Abstract
In Comamonas testosteroni TA441, testosterone is degraded via aromatization of the A ring, which is cleaved by the meta-cleavage enzyme TesB, and further degraded by TesD, the hydrolase for the product of TesB. TesEFG, encoded downstream of TesD, are probably hydratase, aldolase, and dehydrogenase for degradation of 2-oxohex-4-enoicacid, one of the products of TesD. Here we present a new and unique steroid degradation gene cluster in TA441, which consists of ORF18, ORF17, tesI, tesH, ORF11, ORF12, and tesDEFG. TesH and TesI are 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-Δ4(5α)-dehydrogenase, respectively, which work in the early steps of steroid degradation. ORF17 probably encodes the reductase component of 9α-hydroxylase for 1,4-androstadiene-3,17-dione, which is the product of TesH in testosterone degradation. Gene disruption experiments showed that these genes are necessary for steroid degradation and do not have any isozymes in TA441. By Northern blot analysis, these genes were shown to be induced when TA441 was incubated with steroids (testosterone and cholic acid) but not with aromatic compounds [phenol, biphenyl, and 3-(3-hydroxyphenyl)propionic acid], indicating that these genes function exclusively in steroid degradation.
Comamonas testosteroni is able to utilize certain C19 and C21 steroids, as well as a number of aromatic compounds, as sole carbon and energy sources via a complex metabolic pathway involving many steroid-inducible enzymes. The mechanism by which testosterone is degraded in Nocardia restrictus and C. testosteroni was proposed in the 1960s (4, 8, 14). However, the genes involved in this degradation pathway in C. testosteroni have yet to be identified, except for the initial 17β-dehydrogenation (1, 3, 7) and the following Δ1-dehydrogenation (13). In previous studies, we identified the testosterone degradation genes tesB and tesDEFG among some open reading frames (ORFs) which are involved in testosterone degradation in C. testosteroni TA441 (9, 10). tesB encodes the meta-cleavage enzyme for 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (3,4-DHSA) (10), and tesD encodes the hydrolase for 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid (4,9-DSHA) (9). TesEFG are probably the enzymes for degradation of 2-oxohex-4-enoic acid, which is one of the two products of the hydrolysis of 4,9-DSHA by TesD.
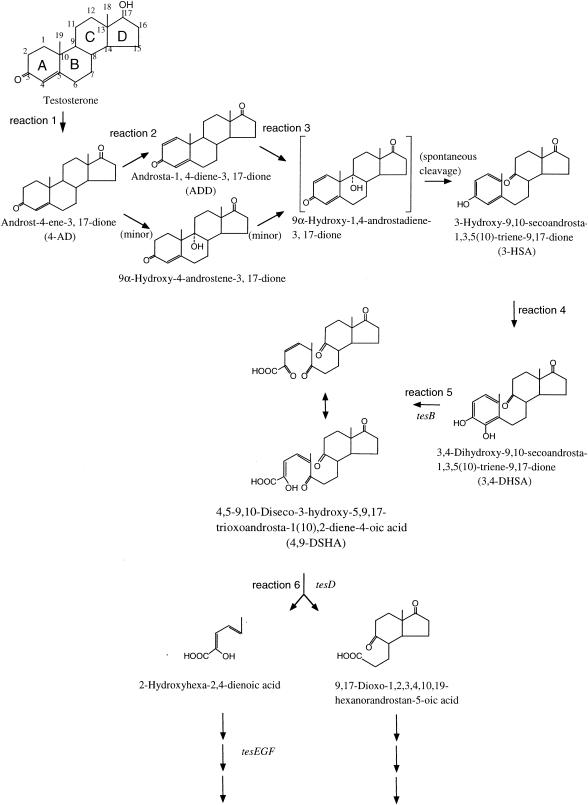
Studies on these genes have shown the probable testosterone degradation pathway of C. testosteroni TA441 to be that presented in Fig. 1. The process is initiated by dehydrogenation of the 17β-hydroxyl group on testosterone to 4-androstene-3,17-dione (4-AD) (Fig. 1, reaction 1), which then undergoes Δ1-dehydrogenation to 1,4-androstadiene-3,17-dione (ADD) (reaction 2), followed by 9α-hydroxylation to produce 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (3-HSA) (reaction 3 and the following spontaneous cleavage). The C-4 of 3-HSA is hydroxylated to yield 3,4-DHSA (Fig. 1, reaction 4), which is cleaved between C-4 and C-5 via a meta-cleavage reaction by TesB (reaction 5) (10). The product of the meta-cleavage reaction of 3,4-DHSA, 4,9-DSHA, is degraded into 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and 2-hydroxyhexa-2,4-dienoic acid by TesD (reaction 6) (9). One of the products, probably 2-hydroxyhexa-2,4-dienoic acid, is degraded by TesEFG.
FIG. 1.
Proposed pathway of testosterone degradation in C. testosteroni TA441.
TesB, -D, -E, -F, and -G show homology to the corresponding enzymes in bacterial aromatic compound degradation, such as BphC, -D, -H, -I, and -J, respectively, in biphenyl degradation. However, the homologies between TesB and BphC and between TesD and BphD are about 40% at most. In the upstream gene region of TesD, ORF11 and ORF12 were identified. ORF11 is oriented in the direction opposite that of ORF12 in relation to tesG. Both of the ORFs are indispensable, are induced in TA441 grown on testosterone, and are suggested to encode oxygenases. The region from ORF11 to tesG was not included in the gene region of approximately 20 kb containing tesB.
