Abstract
Potato cyst nematodes (PCN) are serious pests in commercial potato production, causing yield losses valued at approximately $300 million in the European Community. The nematophagous fungus Plectosphaerella cucumerina has demonstrated its potential as a biological control agent against PCN populations by reducing field populations by up to 60% in trials. The use of biological control agents in the field requires the development of specific techniques to monitor the release, population size, spread or decline, and pathogenicity against its host. A range of methods have therefore been developed to monitor P. cucumerina. A species-specific PCR primer set (PcCF1-PcCR1) was designed that was able to detect the presence of P. cucumerina in soil, root, and nematode samples. PCR was combined with a bait method to identify P. cucumerina from infected nematode eggs, confirming the parasitic ability of the fungus. A selective medium was adapted to isolate the fungus from root and soil samples and was used to quantify the fungus from field sites. A second P. cucumerina-specific primer set (PcRTF1-PcRTR1) and a Taqman probe (PcRTP1) were designed for real-time PCR quantification of the fungus and provided a very sensitive means of detecting the fungus from soil. PCR, bait, and culture methods were combined to investigate the presence and abundance of P. cucumerina from two field sites in the United Kingdom where PCN populations were naturally declining. All methods enabled differences in the activity of P. cucumerina to be detected, and the results demonstrated the importance of using a combination of methods to investigate population size and activity of fungi.
The potato cyst nematodes (PCN) Globodera rostochiensis (Wollenweber) and Globodera pallida (Stone) are serious pests in commercial potato production, causing yield losses valued at approximately $300 million in the European Community (20). Growers are encouraged to adopt an integrated approach to PCN management by using resistant cultivars and nematicides in addition to crop rotation (13). Biological control and other management methods may become more important as alternatives to the use of some nematicides in such integrated control strategies.
The nematophagous fungus Plectosphaerella cucumerina (Lindfors) Gams (22) was isolated from Meloidogyne hapla egg masses from tomato crops in Belgium (27), and recent work has demonstrated that P. cucumerina has potential as a biological control agent against PCN, reducing field populations by up to 60% after incorporation into alginate pellets (15).
Understanding the factors that cause inconsistencies in the biological control of nematodes by nematophagous fungi has been hampered by a lack of useful techniques to determine the presence, abundance, and pathogenicity of these fungi in the rhizosphere and soil. Also, it is important for regulatory purposes to develop methods to monitor the specific fungal biological control agents after their release (4). PCR-based methods to detect mycorrhizal and pathogenic fungi in plant roots have been reported (11, 18). These have relied on primers to recognize specific sequences within the mitochondrial genome or the transcribed (ITS) and nontranscribed (IGS) spacers within the ribosomal DNA genes (25). Recently, PCR detection of a fungus with established nematode biological control potential, Pochonia chlamydosporia (Goddard) Zare and Gams, was achieved by using primers based on the β-tubulin gene (14). In this paper, the design and use of two sets of primers that recognize the ITS region of P. cucumerina are described, one set designed for conventional PCR, and the other set combined with a fluorogenic probe, for real-time PCR quantification.
Although dilution plate methods have significant limitations, the use of selective media that enable the estimation of relative changes in abundance have greatly increased understanding of the ecology of selected nematophagous fungi in soil (16). The use of a selective medium for P. cucumerina, described below, was considered a key step in understanding the dynamics of this fungus in the soil and rhizosphere.
Conventional PCR provides an accurate method for identification of the fungus, and when combined with methods such as serial dilution plating on selective medium and baiting, where it can identify the fungus from colonies or infected eggs, it provides an accurate diagnostic method for the abundance and biological control capability of the fungus (2). Conventional PCR is unable to provide quantitative data on fungal populations, but the development of competitive and real-time PCR has enabled this important advance in molecular diagnostics. Although competitive PCR is quantitative, it requires the construction of a competitor, optimization, and serial dilutions of each sample to derive quantification and has been superseded by real-time PCR. Real-time PCR relies on the combination of a primer set and an additional dual-labeled fluorogenic probe to allow continuous monitoring of amplicon synthesis during thermocycling and requires no post-PCR handling for target quantification (21). It has been used to successfully determine the population levels of a number of fungal species (5, 6, 8, 26). In this paper, the development of molecular tools for the detection and quantification of P. cucumerina is described, and they are compared to more conventional methods, such as soil baiting and serial dilution on a selective medium.
