Abstract
We describe here a dual-labeling technique involving the green fluorescent protein (GFP) and the red fluorescent protein (DsRed) for in situ monitoring of horizontal gene transfer via conjugation. A GFPmut3b-tagged derivative of narrow-host-range TOL plasmid (pWWO) was delivered to Pseudomonas putida KT2442, which was chromosomally labeled with dsRed by transposon insertion via biparental mating. Green and red fluorescent proteins were coexpressed in donor P. putida cells. Cells expressing both fluorescent proteins were smaller in size than cells expressing GFP alone. Donors and transconjugants in mixed culture or sludge samples were discriminated on the basis of their fluorescence by using confocal laser scanning microscopy. Conjugal plasmid transfer frequencies on agar surfaces and in sludge microcosms were determined microscopically without cultivation. This method worked well for in situ monitoring of horizontal gene transfer in addition to tracking the fate of microorganisms released into complex environments. To the best of our knowledge, this is the first study that discusses the coexpression of GFP and DsRed for conjugal gene transfer studies.
Horizontal gene transfer by means of conjugation is considered to be the most important mechanism used by bacteria to rapidly adapt to changing environments (20). Conjugation is likely to play a major role in spreading genetic information in natural environments (2, 3, 14, 23, 24, 27, 28, 29, 30) and can be exploited in bioaugmentation. In natural or anthropogenic environments, bacteria often form biofilms (12, 31). Biofilms may favor conjugation due to the relative stability and close proximity of donors and recipients. Investigations of gene transfer in natural habitats have often been hampered by the fact that only a minor proportion of the bacteria are cultivable by standard microbiology techniques (1). Therefore, quantification of transconjugants by selective plating is either difficult or erroneous. Actual gene transfer frequencies in situ often remain undetermined. It is also not known whether all transconjugants are capable of growing on selective plates (20). However, by using fluorescent proteins such as the green fluorescent protein (GFP) from Aequorea victoria (7, 34, 35) for single-cell detection (16), in situ monitoring of plasmid transfer became possible without the cultivation of transconjugants (18, 19, 20). Application of reporter genes for monitoring gene transfer allowed the quantification of gene transfer frequencies in different environments (agar surfaces, phylloplane of Phaseolus vulgaris, and biofilms) (8, 10, 18, 19, 20, 28, 38).
Recently, a new red fluorescent protein (drFP583, commercially available as DsRed from Clontech Laboratories) was isolated from the Indo-Pacific reef coral, Discosoma sp., that fluoresces brilliantly red (26). Its maximum emission at 583 nm is clearly separated from the 511-nm emission peak of GFPmut3b (11). In spite of its drawbacks, such as long maturation time and aggregation, DsRed has attracted interest as a complementary partner to GFP that would allow simultaneous multicolor imaging of at least two different proteins in living cells (25). Therefore, a combination of GFP and DsRed appears to be promising for dual-labeling studies with negligible cross talk. This approach offers an additional variation to labeling microorganisms with fluorescent proteins for in situ studies.
Dual labeling allows monitoring the fate of donors and their conjugable plasmids released into the environment during bioaugmentation, in addition to quantifying conjugal gene transfer in situ. Before genetically engineered microorganisms (GEMs) with novel metabolic capabilities are released into the environment for biotechnological applications, the fate and effects of novel microorganisms and their genetically altered plasmid or chromosomal DNA on the natural ecosystem must be assessed (5). Fluorescent protein labeling is useful for monitoring the fate of GEMs released into complex environments in GEM-mediated bioaugmentation. Fluorescent proteins are reported to be stable with paraformaldehyde fixation, and cells expressing fluorescent proteins are shown to be hybridizable with fluorescently labeled rRNA-targeted oligonucleotide probes (18-20). Therefore, the combined approach (reporter genes and fluorescence in situ hybridization [FISH] with oligonucleotides) offers the possibility of tracking donors, transconjugants, and thus plasmid transfer in fixed samples of complex natural environments. At the same time, the identity and distribution of indigenous microorganisms that receive catabolic plasmids can be determined.