In the present study, we report a new steroid degradation gene cluster consisting of ORF18, ORF17, tesI, tesH, ORF11, ORF12, and tesDEFG. 3-Ketosteroid-Δ1-dehydrogenase (TesH) and 3-ketosteroid-Δ4(5α)-dehydrogenase (TesI) are encoded in sequence, followed by two ORFs, ORF17 and ORF18, which are indispensable and are induced in TA441 cells grown on steroids. ORF17 probably encodes the reductase component of 9α-hydroxylase for ADD, which is the product of the reaction catalyzed by TesH in testosterone degradation. In contrast to the 3-ketosteroid-Δ1-dehydrogenases in Rhodococcus erythropolis SQ1 (KstD1 and KstD2) (17, 18), TesH is a unique Δ1-dehydrogenase for 4-AD in TA441. The enzymes encoded by ORF11 to ORF18 and by ORF12 to tesG are induced when TA441 is grown with steroids but not when it is grown with aromatic compounds [phenol, biphenyl, and 3-(3-hydroxyphenyl)propionic acid], indicating that the enzymes encoded by this gene cluster are used exclusively for steroid degradation in TA441.
MATERIALS AND METHODS
Culture conditions.
C. testosteroni TA441 and mutant strains were grown at 30°C in either Luria-Bertani (LB) medium, C medium (10), or a mixed LB-plus-C medium (a mixture of equal volumes of LB and C media) with suitable carbon sources when necessary. Steroids and aromatic compounds, except for 3-(3-hydroxyphenyl)propionic acid, were added in a filtered dimethyl sulfoxide solution at a final concentration of 0.1% (wt/vol). 3-(3-Hydroxyphenyl)propionic acid was added as a CH3CN solution at a final concentration of 0.1% (wt/vol).
Construction of plasmids and mutant strains for gene disruption experiments.
The tesH gene was disrupted by insertion of a kanamycin resistance gene into the NotI site in tesH. The resultant plasmid, pTesH-Kmr, was used for inactivation of the tesH gene in TA441 by homologous recombination. The plasmid was introduced into TA441 by electroporation, and a Kmr and carbenicillin-sensitive TA441 mutant was selected. Insertion of the Kmr gene in tesH was confirmed by Southern hybridization using the Kmr gene and tesH as probes. Gene disruption of tesI, ORF17, and ORF18 was performed in the same manner. The restriction site at which the Kmr gene was inserted is indicated in Fig. 2.
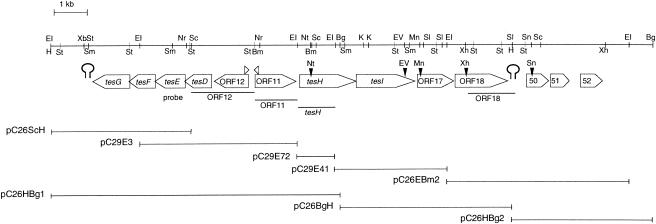
FIG. 2.
Partial restriction map of the DNA fragment containing the steroid degradation genes of C. testosteroni TA441. Large open arrows, genes and putative ORFs; small open arrowheads above large arrows, putative promoters. Putative terminators are located just downstream of tesG and ORF18. Small solid arrowheads above genes and ORFs indicate restriction sites where the Kmr gene was inserted. DNA segments used as probes are shown just below the ORFs. Deletion plasmids and remaining gene segments are also shown below the restriction map. Abbreviations: Bm, BamHI; Bg, BglII; EI, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; Mn, MunI; Nr, NruI; Nt, NotI; Ps, PstI; Sc, SacI; Sl, SalI; Sm, SmaI; Sn, SnaBI; Xb, XbaI; Xh, XhoI.
Growth of TA441 and mutant strains on testosterone.
Growth was monitored by CFU counting, because the culture to which testosterone was added in a dimethyl sulfoxide solution was turbid, which prevented measurement of cell absorbance. Mutants were grown with 0.1% (wt/vol) testosterone or androsterone, and growth was monitored by counting colonies that appeared on LB plates on which appropriate dilutions of the culture had been spread and incubated at 30°C, as previously described (10).
Three-dimensional HPLC.
A volume of methanol equivalent to twice the volume of culture was added to the culture, which was then centrifuged. The supernatant was directly injected into a high-pressure liquid chromatography (HPLC) system. A Waters 2690 HPLC was utilized with an Inertsil ODS-3 column (2.1 by 150 mm), and elution was carried out using a linear gradient from 20% solution A (CH3CN-CH3OH-trifluoroacetic acid [95:5:0.05]) and 80% solution B (CH3CN-H2O-trifluoroacetic acid [95:5:0.05]) to 65% solution A and 35% solution B over 10 min; the latter proportion was maintained for 3 min, and then the solvent was adjusted to 20% solution A for 5 min. The flow rate was 0.21 ml/min.
Isolation of the intermediate product X4 accumulated by the TesH disruption mutant.
For isolation of the intermediate product accumulated by the TesH disruption mutant (the compound X4), a 1,200-ml culture of the tesH disruption mutant incubated in C medium with 0.1% testosterone was twice extracted with the same volume of ethyl acetate. The ethyl acetate layer was concentrated in vacuo, and the residue was dissolved in a small amount of methanol and loaded onto a Waters 600 HPLC (Nihon Waters, Tokyo, Japan) with an Inertsil ODS-3 column (20 by 250 mm; GL Science) and a solvent composed of CH3CN and H2O (1:1), with a flow rate of 1 ml/min, at 40°C. X4 was detected at 245 nm. The fraction containing X4 was collected and studied further.
General experimental procedures.
For gas chromatography-mass spectrometry (GC-MS), an API 2000 LC/GC/MS spectrometer (Perkin-Elmer, Inc., Boston, Mass.) was used. UV spectra were recorded with an Ultrospec 3300 (Amersham Pharmacia Biotech, Piscataway, N.J.). Nuclear magnetic resonance (NMR) (1D, 1H, and 13C, DEPT, PFG-DQFCOSY, PFG-HMQC, PFG-HMBC) spectra were taken on a JNM-ECP500 spectrometer (JEOL, Tokyo, Japan) in a CDCl3 solution with tetramethylsilane at 0 ppm as an internal standard for 1H and 13C chemical shifts.