MATERIALS AND METHODS
P. cucumerina (isolate number 380408) was isolated from PCN cysts obtained from infested soil in Jersey, United Kingdom. The culture was identified by CABI Bioscience (Egham, Surrey, United Kingdom), referenced as described above, and stored in their collection. A culture of the fungus was maintained on potato-dextrose-agar (PDA) plates (Oxoid) at 25°C and stored at 4°C on PDA plates or at −80°C in 15% glycerol stocks. Cultures were also grown in potato dextrose broth (Oxoid) at 25°C in an orbital shaker at 130 rpm for 7 days. Fungal isolates were taken as required from a culture collection maintained as above at Rothamsted Research. P. cucumerina isolates 17748 and 17763 were supplied from the CABI Bioscience culture collection. Isolates Pc1 and Pc2 were isolated from PCN-infected eggs taken from soil collected from a potato field in Spalding, United Kingdom. They were identified by morphological comparison to isolate 380408 before being stored in the Rothamsted culture collection.
Development of selective medium.
The selective medium for the isolation of Paecilomyces lilacinus (19) from soil was modified for the enumeration of P. cucumerina by decreasing the level of pentachloronitrobenzene (PCNB) to 37.5 mg liter−1. The addition of PCNB and benomyl to the medium inhibited the growth of Rhizoctonia sp. and Trichoderma spp. (19).
The selective medium contained 10 g of sodium chloride, 37.5 mg of PCNB (active ingredient: pentachloronitrobenzol [99%]) (Aldrich), 50 mg of benomyl (active ingredient: benlate [50% wt/wt]) (DuPont), 39 g of PDA (Oxoid), and high-quality water (demineralized and deionized) to bring the volume to 1 liter. After the medium was autoclaved for 15 min at 1.01 × 105 N/m2 (15 lb/in2) and cooled to 45 to 50°C, 100 mg of streptomycin sulfate (Sigma), 50 mg of chlorotetracycline hydrochloride (Sigma), and 1 ml of Tergitol NP 10 (Sigma) were added.
Estimation of fungal densities in soil.
The abundance of P. cucumerina in different soils was quantified by assessing the numbers of CFU g−1 of soil by using the selective medium as previously described (9). Three different soils were used: steam-sterilized sandy loam, commercial peat-based potting compost, and a sandy loam soil collected from a field site at Rothamsted, United Kingdom. A spore suspension of P. cucumerina, made by washing a cultured plate with 5 ml of sterile water, was added to 20 g of soil. To reisolate the fungus from soil, 1 g was suspended in 9 ml of sterilized 0.05% technical agar solution, and a serial dilution was prepared. An aliquot (0.2 ml) of each suspension was plated onto duplicate plates of the selective medium and incubated at 25°C for 7 days.
Comparison of the growth of P. cucumerina on the selective medium developed for P. chlamydosporia (9) and P. cucumerina was performed by washing a culture plate of P. cucumerina and preparing a dilution series of the spores. An aliquot of each spore suspension was spread onto two sets of duplicate P. chlamydosporia and P. cucumerina selective media plates and incubated at 25°C for 11 days.
Development of P. cucumerina selective primers.