The aim of the present study was to evaluate the use of a dual-labeled donor strain for the in situ detection of conjugal plasmid transfer in environmental samples. For this purpose, a nalidixic acid-resistant strain, Pseudomonas putida KT2442, was chromosomally tagged with the dsRed gene by transposon insertion via biparental mating. A gfpmut3b-modified plasmid pWWO (10, 37) was electroporated into P. putida KT2442 carrying the dsRed gene. Both the green and red fluorescent proteins were coexpressed in the labeled P. putida cells. Expression of the gfp genes (on plasmids) and dsRed genes (in chromosomes) was the basis for monitoring of donors (red and green fluorescence) and transconjugants (green fluorescence). Plasmid pWWO was used since it is a well-characterized plasmid that codes for the degradation of toluene and benzyl alcohol. Benzyl alcohol was used as a source of carbon in later experiments in sequencing biofilm batch reactors. The newly constructed dual-labeled strain was thoroughly tested for in situ quantification of plasmid pWWO transfer on solid agar surfaces and in a sequencing batch biofilm reactor (SBBR). Donors and transconjugants were discriminated on the basis of their fluorescence by using confocal laser scanning microscopy (CLSM). GFP and DsRed, in combination with CLSM, were successfully used for quantifying conjugal gene transfer and also used to track the fate of the donor strain released into a laboratory SBBR treating synthetic wastewater during the course of bioaugmentation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains, plasmids, and their relevant characteristics are listed in Table 1. The strains were grown either on Luria-Bertani (LB) agar (tryptone, 10 g; yeast extract, 5 g; NaCl, 5 g; agar, 18 g; Milli-Q water, 1,000 ml; pH 7.2) or in Tris medium (24). When required, antibiotics were added at final concentrations of 50 μg ml−1 for kanamycin, 20 μg ml−1 for tetracycline, 35 μg ml−1 for nalidixic acid, and 50 μg ml−1 for ampicillin. Plasmid DNA was isolated according to the alkaline lysis method (21).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or referenceb |
|---|---|---|
| Strains | ||
| Pseudomonas putida | ||
| KT2442 | Rifr | 10 |
| TUM-PP8 | KT2442; Rifr Nalr | This study |
| BBC443 | KT2442 lacIq (pWWO::gfpmut3b); Kmr | 10 |
| TUM-PP10 | KT2442::dsRed; Rifr Nalr Tcr | This study |
| TUM-PP12 | TUM-PP10 (pWWO::gfpmut3b); Rifr Kmr Nalr Tcr | This study |
| Escherichia coli | ||
| S17-1 (λpir) | λ lysogenic S17-1 derivative producing π protein for replication of plasmids carrying oriR6K | K. N. Timmis |
| TUM-EC93 | S17-1 (λpir) (pUT-Tc-mini-Tn5-dsRed); Apr Tcr | This study |
| Other | ||
| Serratia ficaria | ATCC 33105 | |
| Serratia liquefaciens MG1 | 18 | |
| Hydrogenophaga palleronii | DSM63 | |
| Ralstonia eutropha | DSM531 | |
| Acidovorax defluvii | DSM12644 | |
| Acinetobacter calcoaceticus BD413 | ATCC 33305 | |
| Aeromonas hydrophila | DSM30187 | |
| Novospingobium capsulatum | DSM30196 | |
| Comamonas testosteroni | DSM50244 | |
| Plasmids | ||
| pWWO (TOL) | Self-transmissible; approximately 117 kb; narrow host range, Inc | 10, 37 |
| TOL-gfpmut3b | Integration of PA1/04/03::gfpmut3b from pJBA28 into TOL | 10 |
| pUT-Tc-mini-Tn5-dsRed | Mini-transposon delivery vector | 32 |
Rifr, rifampin resistance; Nalr, nalidixic acid resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance.
DSM, Deutsche Sammlung von Microorganismen, Braunschweig, Germany; ATCC, American Type Culture Collection, Rockville, Md.
Strain construction.
A nalidixic acid-resistant derivative of P. putida KT2442 was obtained by plating the strain on increasing concentrations of the antibiotic. The resulting strain, called TUM-PP8, was then subjected to transposon insertion by biparental mating with the conjugative Escherichia coli S17-1 (λpir) (pUT-Tc-mini-Tn5-dsRed) (32). Briefly, 100-μl suspensions of each strain (concentration of 5 × 109 bacteria/ml) were mixed and incubated on an LB agar plate overnight at 30°C. The mated bacteria were resuspended in 0.9% NaCl, and serial dilutions were plated onto LB agar supplemented with nalidixic acid and tetracycline. A few colonies were analyzed for their ability to display a bright red fluorescence and to grow on a mineral medium supplemented with glucose. A representative colony was selected (TUM-PP10) and grown in liquid culture. Purified DNA (5 μl) of the plasmid pWWO::gfpmut3b from P. putida strain BBC443 (10) was then transformed by electroporation into freshly prepared TUM-PP10 cells according to a standard procedure. Recombinant colonies were selected on agar plates supplemented with nalidixic acid, tetracycline and kanamycin, yielding the dually labeled strain TUM-PP12. A summary of the different plasmids and bacterial strains is presented in Table 1.
Coexpression of GFP and DsRed in P. putida cells.