Northern blot analysis.
For the data presented in Fig. 8, TA441 cells incubated in LB medium for about 12 h were collected and added to newly prepared LB medium containing 0.1% testosterone and succinate. Total RNA was purified every 2 h after the start of the incubation and analyzed by Northern hybridization with suitable probes. Succinate was used as a negative control. For the data presented in Fig. 9, TA441 cells incubated in LB medium for about 12 h were collected and added to newly prepared LB medium containing 0.1% of each substrate [testosterone, cholic acid, phenol, biphenyl, 3-(3-hydroxyphenyl) propionic acid, and succinate]. Total RNA was purified at 4 h after the start of the incubation and analyzed by Northern hybridization with suitable probes.
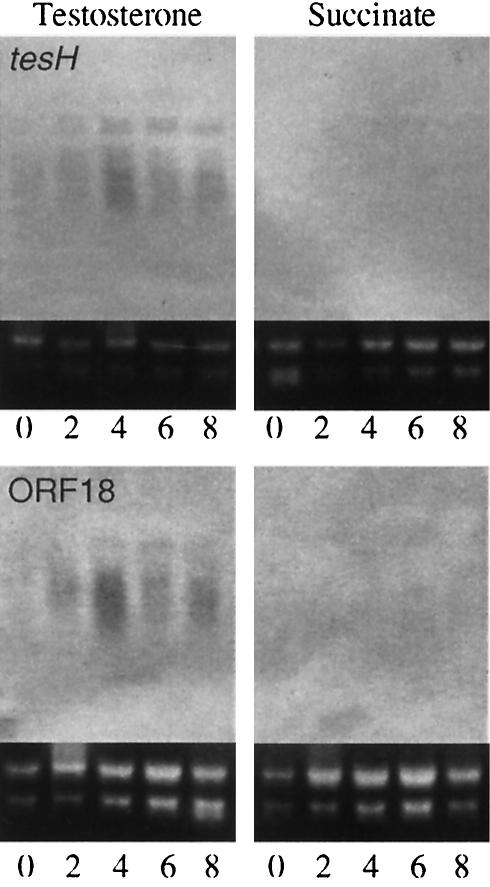
FIG. 8.
Induction of tesH and ORF18 in TA441 incubated with testosterone. Total RNA was purified every 2 h and analyzed by Northern hybridization with suitable probes (indicated in Fig. 2). Succinate was used as a negative control.
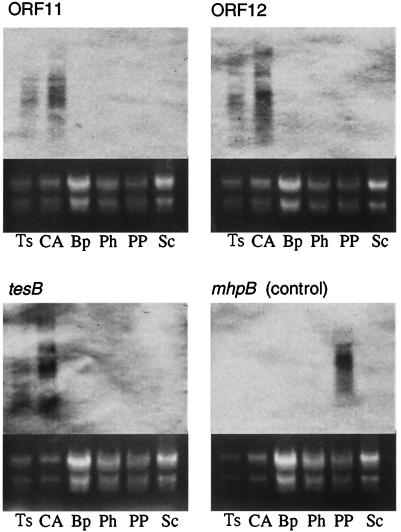
FIG. 9.
Induction of ORF11, ORF12, and tesB in TA441 cells incubated with steroids or aromatic compounds. Cells were collected 4 h after the start of incubation and were subjected to Northern blot analysis with suitable probes (ORF11, ORF12, tesB, and mhpB). mhpB is the gene encoding the meta-cleavage enzyme of TA441 in 3-(3-hydroxyphenyl)propionic acid degradation and was used as a control. Substrates tested were testosterone (Ts), cholic acid (CA), biphenyl (Bp), phenol (Ph), 3-(3-hydroxyphenyl)propionic acid (PP), and succinate (Sc).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with accession number AB076368.
RESULTS
Isolation and sequencing of the gene segment downstream of ORF11.
The two DNA segments of C. testosteroni TA441 containing genes necessary for testosterone degradation have been reported previously. One contains tesB, the gene encoding the meta-cleavage enzyme which converts 3,4-DHSA to 4,9-DSHA (10). In another gene segment, tesD, a gene encoding the hydrolase for 4,9-DSHA, and tesEFG, genes encoding the enzymes for the following reaction (Fig. 2) (9), occur in sequence. ORF11 and ORF12, showing homology to some oxygenases, are located upstream of tesD, and ORF11 is oriented in the direction opposite that of ORF12 in relation to tesG. Downstream of tesG, a putative terminator was found, which indicates that this is one end of this gene cluster.