Primers (PcCF1-PcCR1) (Table 1) were designed by the comparison of P. cucumerina isolate 380408 ITS sequence submitted to the GenBank/EMBL database (accession no. AJ492873) with other ITS sequences. The primers were compared to the database by using FASTA and BLAST to confirm specificity. PCRs of 20 μl contained 0.1 μM each primer, 2 μl of 10× PCR buffer (1.5 mM Mg2+) (Roche), 0.1 μM each dNTP, 1 U Taq polymerase (Roche), and 1 μl of DNA (20 to 60 ng). PCR conditions were optimized as follows: 95°C followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final incubation at 72°C for 5 min. Positive control DNA was prepared from pure culture DNA as described previously (1). DNA was extracted from a range of soil fungi, namely, Pochonia chlamydosporia isolate Vc10, Paecilomyces lilacinus, Penicillium chrysogenum, Aspergillus niger, Aspergillus nidulans, Pythium sp. isolate 31.5, Botrytis cinerea, Metarhizium anisopliae, Verticillium dahliae, Fusarium avenaceum, and Gliocladium roseum, taken from the Rothamsted culture collection. Cultures were incubated at 28°C in 100 ml of Czapek Dox liquid media (Oxoid) and shaken at 130 rpm for 1 week. DNA was extracted as above and was also extracted from P. cucumerina cultures listed in Table 2 and used in a PCR with the P. cucumerina-specific primers. A PCR profile was determined for all the P. cucumerina isolates listed in Table 2 by using ERIC primers as described previously (1).
TABLE 1.
P. cucumerina-specific PCR primers
| Process and primer(s) | Sequence | Target DNA | Size of product (bp) |
|---|---|---|---|
| Conventional PCR | |||
| PcCF1 | GGAGGGATCATTACTGAG TATAC | ITS1 | 440 |
| PcCR1 | CAGGACGCCGAGGACTCC CAA | ITS2 | |
| Real-time PCR | |||
| PcRtF1 | GTGCCCGCCGGTCTC | ITS1 | 72 |
| PcRtR1 | GACAGTTCGCTAAGAACA CTCAGAAGT | ITS1 | |
| Taqman probe, PcRtP1 | TCAGAATCTCTGTTTTCGA ACCCGACGA | ITS1 |
TABLE 2.
Origins of P. cucumerina isolates
| Isolate code | Sourcea | Isolated from: | Location |
|---|---|---|---|
| 380408 | Rothamsted Research | PCN eggs | Jersey, United Kingdom |
| 360147 | CABI Bioscience | Soil | United Kingdom |
| 358508 | CABI Bioscience | Work surface | United Kingdom |
| Pc1 | Rothamsted Research | PCN eggs | Spalding, United Kingdom |
| Pc2 | Rothamsted Research | PCN eggs | Spalding, United Kingdom |
CABI, Commonwealth Agricultural Bureau Institute.
Detection of P. cucumerina from plant roots.
Moist Terra-green (Oil Dri Corporation of America) was added to Magenta boxes (Sigma) and sterilized by autoclaving for 15 min at 1.01 × 105 N/m2 (15 lb/in2). Potato chits (var. Maris Piper) were added aseptically just below the surface of the Terra-green, above five 5-mm agar plugs taken from the edge of a culture of P. cucumerina on a PDA plate. The plants were incubated in a growth chamber at 15°C with 12-h light and dark periods. After 3 weeks, the plants were inoculated with 1,000 G. pallida second-stage juveniles. Control uninoculated potato plants and plants infected only with PCN were included. Extraction of genomic DNA was performed 8 weeks after planting. Plants were removed from pots, and the Terra-green was washed gently from the roots with water. Roots were cut into 1-cm sections and pooled from triplicate plants. DNA extraction was performed from 1 g of pooled roots with the DNeasy Plant mini kit (Qiagen). DNA samples were stored at −20°C.
Detection of P. cucumerina in PCN-infested soils using selective media, baiting, and conventional PCR.
