindIII restriction fragment of plasmid pNC9503 containing the oriV of pNC903 was cloned into EcoRI/HindIII-digested pBBR1MCS-2cos DNA, yielding pOpaCOS (Fig. 1d). Applying a protocol described previously (6), recipient transfer frequencies of 6 × 10−7 for vector pBBRKmNC903 and 5 × 10−6 for vector pOpaCOS were obtained.
Recombinant biosynthesis of polyhydroxyalkanoates (PHAs) in G. polyisoprenivorans.
To analyze the suitability of these plasmids for transfer and heterologous expression of foreign genes in G. polyisoprenivorans, recombinant strains of G. polyisoprenivorans VH2 and Y2K capable of PHA synthesis were constructed. Substrates of PHAMCL synthase (3-hydroxyacyl-coenzyme A) are available from β-oxidation when the cells grow on n-alkanes. Furthermore, PHA biosynthesis was previously reported for various species of the closely related genus Rhodococcus (1, 8) and could be established in recombinant strains of R. opacus. When pAK71 (12) harboring phaC1 from Pseudomonas aeruginosa was introduced into VH2 and Y2K, the recombinant strains accumulated PHAs, contributing up to 8.3 or 13.2%, respectively, of the cell dry matter during cultivation on mineral salts medium under conditions of N starvation on long-chain n-alkanes (20). Gas chromatography (GC) and GC-mass spectrometry (MS) analysis of accumulated PHAs (3) revealed that copolyesters mainly consisting of odd-numbered 3-hydroxyalkanoates (3HHp, 3HHN, 3HUD, and 3HTD; >88 mol%) were synthesized from pentadecane, whereas PHAs mainly consisting of even-numbered 3-hydroxyalkanoates (3HO, 3HD, and 3HDD; ∼75 mol%) were synthesized from hexadecane (Table 2).
TABLE 2.
PHA accumulation by recombinant strains of G. polyisoprenivorans VH2 and Y2K after cultivation in media containing different carbon sourcesa
| G. polyisoprenivorans strain | Plasmid | Carbon source | PHA content (% CDW) | PHA composition (mol%)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3HHx | 3HHp | 3HO | 3HN | 3HD | 3HUD | 3HDD | 3HTD | ||||
| VH2 | pNC9503 | Pentadecane | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| pAK71 | Pentadecane | 8.3 | ND | 19.1 | 6.0 | 32.9 | 5.7 | 23.0 | TR | 13.3 | |
| pNC9503 | Hexadecane | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| pAK71 | Hexadecane | 7.4 | TR | 12.2 | 28.5 | 10.3 | 31.3 | ND | 17.7 | ND | |
| Y2K | pNC9503 | Pentadecane | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| pAK71 | Pentadecane | 13.2 | ND | 18.2 | 6.2 | 32.0 | 4.1 | 24.2 | TR | 13.1 | |
| pNC9503 | Hexadecane | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| pAK71 | Hexadecane | 8.7 | TR | 13.0 | 25.7 | 12.8 | 30.0 | ND | 18.5 | ND | |
The cells were cultivated in 50 or 1,000 ml of MSM containing 0.01% (wt/vol) NH4Cl in the presence of 25 μg of thiostrepton/ml for 96 h at 30°C. Hexadecane and pentadecane were added at concentrations of 0.2% (vol/vol). The content and composition of PHAs were analyzed by GC and coupled GC-MS as described previously (3). ND, not detected; TR, traces (<2.0 mol%); 3HHx, 3-hydroxyhexanoate; 3HHp, 3-hydroxyheptanoate; 3HO, 3-hydroxyoctanoate; 3HN, 3-hydroxynonanoate; 3HD, 3-hydroxydecanoate; 3HUD, 3-hydroxyundecanoate; 3HDD, 3-hydroxydodecanoate; 3HTD, 3-hydroxytridecanoate; CDW, cellular dry weight.
Conclusions.
The present study succeeded in establishing and optimizing two different gene transfer systems for the rubber-degrading, gram-positive bacterium G. polyisoprenivorans strains VH2 and Y2K and several other members of the genus Gordonia based on electroporation. Furthermore, conjugational plasmid transfer with E. coli S17-1 as the donor, enabling the transfer of large constructs as required for the phenotypic complementation of mutants, was established. This is the first description of genetic transfer of DNA and maintenance of foreign plasmids for various species of the genus Gordonia. It will make these bacteria accessible for genetic engineering, complementation of mutants, and heterologous expression of genes to reveal the molecular and biochemical basis of interesting metabolic pathways of Gordonia species. Transformation efficiencies of up to 4 × 105 CFU/μg of plasmid DNA are sufficiently high to comply with the demands of standard genetic techniques and resemble those reported for R. opacus (12), Rhodococcus sp. strain TE1 (21), R. fascians (5), and Clavibacter michiganensis subsp. sepedonicus (15). The application of a heat shock after electroporation increased transformation efficiencies, as reported for Corynebacterium glutamicum (25), and prevented the specific deletion of introduced plasmid DNA. Presumably, both effects were due to the inactivation of a restriction system (19, 25). The newly established electrotransformation protocol was successfully applied to establish a functional active PHA synthase of P. aeruginosa in G. polyisoprenivorans, resulting in PHAMCL biosynthesis from n-alkanes. The E. coli lacZ promoter of pAK71 located upstream of phaC1 was obviously recognized by the G. polyisoprenivorans RNA polymerase. Since PHAMCL biosynthesis did not depend on IPTG (isopropyl-β-d-thiogalactopyranoside) addition, G. polyisoprenivorans obviously does not produce a lac repressor, and lacZ promoter dependent genes are constitutively expressed.