Coexpression of GFP and DsRed in Pseudomonas putida cells was monitored along the growth phase in liquid culture by using both fluorescence and confocal laser scanning microscopy. P. putida was grown in Tris medium. Cells were harvested at periodic time intervals, washed and resuspended in phosphate-buffered saline (PBS; 8 g of NaCl liter−1, 0.2 g of KCl liter−1, 1.44 g of Na2HPO4 liter−1, 0.2 g of NaH2PO4 liter−1 [pH 7.0]). Then, 5 μl of cells suspended in PBS was immobilized between a microscopic slide and a coverslip. Green and red fluorescence was visualized by CLSM. In order to assess the number of cells expressing DsRed and/or GFP, dually tagged P. putida cells grown in liquid culture and on plates were stained with the general nucleic acid stain SYTO 60 (Molecular Probes, Eugene, Oreg.) and imaged by CLSM. SYTO 60 is used as a counterstain to determine total cell counts and, in this regard, has the same function as other commonly used nucleic acid stains such as DAPI (4′,6′-diamidino-2-phenylindole). In the present study, SYTO 60 staining was used to calculate the percentage of P. putida TUM-PP12 exhibiting green and red fluorescence. SYTO 60 can be excited by a laser wavelength of 633 nm and thus can be used with CLSM systems that do not have a UV laser, which is normally necessary for the visualization of DAPI. For SYTO 60 staining, 100 μl of suspended cells was added to 100 μl of SYTO 60 solution (2.5 μl/ml) in a microfuge tube. The contents were mixed and allowed to stand for 15 min at room temperature in the dark. The stained cells were then harvested, washed, and suspended in PBS for microscopic visualization of GFP and/or DsRed and SYTO 60 signals by using confocal microscopy.
Conjugation experiments on agar surfaces with single recipients.
In order to test the double-labeled P. putida TUM-PP12 for estimating conjugal gene transfer frequencies and also to estimate the host range of the TOL plasmid with respect to wastewater microorganisms, conjugation experiments with several recipients relevant to wastewater treatment were performed on LB agar surfaces. Overnight cultures of donor and recipient strains were harvested, washed in PBS, resuspended in 0.9% NaCl, and mixed in equal amounts before applying them as 100-μl drops onto the agar surface of LB plates. The plates were incubated for 24 h at 30°C. The cells were then scraped off the agar plates and suspended in 0.9% NaCl. Suspensions were incubated at room temperature for 12 h before analysis for efficient visualization of DsRed and to avoid the interpretation of donor cells as false transconjugants in gene transfer studies. Next, 5 μl of the mating patch suspension was immobilized between a microscopic slide and a coverslip and analyzed for cells expressing either only green fluorescence (transconjugants) or both red and green fluorescence (donors) by CLSM. To obtain total cell counts, the nucleic acid stains DAPI or SYTO 60 or transmission images were used interchangeably. Each of these approaches allows the determination of total cell numbers. Transmission images were used to obtain total cell numbers in the case of conjugations carried out with pure recipient cultures. A minimum of 10 images were analyzed for each mating experiment. To determine potential recipient cells, the number of donor cells was subtracted from the total cell number. Conjugation frequencies were expressed as the number of transconjugants per recipient cell.
Conjugation on agar surfaces with sludge and biofilm as the recipient mixture.
Conjugation experiments were also carried out with sludge collected from a full-scale municipal wastewater treatment plant and with biofilm collected from a laboratory SBBR. Mixed liquor was collected from an operating municipal wastewater treatment plant (WWTP Grueneck, Munich, Germany). Cells were harvested by centrifugation, washed twice with PBS, and stored as a glycerol culture at −80°C until futher use. For conjugation experiments on agar surfaces, cells were grown overnight in synthetic wastewater at 30°C on a shaker. Donor and sludge samples grown overnight were harvested, washed in PBS, and suspended in 0.9% NaCl. Conjugation was performed on LB agar surfaces as described above for single recipients. Biofilm collected from a laboratory-scale SBBR was homogenized by subjecting it to vortexing at high speed for 15 min and then washed in PBS twice before being used as a recipient mixture for conjugation on an agar surface. Samples suspended in PBS were incubated at room temperature for 12 h before analysis for efficient visualization of DsRed and to avoid the interpretation of donor cells as false transconjugants in gene transfer studies. A minimum of 10 confocal images was obtained for each conjugation experiment and then analyzed for donors and transconjugants. A 10-μg/ml solution of DAPI (Sigma, Deisenhofen, Germany) was used as a counterstain for total cell counts. Conjugation frequency was expressed as the number of transconjugants per original recipient cell number.