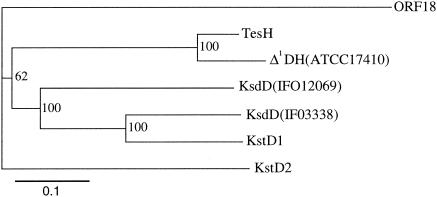
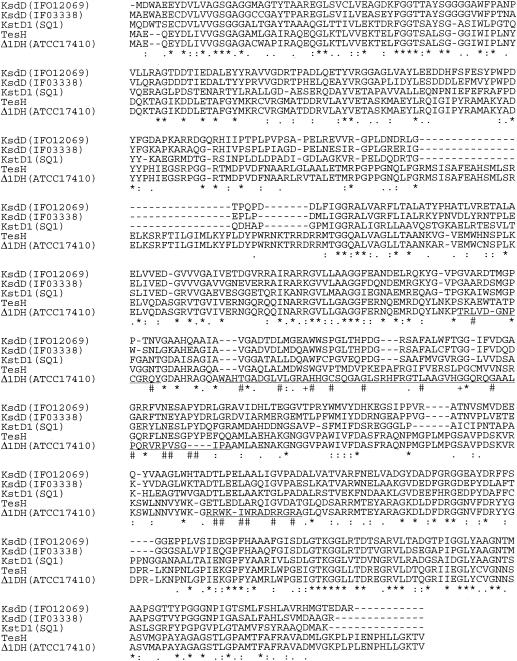
To obtain the gene segment downstream of ORF11, several pUC19-derived plasmids were constructed from pC26 and pC29, SuperCosI-derived plasmids carrying the DNA of TA441 described previously (9) (Fig. 2). Analysis of a 17-kb region downstream of ORF11 revealed nine ORFs (tesH, tesI, and ORFs 17, 18, 50, 51, 52, 53, and 54). Of the 17-kb region sequenced, about 10 kb comprising the region from tesH to ORF52 is shown in Fig. 2, because the following experiments showed that ORFs 50 to 54 are not involved in testosterone degradation in TA441. Two ORFs just downstream of ORF11 are named tesH and tesI, because they encode enzymes with high homologies to 3-ketosteroid-Δ1-dehydrogenase (Δ1DH) (83.0% identity) and 3-ketosteroid-Δ4(5α)-dehydrogenase [Δ4(5α)DH] (75.8% identity), respectively, in C. testosteroni ATCC 17410 (6, 13). TesH also showed identities of about 33, 34, 38, and 33% to the following Δ1DHs, respectively: Arthrobacter simplex IFO12069 KsdD (11), Rhodococcus rhodochrous IFO3338 KsdD (12), and R. erythropolis SQ1 KstD1 and KstD2 (identity between KstD1 and KstD2, 35%) (17, 18). The phylogenetic tree and the alignment of these Δ1DHs are shown in Fig. 3 and 4, respectively (KstD2 is not included in the alignment).
FIG. 3.
Phylogenetic tree for TesH and Δ1DHs. The branching pattern was generated by the neighbor-joining method (CLUSTAL X). The putative amino acid sequence of ORF18 was used as an outgroup. KsdD(IFO12069), Δ1DH of A. simplex IFO12069 (accession number S61889); KsdD(IFO3338), Δ1DH of R. rhodochrous IFO3338 (accession number AB007847); KstD1 and KstD2, Δ1DHs of R. erythropolis SQ1 (accession numbers AF096929 and AY078169, respectively); Δ1DH(ATCC17410), Δ1DH of C. testosteroni ATCC 17410 (accession number M68488).
FIG. 4.
Alignment of TesH and the Δ1DHs. Asterisks indicate amino acids identical among all the proteins; colons and periods indicate amino acids of high and low similarity, respectively. Regions in which the amino acid sequences of TesH and the Δ1DH of ATCC 17410 differ greatly are underlined. In these regions, number signs indicate amino acids identical among all the proteins except the Δ1DH of ATCC 17410, and plus signs indicate amino acids identical among all the proteins except TesH. Abbreviations are the same as for Fig. 3.
The alignment revealed three regions in which the amino acid sequences of TesH and the Δ1DH of ATCC 17410 differ greatly (Fig. 4). These differences are the results of a frameshift caused by the lack of several nucleotides in the gene sequence of the Δ1DH of ATCC 17410 relative to that of tesH. A search of TesI homology revealed significant homology only with the Δ4(5α)DH of ATCC 17410 (75.0% identity). Alignment of TesI and the Δ4(5α)DH of ATCC 17410 also revealed three significantly different regions as a result of a frameshift.
Downstream of tesI, two ORFs, ORF17 and ORF18, followed by a putative terminator, were found. The putative amino acid sequence of ORF17 showed homology with several kinds of ferredoxin reductases and possessed three conserved motifs of ferredoxin reductase: (i) the consensus sequence (RXFS), which is considered to be the binding region of a flavin adenine dinucleotide-isoalloxazine ring, (ii) the G-rich region, which is considered to be the binding region of an NAD(P)-ribose, and (iii) the consensus sequence (CXXXXCXXC-any 29 amino acids-C) of the chloroplast type Fe-S cluster (Fig. 5). The putative amino acid sequence of ORF17 showed the highest homology, about 46.0%, with KshB, the ferredoxin reductase component of R. erythropolis SQ1 3-ketosteroid-9α-hydroxylase (KSH) (16). All of this evidence indicates that ORF17 encodes the ferredoxin reductase component of 9α-hydroxylase for ADD in testosterone degradation in C. testosteroni TA441. However, none of the putative proteins that we cloned showed significant homology to KshA, the terminal oxygenase component of R. erythropolis SQ1 KSH (16). ORF18 showed the maximum homology, about 45%, to several putative acid-coenzyme A (CoA) ligases, about 30% homology to long-chain fatty acid-CoA ligases, and lower homologies to other CoA ligases.
FIG. 5.
Alignment of the putative amino acid encoded by ORF17 with R. erythropolis SQ1 KshB. KshB is the ferredoxin reductase component of the two-component class IA monooxygenase KSH. Asterisks indicate amino acids identical in the two proteins; colons and periods indicate amino acids of high and low similarity, respectively. Three conserved motifs of ferredoxin reductase are boxed.
Downstream of ORF18, several putative ORFs (ORF50 to ORF54) were found. Because the growth of the gene disruption mutants of these ORFs on testosterone was almost equal to that of TA441 (data not shown), these ORFs are not considered to be involved in testosterone degradation.
Growth of tesH, tesI, ORF17, and ORF18 disruption mutants on testosterone.