Soil was collected from two fields in the United Kingdom reported to have PCN populations that are naturally declining. One site was in Ely, Cambridgeshire, United Kingdom (A. D. Barker, personal communication) and the other in Spalding, Lincolnshire, United Kingdom (7). Soil was sieved through a 5-mm sieve, and 1 kg was put into a 12-cm-diameter pot for each treatment. Treatments were duplicated. A serial dilution series was prepared using 1 g of soil from a thoroughly mixed sample from each of the pots. Aliquots (0.2 ml) of each suspension (10−2 to 10−4 dilutions) were spread onto the selective medium for P. cucumerina. Ely and Spalding soils were baited with 25 G. pallida cysts. Baits were prepared by containing 25 G. pallida cysts in a nylon mesh of 60-μm pore size (Lockertex) and held in place by a slide mount (Gepe) as described previously (2, 23). The slide mounts were pushed just below the surface of the soil, and the pots were incubated at 21°C for 2 weeks with light watering every 2 days. Baits were collected, and the cysts were crushed with a glass rod, and eggs were collected by standard techniques (7). Eggs were plated out onto water agar containing antibiotics (0.5 g of technical agar, 50 mg of streptomycin, 50 mg of chloramphenicol, and 50 mg of chlortetracycline per liter) and incubated at 25°C for 2 days. A bait was added to sterilized sandy loam soil and incubated as above as a control. This control provided the level of background infection in the cysts added as bait. Infected eggs were counted after 2 days, and the percentage of infection was calculated. The proportion of eggs infected from each soil was compared to the control and analyzed by one-way analysis of variance by using the Genstat program (12). Fifty infected eggs (infected eggs were classified as eggs with mycelia growing from them) were taken from each bait experiment, and individual eggs were added to 10 ml of Czapek Dox liquid medium and incubated at 25°C for 2 weeks. DNA was extracted from the fungal culture that developed from each infected egg by an alkaline lysis method (17) and subjected to PCR amplification with the P. cucumerina-specific primers PcCF1 and PcCR1. DNA was also extracted from four replicate 0.25-g sieved soil samples from both field sites by using a soil DNA extraction kit (MoBio Laboratories, Solana Beach, Calif.), and an aliquot (20 ng) from each was used in a PCR containing the P. cucumerina primer set PcCF1-PcCR1.
Real-time PCR quantification of P. cucumerina from field sites.
The Primer Express software (PE Applied Biosystems) was used to design primers and Taqman probes (PcRtP1) based on the original ITS sequence to develop real-time quantitative PCR. A series of primer and probe sets was compared to DNA databases and alignments of fungal ITS sequences to identify the most specific set for P. cucumerina. The original primers (PcCF1-PcCR1) developed were not used because real-time PCR requires small amplicons of 50 to 100 bp in length to yield consistent results (PE Applied Biosystems). The fluorogenic probe (PcRtP1) was labeled at the 5′ end with the fluorescent reporter dye FAM (6-carboxy-fluorescein), and the 3′ end was modified with the quencher dye TAMRA (6-carboxy-tetramethylrhodamine) (PE Applied Biosystems). Primer and probe sequences are described in Table 1.
Inoculation of soil for standard curve analysis for real-time PCR.
An aliquot of sandy loam soil (10 g) was inoculated with 108 conidiospores of P. cucumerina. The fungus had not been detected in the soil by using selective media or PCR methods prior to inoculation. DNA was extracted from the 10-g sample with a mega soil DNA extraction kit (MoBio). The resulting DNA preparation represented 107 P. cucumerina conidiospores per g of soil. Triplicate serial dilutions were prepared in ultrapure dH2O (high-performance liquid chromatography grade; Sigma) so that 1 μl represented 1 × 106, 5 × 105, 1 × 105, and 5 × 104 spores g−1 soil to a final dilution of 5 spores g soil−1. DNA prepared from the uninoculated soil was used as the negative control.
Real-time quantification PCR was performed in MicroAmp optical 96-well plates with the automated ABI Prism 7700 sequence detector (PE Applied Biosystems). Reaction components were made to 25 μl from the Taqman universal PCR master mix (PE Applied Biosystems). Primer and probe concentrations were optimized previously according to the manufacturer's guide. Primers PcRtF1 and PcRtR1 were included at a final concentration of 900 and 300 nM, respectively, and the Taqman probe (PcRtP1) was used at 225 nM. The manufacturer's recommended universal thermal cycle protocol (PE Applied Biosystems) was used for PCR amplification: stage 1, 50°C for 5 s; stage 2, 95°C for 10 min (activation of Taq DNA polymerase); and stage 3, 50 cycles of 95°C for 15 s and 60°C for 1 min. Reactions were performed in triplicate. An aliquot (1 μl) from each spore dilution was used to calculate the standard curve. Aliquots (20 ng) of DNA were used from each of the soil DNA preparations taken from the field sites at Spalding and Ely. The CT (cycle threshold) value (defined as the cycle number at which a statistically significant increase in the reporter fluorescence can be detected) for each PCR was automatically calculated and analyzed by the ABI Prism sequence detection software (version 1.5). To determine if the PCR was affected by soil type, a serial dilution of P. chlamydosporia DNA was added to DNA extracted from the two field sites. This fungus had not been detected at either field site. The presence of P. chlamydosporia was detected by PCR with the specific primers designed by Hirsch et al. (14). To compare the conventional PCR primers to the real-time primers, a conventional PCR was performed with the soil DNA extractions as described above.