Conjugal gene transfer in a sequencing batch biofilm reactor. A biofilm reactor with a 1.0-liter working volume was operated in a sequencing batch mode. The cycle length was 8 h, consisting of a 16-min fill period, a 450-min reaction period, and a 14-min draw period with 100% water exchange. Glass beads with an average diameter of 8.5 mm were used as carrier material for biofilm formation. The reactor was fed with synthetic wastewater containing the following (in mg liter−1): sodium acetate, 61.7; citric acid, 66.7; d-(+)-glucose, 46.7; sodium gluconate, 61.7; benzyl alcohol, 108.2; yeast extract, 1.0; NH4Cl, 26.7; NaH2PO4 · 2H2O, 75.5; MgSO4 · 2H2O, 90; CaCl2, 14; NaHCO3, 275.4; FeCl3 · 6H2O, 0.45; H3BO3, 0.045; CuSO4 · 5H2O, 0.009; KI, 0.054; MnCl2 · 4H2O, 0.036; Na2MoO4 · 2H2O, 0.018; ZnSO4 · 7H2O, 0.036; CoCl2 · 6H2O, 0.045; and disodium EDTA, 3.0. To develop a mixed-culture biofilm, the reactor was inoculated with wastewater collected from the municipal wastewater treatment plant mentioned above. After a mixed culture biofilm was established on carrier material, the reactor was inoculated with an overnight culture of the donor strain (3 × 105 cells/ml) added to the feed. Biofilm samples collected after 24 h of donor addition were analyzed for donors and transconjugants by using CLSM. Again, samples suspended in PBS were incubated at room temperature for 12 h before analysis for efficient visualization of DsRed to avoid interpreting donor cells falsely as transconjugants. For total cells, a 5-μl sample stained with DAPI was immobilized between a microscopic slide and a coverslip. A minimum of 10 fields were counted with an epifluorescence microscope (Leica, Wetzlar, Germany) and analyzed for donors and transconjugants. Conjugation frequency was expressed as the number of transconjugants per original recipient cell number. Biofilms sampled from the reactor before inoculation with the donor strain and also from a parallel control reactor (without any donors added) were analyzed for autofluorescence.
Microscopy and image analysis.
CLSM was performed with an LSM 410 (Carl Zeiss, Jena, Germany) instrument equipped with an Axiovert inverted microscope to visualize cells expressing green and red fluorescent proteins. A ×63 water immersion lens with a numerical aperture of 1.4 was used for obtaining all images. The 488-nm laser line and a 515- to 545-nm band-pass emission filter, and 543-nm laser line and a 570-nm long-pass emission filter were used to detect GFPmut3b and DsRed, respectively. The 633-nm laser line and a 665-nm emission filter was used to detect SYTO 60. Cells expressing green and red fluorescent proteins were visualized by Carl Zeiss fluorescence microscopy equipped with no. 9 filter set for GFPmut3b and a no. 15 filter set for DsRed. Images were analyzed for the number of donors, transconjugants, and recipients by using Photoshop 5.5 image software (Adobe Systems, Inc., San Jose, Calif.).
RESULTS
Coexpression of GFP and DsRed in P. putida cells.
The expression of fluorescent proteins in P. putida cells was monitored by using both epifluorescence and confocal microscopy. Observations of cells from a liquid culture by using an epifluorescent microscope revealed a high degree of variation, particularly in the expression levels of GFP. When cells were excited with blue light, a heterogeneous population (in terms of fluorescence) of cells appearing yellow, orange, and green were observed with the epifluorescence microscope. A few bright green cells were observed, in which weak red fluorescence was detected. When excited with green light, brightly and weakly fluorescing red cells were observed. A few elongated cells with bright green fluorescence associated with weak red fluorescence were also observed upon green excitation. This may be due to the spectral overlap between GFP and DsRed signals and not necessarily to differential expression. We found, in contrast to our results with the epifluorescence microscope, that we were able by CLSM to separate GFP and DsRed signals with minimal or negligible cross talk in coexpressing cells by using differential excitation and emission filters.
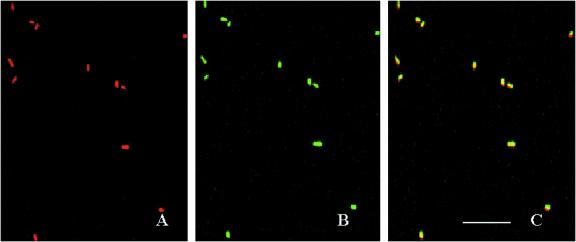
Differential excitation and emission were used to detect GFP and DsRed signals in bacterial cells using CLSM. Figure 1 shows the expression of green and red fluorescent proteins and their overlay in P. putida TUM-PP12 cells. With the settings used for imaging of cells by using confocal microscopy, no GFP fluorescence was detected in the red channel. The crossover of DsRed into the GFP channel was negligible. Therefore, cells expressing either GFP or DsRed or both could be separated easily without any image processing. GFP and DsRed signals were detected by CLSM 4 and ∼12 h after inoculation, respectively, in liquid culture. DsRed fluorescence was weak in rapidly growing cells, whereas stationary-phase cells were bright red fluorescent. No such correlation could be determined for GFP, since GFP expression was extremely heterogeneous. Determination of the percentage of cells expressing GFP and/or DsRed revealed that almost all cells (99%) were both red and green fluorescent.