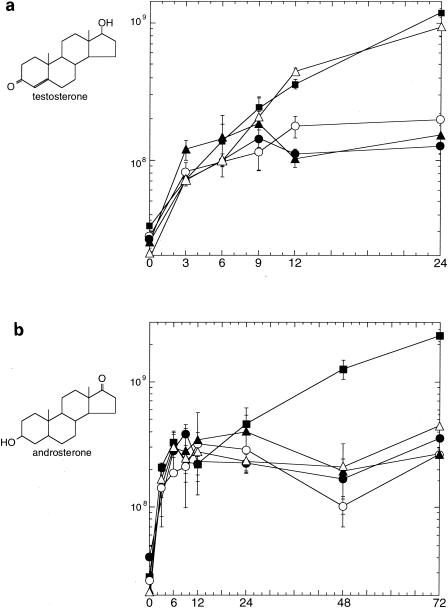
tesH, tesI, ORF17, and ORF18 in TA441 were individually disrupted with a Kmr gene by homologous recombination, and the resultant mutant strains were designated TesH−, TesI−, ORF17−, and ORF18−, respectively. Figure 6a shows a comparison of the growth of each mutant strain and TA441 on testosterone as the sole carbon source. The growth of each strain until around 6 h after the start of the incubation was probably caused by the nutrients carried by the cells from preculture on LB medium. After that, growth of the TesH−, ORF17−, and ORF18− mutants on testosterone as the sole carbon source was limited, indicating that these genes are necessary for testosterone degradation in TA441. The growth of the TesI− mutant was almost the same as that of TA441. This result is consistent with the function of TesI as Δ4(5α)DH; because testosterone has a double bond at the Δ4(5α) position, Δ4(5α)DH is not necessary for testosterone degradation. To confirm the function of TesI as Δ4(5α)DH, we also examined the growth of these mutants and TA441 on androsterone, whose Δ4(5α) bond is single (Fig. 6b). Only TA441 showed significant growth on androsterone; the TesI− mutant, as well as the other mutants, showed limited growth.
FIG. 6.
Growth of TA441 and gene disruption mutants on testosterone (a) or androsterone (b) as the sole carbon source. The genes were disrupted by insertion of a Kmr gene. Each strain was grown in LB medium, washed twice, and inoculated into 10 ml of C medium supplemented with 0.1% (wt/vol) testosterone or androsterone. Symbols: ▪, TA441; ▴, TesH disruption mutant; ▵, TesI disruption mutant; •, ORF17 disruption mutant; ○, ORF18 disruption mutant. Growth is expressed in CFU. Data are averages of more than three experimental determinations.
HPLC analysis of the culture media of the mutants.
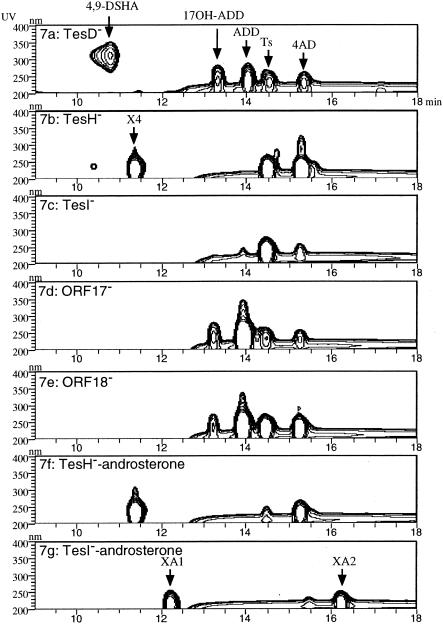
The TesH−, TesI−, ORF17−, and ORF18− gene disruption mutants were incubated in a mixed LB-plus-C medium with testosterone, and after about 40 h the culture media were analyzed by HPLC. The mixed LB-plus-C medium is a mixture of equal volumes of LB and C medium and was used because TA441 mutants accumulated larger amounts of intermediate compounds in this medium than in C medium (9). Figure 7 shows a three-dimensional HPLC chart of the culture of each mutant. The chart of the culture of the TesD disruption mutant, characterized in our previous study (9), is presented as authentic (Fig. 7a). In the culture of the TesH− mutant, 4-AD and an intermediate compound named X4 accumulated (Fig. 7b). Compound X4 was not detected in the culture of any other mutants. The identification of X4 is described in the next section. The TesI− mutant did not accumulate a significant amount of any intermediate (Fig. 7c), a result usually observed in the culture of TA441 or a well-growing mutant strain. The ORF17− and ORF18− mutants accumulated 4-AD, ADD, and 17β-hydroxylated ADD (Fig. 7d and e). These intermediate compounds are detected with most gene disruption mutants of steroid degradation genes (9, 10; also unpublished data). Figure 7f and g show the culture of the TesH− and TesI− mutants, respectively, incubated with androsterone. In the culture of the TesH− mutant incubated with androsterone, 4-AD and X4 accumulated, the same result as for incubation with testosterone. In the culture of the TesI− mutant, two intermediate compounds (XA1 and XA2) accumulated; these were not detected in the culture of any other mutant.
FIG. 7.
Intermediate compounds accumulating in the cultures of the gene disruption mutants of TA441 incubated with testosterone (a through e) or androsterone (f and g). Data are represented in three-dimensional HPLC charts (the vertical axis indicates wavelength, the horizontal axis indicates reaction time, and the UV absorbance of each compound is represented in contour). (a) HPLC chart of the culture of the TesD disruption mutant grown on the mixed LB-plus-C medium with testosterone (presented as authentic). (b through g) HPLC charts showing cultures of the gene disruption mutants of tesH (b), tesI (c), ORF17 (d), and ORF18 (e) incubated with testosterone and of the gene disruption mutants of tesH (e) and tesI (g) incubated with androsterone. Ts, testosterone; 17OH-ADD, 17-hydroxy-1,4-androstadiene-3-one.
Purification and identification of compound X4.