REFERENCES
- 1.Alvarez, H. M., R. Kalscheuer, and A. Steinbüchel. 1997. Accumulation of storage lipids in species of Rhodococcus and Nocardia and effects of inhibitors and polyethylene glycol. Fett/Lipid 99:239-246.
- 2.Arenskötter, M., D. Baumeister, M. M. Berekaa, G. Pötter, R. M. Kroppenstedt, A. Linos, and A. Steinbüchel. 2001. Taxonomic characterization of two rubber-degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hypervariable regions of 16S rDNA sequences. FEMS Microbiol. Lett. 205:277-282. [DOI] [PubMed] [Google Scholar]
- 3.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: high efficiency plasmid transformating recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 5.Desomer, J., P. Dhaese, and M. van Montagu. 1990. Transformation of Rhodococcus fascians by high-voltage electroporation and development of R. fascians cloning vectors. Appl. Environ. Microbiol. 56:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich, B., C. Hogrefe, and H. G. Schlegel. 1981. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert, S. C., J. Morton, S. Buchanan, C. Oldfield, and A. McRoberts. 1998. Isolation of a unique benzothiophene-desulphurizing bacterium, Gordona sp. strain 213E (NCIMB 40816), and characterization of desulphurization pathway. Microbiology 144:2545-2553. [DOI] [PubMed] [Google Scholar]
- 8.Haywood, G. W., A. J. Anderson, D. Williams, E. A. Dawes, and D. Ewing. 1991. Accumulation of a poly(hydroxyalkanoate) copolymer containing primilary 3-hydroxyvalerate from simple carbohydrate substrates by Rhodococcus ruber NCIMB 40126. Int. J. Biol. Macromol. 13:83-88. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez-Perez, G., F. Fayolle, and J.-P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 10.Hirasawa, K., Y. Ishii, M. Kobayashi, K. Koizumi, and K. Maruhashi. 2001. Improvement of desulfurization activity in Rhodococcus erythropolis KA2-5-1 by genetic engineering. Biosci. Biotechnol. Biochem. 65:239-246. [DOI] [PubMed] [Google Scholar]
- 11.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 12.Kalscheuer, R., M. Arenskötter, and A. Steinbüchel. 1999. Establishment of a gene transfer system for Rhodococcus opacus PD630 based on electroporation and its application for recombinant biosynthesis of poly(3-hydroxyalkanoic acids). Appl. Microbiol. Biotechnol. 52:508-515. [DOI] [PubMed] [Google Scholar]
- 13.Klatte, S., F. A. Rainey, and R. M. Kroppenstedt. 1994. Transfer of Rhodococcus aichiensis Tsukamurella 1982 and Nocardia amarae Lechevalier and Lechevalier 1974 to the genus Gordona as Gordona aichiensis comb. nov. and Gordona amarae comb. nov. Int. J. Syst. Bacteriol. 44:769-773. [DOI] [PubMed] [Google Scholar]
- 14.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 15.Laine, M. J., H. Nakhei, J. Dreier, K. Lehtilä, D. Meletzus, R. Eichenlaub, and M. C. Metzler. 1996. Stable transformation of the gram-positive phytopathogenic bacterium Clavibacter michiganensis subsp. spedonicus with several cloning vectors. Appl. Environ. Microbiol. 62:1500-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linos, A., A. Steinbüchel, C. Spröer, and R. M. Kroppenstedt. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tire. Int. J. Syst. Bacteriol. 49:1785-1791. [DOI] [PubMed] [Google Scholar]
- 17.Rauzier, J., J. Moniz-Pereira, and B. Gicquel-Sanzey. 1988. Complete nucleotide sequence of pAL5000, a plasmid from Mycobacterium fortuitum. Gene 71:315-321. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Schäfer, A., J. Kalinowski, and A. Pühler. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl. Environ. Microbiol. 60:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222. [PubMed] [Google Scholar]
- 21.Shao, Z., W. A. Dick, and R. M. Behki. 1995. An improved Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus ssp. using electroporation. Lett. Appl. Microbiol. 21:261-266. [DOI] [PubMed] [Google Scholar]
- 22.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 23.Szostková, M., and D. Horáková. 1998. The effect of plasmid DNA size and other factors on electrotransformation of Escherichia coli JM109. Bioelectrochem. Bioenerg. 47:319-323. [Google Scholar]
- 24.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 25.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]