FIG. 1.
Dually labeled P. putida TUM-PP12 cells expressing GFP and DsRed were imaged by CLSM. (A) DsRed expression; (B) GFP expression; (C) overlay of DsRed- and GFP-induced fluorescence. Bar, 20 μm.
Plasmid transfer on agar surfaces.
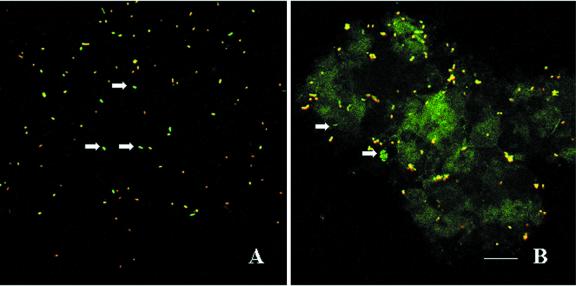
Conjugation experiments between the donor bacterium P. putida TUM-PP12 and several recipients performed on LB agar surfaces for 24 h at 30°C revealed that recipients that had received the plasmid could be easily distinguished from donors since they expressed only green fluorescence. Donors could be easily visualized by superimposing GFP and DsRed fluorescence results (Fig. 1C). Figure 2A illustrates the detection of plasmid pWWO transfer into recipient cells having the same genetic background as the donor but no plasmid, based on green fluorescence. In situ detection of GFP and DsRed fluorescence by confocal microscopy revealed that the plasmid pWWO was preferentially transferred to only a few recipients in the study (Table 2). In control experiments in which donors (Fig. 1C) and recipients were imaged separately, no green fluorescent cells (which could be falsely interpreted as transconjugants) could be detected. After the dually labeled donor strain was tested by monitoring conjugal transfer to pure recipient cultures on agar surfaces, it was used to monitor plasmid transfer to biofilm and sludge as recipient mixtures on agar surfaces. Transfer frequencies in the order of 10−3 and 10−4 transconjugants per recipient were observed in sludge and biofilm matings (Table 3).
FIG. 2.
In situ visualization of donors (yellow) and transconjugants (green [white arrows]) by CLSM. (A) Conjugation mixture with P. putida as recipient; (B) conjugal transfer of plasmid in the biofilm of SBBR. Bar, 20 μm.
TABLE 2.
Conjugal transfer of plasmid pWWO from P. putida TUM-PP12 on agar surfacesa
| Recipient | Conjugation frequencyb |
|---|---|
| P. putida KT2442 | (1.8-3.5) × 10−1 |
| Serratia ficaria | (7.9-10.0) × 10−5 |
| Serratia liquefaciens MG1 | (3.0-9.2) × 10−3 |
| Hydrogenophaga palleronii | (5.1-9.5) × 10−3 |
| Ralstonia eutropha | ND |
| Acidovorax defluvii | ND |
| Acinetobacter calcoaceticus | ND |
| Aeromonas hydrophila | ND |
| Novospingobium capsulatum | ND |
| Comamonas testosteroni | ND |
The donor cell density was 3 × 108 to 6 × 108 cells/ml.
Conjugation frequency was expressed as the number of transconjugants per recipient. The data represent the range of four independent experiments. ND, not detected.
TABLE 3.
Transfer of plasmid pWWO from P. putida TUM-PP12 to indigenous wastewater bacteriaa
| Recipient mixture | Mating environment | No. of recipient cellsb (cells/ml) | Conjugation frequencyc |
|---|---|---|---|
| Sludge | Agar surface | 6 × 109-9 × 109 | 7.8 × 10−4-1.4 × 10−3 |
| Biofilm | Agar surface | 3 × 109-7 × 109 | (1.5-9.8) × 10−4 |
| Biofilm in an SBBR | Biofilm reactor | 2 × 108-6 × 108 | (1.8-5.4) × 10−2 |
The donor cell density (agar surface matings) was 3 × 108 to 6 × 108 cells/ml.
The number of recipient cells was determined by enumeration of the total cells prior to conjugation.
Conjugation frequency was expressed as the number of transconjugants per original recipient cell number. Data represent the range of three independent experiments.
Plasmid transfer in a sequencing batch biofilm reactor.