For isolation of X4, the TesH− mutant was incubated with testosterone, and the culture was extracted with ethyl acetate. The ethyl acetate layer was concentrated, and the residue was dissolved in a small amount of methanol. The solution was loaded onto the HPLC column, and the fraction containing X4 was collected from the eluent. The purified X4 gave a molecular ion at m/z 325 (M + Na+) and m/z 303 (M + H+) by direct MS analysis, indicating its molecular formula to be C19H26O3. Compound X4 showed maximum absorbance at 241.5 nm (ɛ = 16,000) in methanol solution, which agreed well with the value obtained by Dodson and Muir (5) for 9α-hydroxyandrost-4-ene-3,17-dione. Because complete NMR data were not available from previous studies of 9α-hydroxyandrost-4-ene-3,17-dione, complete NMR assignments were performed (Table 1). The 13C NMR spectrum confirmed the presence of 19 carbons including 2 methyls at 12.7 and 19.9 ppm and 2 carbonyl carbons at 198.8 and 219.9 ppm. One hydroxy signal was observed at 76.7 ppm. The 1H NMR spectrum confirmed two methyl protons at 0.93(s) and 1.35(s) ppm and one olefinic methine proton at 5.90(d) ppm. These characteristic signals of 1H NMR spectra are consistent with the partial 1H NMR spectral data of 9α-hydroxyandrost-4-ene-3,17-dione reported by Bell et al. (2), in which 9α-hydroxyandrost-4-ene-3,17-dione was prepared by the conversion of 4-AD by the fungus Diaporthe celastrina. Because the measured value of compound X4 correlated well with the MS, UV spectrum, and partial NMR analysis data reported in previous studies, X4 was identified as 9α-hydroxyandrost-4-ene-3,17-dione.
TABLE 1.
NMR data for X4
| Carbon no. | 13C (ppm) | 1H NMR (δ[ppm])a |
|---|---|---|
| 1 | 28.6 | 2.43 m, 1.75 m |
| 2 | 33.9 | 2.42 m |
| 3 | 198.8 | |
| 4 | 127.2 | 5.90 d (1.8) |
| 5 | 167.7 | |
| 6 | 31.5 | 2.51 ddd (15.1, 6.4, 1.8), 2.36 ddd (15.1, 5.1, 1.8) |
| 7 | 24.2 | 1.75 m, 1.54 m |
| 8 | 37.2 | 2.11 ddd (12.0, 11.5, 3.7) |
| 9 | 76.7 | |
| 10 | 44.5 | |
| 11 | 26.2 | 1.78 ddd (14.2, 13.3, 4.1), 1.65 ddd (14.2, 4.1, 2.3) |
| 12 | 27.0 | 1.69 ddd (13.3, 4.1, 2.3), 1.56 m |
| 13 | 47.2 | |
| 14 | 44.3 | 1.86 ddd (12.0, 6.0, 6.0) |
| 15 | 21.6 | 1.91 m, 1.56 m |
| 16 | 35.7 | 2.48 br.dd (18.8, 8.7), 2.13 ddd (18.8, 8.7, 8.7) |
| 17 | 219.9 | |
| 18 | 12.7 | 0.93 s |
| 19 | 19.9 | 1.35 s |
Coupling constants (J values in hertz) are given in parentheses.
Induction of the steroid degradation genes in TA441.
Total RNA of TA441 incubated in LB medium with testosterone or succinate (negative control) was purified every 2 h for 8 h and subjected to Northern blot analysis. LB medium, which has been shown not to inhibit the induction of testosterone degradation genes in TA441 (9), was used to obtain almost the same volume of cells from cultures of all the compounds tested. Northern blot analysis showed that tesH and ORF18 were induced in testosterone-grown cells (Fig. 8). Induction of ORF11, ORF12, and tesB in TA441 cells incubated with steroids (testosterone and cholic acid) or with aromatic compounds [phenol, biphenyl, and 3-(3-hydroxyphenyl)propionic acid] at 4 h after the start of the incubation is shown in Fig. 9. Cholic acid was tested because TA441 shows good growth on it. Induction of ORF11, ORF12, and tesB was examined because these genes are located at the very beginning in each sequence of genes (putative operon). ORF11, ORF12, and tesB were induced during growth on steroids but not during growth on aromatic compounds. The mhpB gene, which encodes the meta-cleavage enzyme for 3-(3-hydroxyphenyl)propionic acid degradation, was used as a control. In contrast to ORF11, ORF12, and tesB, mhpB was induced in cells incubated with 3-(3-hydroxyphenyl)propionic acid but not with steroids.
DISCUSSION
In this paper, we reported a bacterial steroid degradation gene cluster with a unique combination of genes that has not been reported before. It consists of ORF18, ORF17, tesI, tesH, ORF11, ORF12, and tesDEFG. TesDEFG, reported in our previous study (9), are the hydrolase for 4,9-DSHA (TesD) and enzymes for the subsequent reaction (TesEFG). The present study revealed the presence of a Δ1DH gene and a Δ4(5α)DH gene, tesH and tesI, followed by two ORFs, ORF17 and ORF18, in the region upstream of tesD. All these genes are involved and induced in testosterone degradation in TA441. Involvement of TesH and TesI in androsterone degradation, as well as induction of isolated testosterone degradation genes in TA441 cells incubated with cholic acid, indicates that these genes are targeted not only to testosterone but also to various kinds of steroids. Putative terminator sequences were found just next to the end of ORF18 and just next to the end of tesG, and ORFs located downstream of ORF18 were not necessary for steroid degradation in TA441, indicating that this cluster consists of ORF18 through tesG. Genes tesH, tesI, and probably ORF17 encode enzymes for steroids with an androstane structure, and ORF11, ORF12, and tesDEFG encode enzymes for the aromatized A ring of seco-steroids. (The roles of ORF11 and ORF12 will be described in a subsequent paper.) Gene disruption experiments and analysis of the culture media of the mutants showed that all these enzymes are involved in steroid degradation. The induction of these genes by steroids (testosterone and cholic acid) but not by aromatic compounds [phenol, biphenyl, and 3-(3-hydroxyphenyl)propionic acid] was confirmed by Northern blot analysis. These results clearly show that tesDEFG developed exclusively for steroid degradation in TA441 in spite of their similarity to aromatic compound degradation genes. From the evolutionary viewpoint, the relationship between the tes genes and aromatic compound degradation genes is particularly interesting.