RESULTS
Enumeration of P. cucumerina on selective media.
P. cucumerina was reisolated from all inoculated soil types investigated (Table 3) using the selective medium. No P. cucumerina was recovered from the control soils. The fungus grew on both selective media tested, but significantly more colonies grew on the P. cucumerina selective medium than on the P. chlamydosporia selective medium (Table 4).
TABLE 3.
Recovery of P. cucumerina after addition to different soils
| Treatment | CFU g soil−1 in:
|
|
|---|---|---|
| P. cucumerina-seeded soila | Control | |
| Steam sterilization | 1.04 × 106 ab | <102 |
| Composting | 1.33 × 106 a | <102 |
| None | 1.97 × 106 b | <102 |
In each column, values followed by the same letter do not differ significantly according to Tukey's test (P < 0.05).
TABLE 4.
Mean P. cucumerina CFU counts on P. cucumerina and P. chlamydosporia selective media (n = 4)a
| P. cucumerina selective medium |
P. chlamydosporia selective medium
|
||
|---|---|---|---|
| Dilution | Mean CFU ± SE | Dilution | Mean CFU ± SE |
| 0 | 898 ± 28 | 0 | 318 ± 3 |
| 10−1 | 85 ± 10 | 10−1 | 63 ± 8 |
| 10−2 | 23 ± 13 | 10−2 | 10 ± 0 |
| 10−3 | 3 ± 3 | 10−3 | 5 ± 5 |
Significant differences at the 5% level of significance were recorded between the two media at the undiluted state (F pr, 0.033; standard errors of the differences, ± 30) when analyzed by analysis of variance.
Specificity of P. cucumerina primers for conventional PCR.
The primer set PcCF1-PcCR1, specific for P. cucumerina, generated a single PCR product of 440 bp specific to DNA extracted from P. cucumerina and did not generate any products from DNA extracted from other fungal isolates. This 440-bp product was seen with all the P. cucumerina isolates tested (Table 2). Enterobacterial repetitive intergenic consensus PCR fingerprints differentiated four of the five P. cucumerina isolates listed in Table 2 and showed that Pc1 and Pc2 were the same isolate (gel not shown).
Detection of P. cucumerina from plant roots by conventional PCR.
A single band of the same size as the control of P. cucumerina DNA was generated from potato roots inoculated with P. cucumerina, and no product was generated when PCR was performed with DNA extracted from potato roots alone or potato roots infected only with PCN, using primer set PcCF1-PcCR1 (gel not shown).
Detection of P. cucumerina in PCN-infected soil.
The primer set PcCF1-PcCR1 was able to detect P. cucumerina in all four of the soil samples taken from the Spalding field and in two of the four samples taken from the Ely site. The presence of P. chlamydosporia was detected in the DNA-seeded soil extractions from both sites but not from the unseeded controls. Detection was not observed after a 1,000-fold dilution of the DNA at either site. This demonstrated that the PCR was not affected by the soil type. The number of fungal CFU in the Spalding soil was estimated to be 700 CFU g soil−1, whereas no colonies were detected on the selective medium from plates inoculated with soil from Ely, suggesting population densities lower than 102 CFU g soil−1 (Table 5). The baits provided a means of isolating from soil fungi that could attack cyst nematodes. Serial plate dilution provides information on the number of viable propagules of P. cucumerina in the soil but does not provide information on how their presence can affect the PCN population, data which were provided here by the level of infection in the baits. When the baiting technique was combined with PCR with selective primers, the level of infection of the eggs by P. cucumerina could be assessed. The proportion of infected eggs in cysts incubated in the soil from Ely was 15% (± 1.9) (n = 8), and in the soil from Spalding, the level of infection of the encysted eggs added as a bait was similar, at 14% (± 1.6) (n = 8). Both these results were significantly greater than the 9% (± 1.09) (n = 6) infection of eggs observed in the control (the probability value corresponding to the variance ration [F pr] was 0.015 [standard errors of differences, 2.7] at Ely and 0.021 [standard errors of differences, 2.74] at Spalding). This is assumed to represent a background level of fungal infection in the cysts used for the assays. Of the 50 infected eggs taken from the Ely baits, four colonies (8%) were identified by PCR as P. cucumerina, whereas 14 colonies (28%) were identified as P. cucumerina from the Spalding soil.