Introduction of donor cells into a biofilm reactor microcosm resulted in the appearance of green fluorescent cells after 12 h, indicating that the plasmid was transferred to indigenous bacteria. Figure 2B shows the in situ visualization of donors and tranconjugants as determined by CLSM in a biofilm sample collected after 24 h of donor addition. To estimate the frequency of plasmid transfer, the numbers of donors, transconjugants, and total cells were determined. A conjugation frequency on the order of 10−2 transconjugants per recipient cell was observed (Table 3).
DISCUSSION
In the present study, transfer of gfp-labeled plasmid pWWO was investigated in situ by using a chromosomally DsRed-tagged P. putida donor strain KT2442. This approach allowed for the differentiation of donors and transconjugants on the basis of their fluorescence without the need for additional labels or selective plating conditions. Epifluorescent microscopic monitoring of dually labeled cells revealed a heterogeneous population of cells with different fluorescent intensities with both blue-green and green-red excitation and emission filter sets. The observed differences may reflect variations in the expression efficiency of the reporter genes but most likely are due to the spectral overlap between GFP and DsRed signals when they are observed in the epifluorescent mode. However, using the correct settings on the CLSM, we were able to separate GFP and DsRed signals with minimal or negligible cross talk in coexpressing P. putida TUM-PP12 cells (Fig. 1). Jakobs et al. (22) have already reported that E. coli cells expressing either enhanced GFP (EGFP) or DsRed could be easily discriminated by using differential excitation without any further image processing. This is an advantage over the frequently used GFP and its variants (13), which require image processing for quantitative separation after simultaneous excitation (17, 39). Confocal microscopy with two excitation wavelengths allows simultaneous discrimination of cells expressing either or both of the fluorescent proteins in a mixed cell population.
Conjugation experiments between the donor bacterium P. putida TUM-PP12 and P. putida KT2442 as the recipient performed on agar surfaces for 24 h at 30°C on LB plates showed conjugation frequency in the order of 10−1 per recipient (Table 2). This conjugation frequency is in accordance with the reported transfer frequency of plasmid pWWO to pseudomonads observed by selective plating (18, 29). Transfer of pWWO plasmid to Hydrogenophaga palleronii was observed in the order of 10−3 transconjugants per recipient in our experiments (Table 2). The conjugation frequency with Serratia strains as recipients was between 10−5 and 10−3 (Table 2). The observations are in agreement with other studies (18) in which pWWO plasmid transfer, as determined on selective agar plates, from a P. putida donor to Serratia strains, ranged from 10−3 to 10−5 transconjugants per recipient. With other strains under study, no transfer of plasmid was observed. These results are also comparable to those found in plate matings (18).
The plasmid used in the present study is a narrow-host-range plasmid, and thus the culturable recipient range is expected to be limited. This plasmid codes for the degradation of toluene and benzyl alcohol and was selected for the following reasons: it is a well-characterized plasmid able to replicate in the donor strain that was chosen for our experiments, namely, P. putida. Benzyl alcohol was used as a carbon source when experiments were carried out in reactors to answer questions regarding the success of bioaugmentation. For these reasons pWWO was selected, even though it is a narrow-host-range plasmid.
Conjugal transfer of the TOL plasmid has been investigated previously (8, 10, 18). Christensen et al. (8) used the gfp gene successfully for monitoring plasmid pWWO transfer between P. putida KT2442 strains on nutrient agar surfaces. In another study (10), P. putida donor cells carrying a chromosomally integrated the lacIq repressor gene and a Plac-gfp-tagged TOL plasmid, resulting in the repression of gfp expression from the plasmid in donor cells, were used in gene transfer studies in a biofilm community. The expression of the gfp gene was restored upon transfer of the TOL plasmid to a recipient. Using this elegant approach in combination with FISH, the authors of that study were able to determine the spatial distribution of bacterial cells in flow chamber biofilms. This approach is certainly superior if donors are to be tracked by means than other fluorescence protein labeling. In contrast, the dual-labeling method allows the differentiation of both donors and transconjugants without any additional stains. Geisenberger et al. (19) used also a combined approach, a gfp gene for the monitoring of plasmid RP4 transfer and FISH for the characterization of transconjugants. In the present study, we used dual labeling for monitoring donor and transconjugant cells. This approach allows direct microscopic visualization of donor and transconjugant cells in environmental samples and shows the compatibility of using both GFP and DsRed in one cell.
In the present study, P. putida donors were introduced into a sequencing batch biofilm reactor to monitor gene transfer and to track donor survival and establishment. Most bacteria found in nature are attached to surfaces and grow as surface-attached biofilms. It has been assumed that plasmid transfer in biofilms is usually greater because biofilms in nature can support dense, diverse, and active microbial communities. Since little information is available on the dynamics of gene transfer to and between biofilm organisms, improved knowledge of gene transfer events in biofilms will contribute to the understanding of the effect of GEMs in natural ecosystems (5).