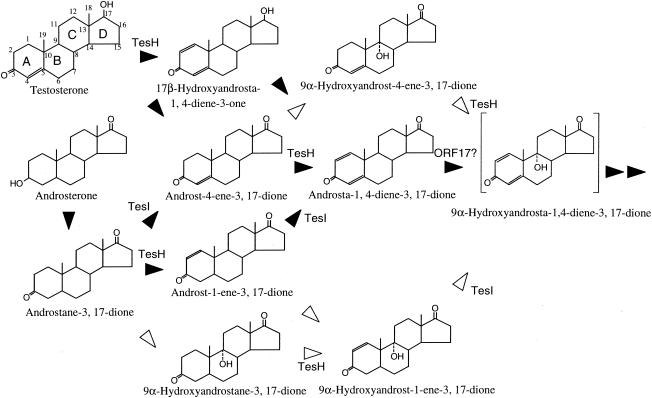
TesH showed about 83 and 38% identity with C. testosteroni ATCC 17410 Δ1DH (13) and R. erythropolis SQ1 KstD1 (17, 18), respectively. R. erythropolis SQ1 has two Δ1DHs, KstD1 and KstD2, and the kstD1 disruption mutant of SQ1 still possessed the ability to grow on testosterone. The tesH disruption mutant of TA441 did not show significant growth on testosterone and accumulated 4-AD and 9α-hydroxyandrost-4-ene-3,17-dione, indicating that TesH is a unique Δ1DH in TA441. 4-AD and ADD were detected in the cultures of most of the mutants incubated with testosterone and in the culture of TA441 in the early phase of growth on testosterone, while 9α-hydroxyandrost-4-ene-3,17-dione was detected only in the culture of the tesH disruption mutant. These results indicate that in TA441, Δ1-dehydrogenation of 4-AD to ADD is faster and more significant than 9α-hydroxylation of 4-AD. Figure 10 shows proposed testosterone and androsterone conversion in TA441.
FIG. 10.
Conversion of testosterone and androsterone in C. testosteroni TA441. Solid arrowheads indicate the probable main degradation pathway. Open arrowheads indicate minor reactions.
TesI, encoded just next to TesH, showed 75.8% identity with C. testosteroni ATCC 17410 Δ4(5α)DH, and the tesI disruption mutant grew quite well on testosterone [in which the Δ4(5α) bond is double] but failed to show significant growth on androsterone [in which the Δ4(5α) bond is single], indicating that TesI is a unique Δ4(5α)DH for steroid degradation in TA441. In the culture of the tesI disruption mutant incubated with androsterone, two intermediate compounds (XA1 and XA2), which were not detected in the cultures of other mutants, were detected by HPLC. By comparison with peaks detected in the tesH disruption mutant incubated with androsterone, these compounds are believed to be 9α-hydroxyandrost-1-ene-3,17-dione and androst-1-ene-3,17-dione.
The putative amino acid sequence of ORF17 possessed conserved motifs of a ferredoxin reductase component of hydroxylases and showed similarity to ferredoxin reductases; the highest identity, about 48%, was to R. erythropolis SQ1 KshB (16). Since KshB is the ferredoxin reductase component of SQ1 KSH, there is a high possibility that ORF17 encodes a ferredoxin reductase component of KSH. This is supported by the result of HPLC analysis of the culture of the ORF17 disruption mutant incubated with testosterone: most of the testosterone added was converted to ADD. KshA is the terminal oxygenase of SQ1 KSH, though none of the putative proteins that were identified in TA441 as involved in steroid degradation showed significant homology to KshA. Examination of the function of ORF17 and a search for the probable terminal oxygenase component are now under way. ORF18 is also necessary and induced in testosterone degradation and may encode an acid-CoA ligase. CoA-ligase adds CoA to a carboxyl group, but its role in steroid degradation is unclear.
Another gene cluster isolated from TA441 contains tesB, encoding a meta-cleavage enzyme for 3,4-DHSA, followed by three putative ORFs, ORF1 to ORF3, all of which are necessary for testosterone degradation in TA441. Recently, a testosterone-inducible biphenyl-2,3-diol-1,2-dioxygenase of C. testosteroni ATCC 11996, TIP1, which is almost identical to TesB (only two amino acids are different), was reported (15). The tip1 gene is followed by ORF1, tip6, ORF2, and ORF3, which are almost identical to ORFs 1 to 3 in TA441. (The ORF2 of ATCC 11996 presented in the report of Skowasch et al. is shorter than ORF2 in TA441. This is probably the result of an error in DNA sequence determination, which resulted in the division of an ORF, which corresponds to ORF2 in TA441, into tip6 and ORF2, because the estimated molecular weight of TIP6 in their sodium dodecyl sulfate-polyacrylamide gel electrophoresis experiment was not consistent with the molecular weight of the putative protein encoded by tip6 but was consistent with ORF2 of TA441.) Extradiol dioxygenase, another name of the meta-cleavage enzyme, is well known for its wide substrate specificity, so the activity on 2,3-dihydroxy biphenyl and the approximately 40% homology to BphC's are not considered to be enough evidence to conclude that it is biphenyl-2,3-diol-1,2-dioxygenase. To identify it as biphenyl-2,3-diol-1,2-dioxygenase, it is necessary to examine the growth of the gene disruption mutant on biphenyl and its induction in biphenyl-grown cells. TA441 does not grow on biphenyl. Northern blot analysis showed that tesB is induced in TA441 incubated with steroids (testosterone and cholic acid) but not with aromatic compounds [phenol, biphenyl, and 3-(3-hydroxyphenyl)propionic acid]. These results showed that TesB is an enzyme only for steroid degradation. The gene cluster containing tesB must be another steroid degradation gene cluster in TA441. Analysis of genes in this cluster is also necessary in order to clarify the entire degradation pathway of steroids in C. testosteroni.