TABLE 5.
Quantification of P. cucumerina by serial plate dilution and real-time PCR in field samples taken from Ely and Spalding
| Field site | Results of quantification by:
|
|
|---|---|---|
| Plate dilution (CFU g soil−1) | Real-time PCR (no. of target sequences g soil−1)a | |
| Ely | <100 | 17,513 ± 5,908 |
| Spalding | 700 | 283,581 ± 49,954 |
Mean ± standard error from triplicate replicate of three samples taken from each site.
Real-time quantification of P. cucumerina from field sites.
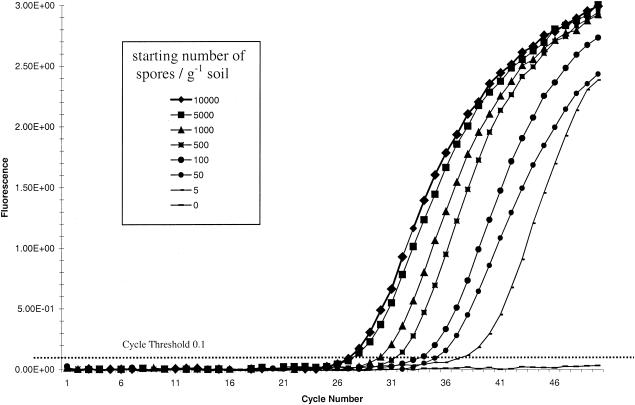
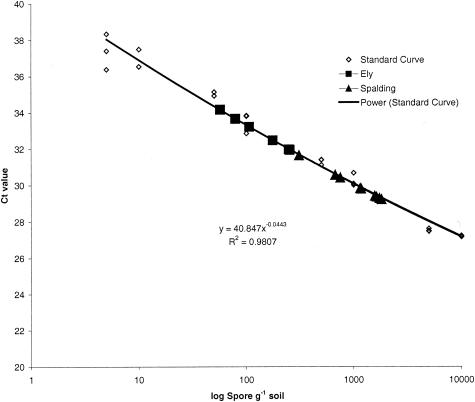
The primers PcTRF1 and PcRTR1 generated a PCR product of 72 bp. No fluorescent signal was recorded from negative controls or from unseeded soil-extracted DNA samples. The CT was 0.1, measuring amplification during the logarithmic phase of the PCR (Fig. 1). A standard curve for P. cucumerina was generated by using data derived from the serial dilution of P. cucumerina spores in soil (Fig. 2). The linear correlation coefficient of the standard curve was r2 = 0.981, demonstrating the accuracy of the PCR-based quantification. Based on three sample replications, the ABI Prism fluorescence detection system automatically calculated the starting concentrations of P. cucumerina DNA by a comparison of the CT values from the unknown samples with those of the standard curve (Fig. 2). By using this method, it was possible to consistently detect populations as low as 5 spores g soil−1. The conventional PCR primers could detect the presence of P. cucumerina in soils inoculated with 50 spores g soil−1 and above. Below this level the fungus was not detected. P. cucumerina was detected in two of the three samples taken from Ely and in all three of the samples from Spalding. The CT values from the Ely samples were lower than those from Spalding, indicating that the fungal population was smaller at Ely (Fig. 2). To determine the exact level of P. cucumerina target ITS sequences in each soil sample, the CT value was compared to the standard curve and then adjusted to determine P. cucumerina target sequences per gram of soil (Table 5). This level was recorded as the number of fungal target sequences per gram of soil or as the number of ITS repeats per genome or per nucleus; whether the target sequence occurred in uninucleate spores or multinucleate mycelia is unknown.