The GFP system has already been used for studying plasmid transfer in different complex environments. However, there are potential pitfalls associated with the use of GFP and/or DsRed as a marker that need to be evaluated for individual environments. GFP has been used as a reporter protein and an in situ cell marker in eukaryotic and prokyryotic cells. GFP has also been applied in studies addressing gene transfer in bacterial communities such as activated sludge (19) or biofilms (8, 9, 10, 28, 38). Even though GFP has major advantages, such as its lack of requirements for cofactors or substrates (except for oxygen), its expression may be variable. In addition, the use of promoterless gfp transposon marker systems for the introduction of gfp into the chromosomes of strains of interest may affect the survival and metabolism of the tagged strains (15). GFP fluorescence based on gfp-labeled plasmids should be evaluated in each specific case. Banning et al. (4) have shown that the plasmid pEGFP and the fluorescence mediated by this plasmid were stable in E. coli DH5α for 100 generations of growth in LB broth under nonselective conditions. Environmental conditions may also influence gfp fluorescence. Chromosomally gfp-tagged Pseudomonas fluorescens (33, 36) and Salmonella enterica serovar Typhi (9) retained GFP fluorescence under starvation conditions. Banning et al. (4) noted a decrease in the total of pEGFP-labeled E. coli DH5α cells during a period of energy starvation. These authors attributed this to GFP degradation by live cells or to a loss of GFP from nonviable cells. The nature of the GFP gene expression and/or sufficient fluorescence of the expressed protein in all possible recipients, particularly in uncultivated bacteria, may also underestimate real plasmid transfer frequencies. The influence of fluorescent proteins on bacterial cell physiology and conjugal plasmid transfer needs further study.
In the present study, the fluorescent proteins GFP and DsRed were simultaneously expressed in a P. putida strain. Even though DsRed expression could be visualized only after ∼12 h in liquid culture, the two fluorescent proteins were distinguished by differential excitation and emission by CLSM. Dual labeling with recently available rapidly maturing variants of DsRed (6) should eliminate the need for preincubation of samples and allow in situ visualization of donors and transconjugants within a few hours. In our study, cells expressing DsRed and GFP were smaller than cells expressing GFP alone or untagged cells. This finding is in accordance with the observed size difference in E. coli cells expressing either EGFP or DsRed (22) and may indicate possible toxicity or metabolic burden exerted onto DsRed-expressing cells. In summary, in situ detection of gene transfer on the basis of fluorescent proteins allows more direct assessment than is possible where selective plating is required. Selective plating assumes that all transconjugants are viable and can grow and divide on selective media. In addition, selective plating may underestimate transconjugants present in clusters. On the other hand, the sample handling used in the present study (centrifugation and washing process) may introduce some potential for mating in test tubes. However, this bias is probably negligible considering the limited amount of time during sample processing. Another weak point in microscopy-based quantification of plasmid transfer is the detection limit, if transfer rates are low. Thus, it is necessary to collect a sufficient number of images for analysis in order to assure that low transfer rates may be determined. Nevertheless, the use of the dual-labeled strain in the present study allowed the tracking of donor P. putida TUM-PP12 cells released into a complex environment such as a sequencing batch biofilm reactor, along with the detection of transconjugants and thus monitoring of plasmid transfer. In addition, this approach should also prove useful in experiments designed to monitor the fate of organisms in bioaugmentation studies.
Acknowledgments
We thank S. Molin and L. Eberl for the donation of a strain carrying the pUT-mini-Tn5-DsRed-tet transposon. Strain S-17 λpir was kindly donated by K. N. Timmis.