Acknowledgments
We thank Hiroyuki Koshino for practical advice on identification of purified intermediate compounds. We thank Y. Ichikawa and R. Nakazawa (Biodesign DNA Sequence Facility, RIKEN) for determination of the nucleotide sequence.
This work was partly supported by a grant from the Eco Molecular Sciences Research Program of RIKEN. M.H. was supported by a grant from the Special Postdoctoral Researchers Program of RIKEN.
REFERENCES
- 1.Abalain, J. H., S. Di Stefano, Y. Amet, E. Quemener, M. L. Abalain-Colloc, and H. H. Floch. 1993. Cloning, DNA sequencing, and expression of (3-17)β-hydroxysteroid dehydrogenase from Pseudomonas testosteroni. J. Steroid Biochem. Mol. Biol. 44:133-139. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. M., A. D. Boul, E. R. Jones, G. D. Meakins, J. O. Miners, and A. L. Wilkins. 1975. Microbiological hydroxylation. Part XVII. Introduction of 16α-, 9α-, and 3α-hydroxy-groups into dioxygenated 5α-androstanes by the fungus Diaporthe celastrina. J. Chem. Soc. 14:1364-1366. [DOI] [PubMed] [Google Scholar]
- 3.Cabrera, J. E., J. L. Pruneda Paz, and S. Genti-Raimondi. 2000. Steroid-inducible transcription of the 3β/17β-hydroxysteroid dehydrogenase gene (3β/17β-hsd) in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 73:147-152. [DOI] [PubMed] [Google Scholar]
- 4.Coulter, A. W., and P. Talalay. 1968. Studies on the microbial degradation of steroid ring A. J. Biol. Chem. 243:3238-3247. [PubMed] [Google Scholar]
- 5.Dodson, R. M., and R. D. Muir. 1958. Microbiological transformations. II. Microbiological aromatization of steroids. J. Am. Chem. Soc. 80:5004-5005. [Google Scholar]
- 6.Florin, C., T. Kohler, M. Grandguillot, and P. Plesiat. 1996. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: gene and protein characterization. J. Bacteriol. 178:3322-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genti-Raimondi, S., M. E. Tolmasky, L. C. Patrito, A. Flury, and L. A. Actis. 1991. Molecular cloning and expression of the β-hydroxysteroid dehydrogenase gene from Pseudomonas testosteroni. Gene 105:43-49. [DOI] [PubMed] [Google Scholar]
- 8.Gibson, D. T., K. C. Wang, C. J. Sih, and H. Whitlock, Jr. 1966. Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Biol. Chem. 241:551-559. [PubMed] [Google Scholar]
- 9.Horinouchi, M., T. Hayashi, H. Koshino, T. Yamamoto, and T. Kudo. 2003. Gene encoding the hydrolase for the product of the meta-cleavage reaction in testosterone degradation by Comamonas testosteroni. Appl. Environ. Microbiol. 69:2139-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horinouchi, M., T. Yamamoto, K. Taguchi, H. Arai, and T. Kudo. 2001. Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 147:3367-3375. [DOI] [PubMed] [Google Scholar]
- 11.Molnar, I., K. P. Choi, M. Yamashita, and Y. Murooka. 1995. Molecular cloning, expression in Streptomyces lividans, and analysis of a gene cluster from Arthrobacter simplex encoding 3-ketosteroid-Δ1-dehydrogenase, 3-ketosteroid-Δ5-isomerase and a hypothetical regulatory protein. Mol. Microbiol. 15:895-905. [DOI] [PubMed] [Google Scholar]
- 12.Morii, S., C. Fujii, T. Miyoshi, M. Iwami, and E. Itagaki. 1998. 3-Ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous: sequencing of the genomic DNA and hyperexpression, purification, and characterization of the recombinant enzyme. J. Biochem. 124:1026-1032. [DOI] [PubMed] [Google Scholar]
- 13.Plesiat, P., M. Grandguillot, S. Harayama, S. Vragar, and Y. Michel-Briand. 1991. Cloning, sequencing, and expression of the Pseudomonas testosteroni gene encoding 3-oxosteroid Δ1-dehydrogenase. J. Bacteriol. 173:7219-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sih, C. J., S. S. Lee, Y. Y. Tsong, and K. C. Wang. 1966. Mechanisms of steroid oxidation by microorganisms: 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione, an intermediate in the microbiological degradation of ring A of androst-4-ene-3,17-dione. J. Biol. Chem. 241:540-550. [PubMed] [Google Scholar]
- 15.Skowasch, D., E. Möbus, and E. Maser. 2002. Identification of a novel Comamonas testosteroni gene encoding a steroid-inducible extradiol dioxygenase. Biochem. Biophys. Res. Commun. 294:560-566. [DOI] [PubMed] [Google Scholar]
- 16.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45:1007-1018. [DOI] [PubMed] [Google Scholar]
- 17.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid-Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197-202. [DOI] [PubMed] [Google Scholar]
- 18.van der Geize, R., G. I. Hessels, R. van Gerwen, J. W. Vrijbloed, P. van Der Meijden, and L. Dijkhuizen. 2000. Targeted disruption of the kstD gene encoding a 3-ketosteroid-Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl. Environ. Microbiol. 66:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]