FIG. 1.
Real-time amplification plot of P. cucumerina ITS DNA from serially diluted sandy loam soil gDNA (10 g) inoculated with 108 conidiospores of. P. cucumerina-specific ITS real-time primers PcRtF1 and PcRtR1 and probe PcRtP1.
FIG. 2.
Real-time quantification of P. cucumerina from Ely and Spalding by plotting CT values against the P. cucumerina calibration curve.
DISCUSSION
This paper clearly demonstrates the effective use of a variety of methods used to determine the presence, viability, population size, and pathogenicity of a nematophagous fungus, P. cucumerina, a potential biological control agent against PCN. It also demonstrates the necessity for a range of techniques to be developed to assess fungal biocontrol agents in the environment.
The baiting method demonstrated that P. cucumerina in the soil was viable and able to parasitize cysts. When combined with PCR, this technique provided a method for direct identification of the fungus from infected nematode eggs.
Both PCR primer sets designed in this study have been shown to be species specific for P. cucumerina and were able to detect the fungus at low population levels, indicating the sensitivity of PCR-based methods. Real-time PCR provided a sensitive method for the quantification of P. cucumerina in soil, being able to detect 5 spores g soil−1, whereas the conventional PCR method was less sensitive, detecting only populations 10-fold greater. Both methods were more sensitive than serial plating of soil onto selective media, as they were able to detect the presence of P. cucumerina in soil from Ely when it was not isolated on the selective plates. As shown in other studies (8), the presence of multiple copies of the ITS region in the genome allows for detection of low concentrations of target DNA, providing a very sensitive diagnostic method.
Real-time PCR provides a method for determining the population of the fungus in the soil, whereas conventional PCR is able only to determine the presence or absence of the fungus and relies on information from serial dilutions on selective media to provide an estimate of the population size. In studies where the ITS copy number has been determined in fungi (10, 24), it has been around 100 per haploid genome; therefore, the estimate of the population size in both soils by real-time PCR could be 100-fold more than the number of nuclei, bringing the number of propagules closer to the CFU levels reported in Table 5. Further work is necessary to determine the copy number of the ITS gene in P. cucumerina to prove this assumption. Real-time PCR will provide a valuable tool in monitoring fungal fluctuations over time in response to exogenous inputs, including changes in crops, farm practices, chemical inputs, and the presence of nematodes on roots and in soil. This will provide a more accurate picture of the influence of the environment on the activity of a biological control agent. Although real-time PCR provides a quantitative estimate, enabling comparisons of the fungal population at the two soil sites, it was not possible to directly relate the value to the numbers of fungal propagules. PCR will amplify DNA from both nonviable as well as viable propagules, and therefore quantification is more closely related to number of fungal nuclei per g of soil than to the number of viable propagules, which, therefore, may be overestimated. The extraction of RNA from soil, combined with real-time PCR quantification, would provide a more meaningful assessment of cellular viability in the soil, and further work is necessary to develop the protocols necessary for this process. Nevertheless, real-time PCR provided a more sensitive method of quantification than that provided by serial dilution on selective media and was able to detect P. cucumerina organisms in Ely soils which were nondetectable using plating methods.
The selective medium developed for the isolation of P. cucumerina was significantly better at isolating the fungus than the medium developed for the isolation of P. chlamydosporia, particularly with greater populations of the fungus (Table 4). The selective medium allowed quantification of viable propagules of the fungus in the soil, which was not provided by the other methods detailed in the paper. Although dilution plate methods have significant limitations (16), the use of selective media that enable the estimation of relative changes in abundance have greatly increased our understanding of the ecology of selected fungi in soil. The development of a selective medium for P. cucumerina is a significant advance in the understanding of the ecology of this fungus.
The combination of baiting, plate dilution on selective medium, and PCR identification and quantification are powerful tools in the monitoring, detection, and quantification of P. cucumerina.