Y.V.N. gratefully acknowledges Deutsches Zentrum für Luft- und Raumfahrt for providing a fellowship as part of the Indo-German bilateral programme. S.B. was supported by German Research Foundation grant Ha3264/2-1 awarded to M.H. and S.W. S.V.M. acknowledges a scholarship from the Humboldt Foundation. M.H. received a scholarship from the Technical University of Munich as part of the HWP-Programme.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., J. C. Fry, and M. A. Learner. 1998. Plasmid transfer between strains of Pseudomonas putida, and their survival, within a pilot scale percolating-filter sewage treatment system. FEMS Microbiol. Ecol. 18:15-26. [Google Scholar]
- 3.Bale, M. J., J. C. Fry, and M. J. Dry. 1987. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J. Gen. Microbiol. 133:3099-3107. [DOI] [PubMed] [Google Scholar]
- 4.Banning, N., S. Toze, and B. J. Mee. 2002. Escherichia coli survival in groundwater and effluent using a combination of propidium iodide and the green fluorescent protein. J. Appl. Microbiol. 93:69-76. [DOI] [PubMed] [Google Scholar]
- 5.Beaudoin, D. L., J. D. Bryers, A. B. Cunningham, and S. W. Peretti. 1998. Mobilization of broad host range plasmid from Pseudomonas putida to established biofilm of Bacillus azotoformans. I. Experiments. Biotechnol. Bioeng. 57:272-278. [PubMed] [Google Scholar]
- 6.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 30:83-87. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for protein expression. Science 263:802-804. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, and S. Molin. 1996. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene 173:59-65. [DOI] [PubMed] [Google Scholar]
- 9.Cho, J. C., and S. J. Kim. 1999. Green fluorescent protein based direct viable count to verify a viable but non-culturable state of Salmonella typhi in environmental samples. J. Microbiol. Methods 36:227-235. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Cowan, S. E., E. Gilbert, A. Khlebnikov, and J. D. Keasling. 2000. Dual labeling with green fluorescent proteins for confocal microscopy. Appl. Environ. Microbiol. 66:413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlberg, C., M. Bergström, and M. Hermansson. 1988. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dandie, C., E., S., M., Thomas, and N., C., McClure. 2001. Comparison of a range of green fluorescent protein-tagging vectors for monitoring a microbial inoculant in soil. Lett. Appl. Microbiol. 32:26-30. [DOI] [PubMed] [Google Scholar]
- 16.Eberl, L., A. Ammendola, O. Geisenberger, R. Schulze, M. Givskov, C. Sternberg, S. Molin, and K. H. Schleifer. 1998. Use of green fluorescent protein for online, single cell detection of bacteria introduced into activated sludge microcosms. Biofilm 3, vol. 3. [Online.] http://www.bdt.org.br/bioline/bf.
- 17.Ellenberg, J., J. Lippincott-Schwartz, and J. F. Presley. 1999. Dual color imaging with GFP variants. Trends Cell Biol. 9:52-56. [DOI] [PubMed] [Google Scholar]
- 18.Geisenberger, O. 1997. Molekularökologische Untersuchungen zum horizontalen Gentransfer unter verwendung GFP-markierter Plasmide. M.Sc. thesis. Technical University of Munich, Munich, Germany.
- 19.Geisenberger, O., A. Ammendola, B. B. Christensen, S. Molin, K. H. Schleifer, and L. Eberl. 1999. Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants. FEMS Microbiol. Lett. 174:9-17. [DOI] [PubMed] [Google Scholar]
- 20.Hausner, M., and S. Wuertz. 1999. High rates of conjugantion in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ish-Horowicz, D., and J. F. Burke. 1981. Rapid and efficient cosmid cloning. Nucleic Acids Res. 9:2989-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobs, S., V. Subramaniam, A. Schönle, T. M. Jovin, and S. W. Hell. 2000. EGFP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Lett. 479:131-135. [DOI] [PubMed] [Google Scholar]
- 23.Kroer, N., T. Barkay, S. Sorensen, and D. Weber. 1998. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant. FEMS Microbiol. Ecol. 25:375-384. [Google Scholar]
- 24.Lilley, A. K., and M. J. Bailey. 1997. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environment conditions. Appl. Environ. Microbiol. 63:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maksimow, M., K. Hakkila, M. Karp, and M. Virta. 2002. Simultaneous detection of bacteria expressing gfp and dsred genes with a flow cytometer. Cytometry 47:243-247. [DOI] [PubMed] [Google Scholar]
- 26.Matz, M. V., A. F. Fradkov, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969-973. [DOI] [PubMed] [Google Scholar]
- 27.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed]
- 28.Normander, B., B. B. Christensen, S. Molin, and N. Kroer. 1998. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 64:1902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos-Gonzalez, M. I., E. Duque, and J. L. Ramos. 1991. Conjugal transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl. Environ. Microbiol. 57:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravatn, R., A. J. B. Zehnder, and J. R. van der Meer. 1998. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in a laboratory-scale activated-sludge microscosms. Appl. Environ. Microbiol. 64:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutherland, I. W. 2001. The biofilm matrix: an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 32.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tombolini, R., A. Unge, M. E. Davey, F. J. deBruijn, and J. K. Jansson. 1997. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol. Ecol. 22:17-18. [Google Scholar]
- 34.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 35.Tsien, R. Y. 1999. Rosy dawn for fluorescent proteins. Nat. Biotechnol. 17:956-957. [DOI] [PubMed] [Google Scholar]
- 36.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams, P. A., and K. Murray. 1974. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120:416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wuertz, S., L. Hendrickx, and M. Hausner. 2001. In situ quantification of gene transfer in biofilms. Methods Enzymol. 336:129-143. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann, T., and F. Siegert. 1998. Simultaneous detection of two GFP spectral mutants during in vivo confocal microscopy of migrating Dictyostelium cells. BioTechniques 24:458-461. [DOI] [PubMed] [Google Scholar]