The fungus was detected and isolated from two sites in the United Kingdom where PCN populations have been shown to be on the decline. The causative agents for this PCN decline have not been established in either of the two field sites tested, and the role that P. cucumerina may play in the regulation of populations of this nematode pest needs further investigation. P. cucumerina has, however, been shown in previous studies to have potential as a biological control agent against a broad range of nematode species, including PCN (3, 15), and has also been isolated from other nematode hosts (27). Therefore, the methods outlined in this paper for the monitoring of P. cucumerina are applicable to further investigations into the potential of this fungal species for biological control of several nematode pests.
Acknowledgments
Rothamsted Research receives grant-aided support from the Biological Sciences Research Council of the United Kingdom. This work was funded by the United Kingdom Department for Environment, Food and Rural Affairs (Defra).
REFERENCES
- 1.Arora, D. K., Hirsch, P. R., Kerry, B. R. 1996. PCR-based molecular discrimination of Verticillium chlamydosporium isolates. Mycol. Res. 7:801-809. [Google Scholar]
- 2.Atkins, S. D., D. Sosnowska, V. J. Evans, I. M. Clark, P. R. Hirsch, and B. R. Kerry. 2002. Investigation of three nematophagous fungi in two potato cyst nematode suppressive soils. IOBC/WPRS Bull., in press.
- 3.Atkins, S. D., I. M. Clark, D. Sosnowska, and B. R. Kerry. 2002. Initial testing of potential fungal biological control agents of potato cyst nematodes, p. 79-84. British Crop Protection Council, Brighton, United Kingdom.
- 4.Avis, T. J., R. C. Hamelin, and R. R. Belanger. 2001. Approaches to molecular characterization of fungal biocontrol agents: some case studies. Can. J. Plant Pathol. 23:8-12. [Google Scholar]
- 5.Bates, J. A., E. J. A. Taylor, D. M. Kenyon, and J. E. Thomas. 2001. The application of real-time PCR to the identification, detection and quantification of Pyrenophora species in barley seed. Mol. Plant Pathol. 2:49-57. [DOI] [PubMed] [Google Scholar]
- 6.Bohm, J., A. Hahn, R. Schubert, G. Bahnweg, N. Adler, J. Nechwatal, R. Oehlann, and W. Oβwald. 1999. Real-time quantitative PCR: DNA determination in isolated spores of the mycorrhizal fungus Glomus mosseae and monitoring of Phytophthora infestans and Phytopthora citricola in their respective host plants. J. Phytopathol. 147:409-416. [Google Scholar]
- 7.Crump, D. H. 1998. Biological control of potato and beet cyst nematodes. Asp. Appl. Biol. 52:383-386. [Google Scholar]
- 8.Cullen, D. W., A. K. Lees, I. K. Toth, and J. M. Duncan. 2001. Conventional PCR and real-time PCR detection of Helminthosporium solani in soil and on potato tubers. Eur. J. Plant Pathol. 107:387-398. [Google Scholar]
- 9.De Leij, F. A. M. M., and B. R. Kerry. 1991. The nematophagous fungus Verticillium chlamydosporium as a potential biological control agent for Meloidogyne arenaria. Rev. Nematol. 14:157-164. [Google Scholar]
- 10.Garber, R. C., B. G. Turgeon, E. U. Selker, and O. C. Yoder. 1988. Organization of ribosomal RNA genes in the fungus Cochliobolus heterostrophus. Curr. Genet. 14:573-582. [DOI] [PubMed] [Google Scholar]
- 11.Gardes, M., T. J. White, J. A. Fortin, T. D. Bruns, and J. W. Taylor. 1991. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae. Can. J. Bot. 69:180-190. [Google Scholar]
- 12.Genstat 5 Committee. 1993. Genstat 5, release 3 reference manual. Clarendon Press, Oxford, United Kingdom.
- 13.Haydock, P. P. J., and K. Evans. 1998. Integrated crop management (ICM) protocols and the management of potato cyst nematodes. Asp. Appl. Biol. 52:361-366. [Google Scholar]
- 14.Hirsch, P. R., T. H. Mauchline, T. A. Mendum, and B. R. Kerry. 2000. Detection of the nematophagous fungus Verticillium chlamydosporium in nematode-infested plant roots using PCR. Mycol. Res. 104:435-439. [Google Scholar]
- 15.Jacobs, H. 2000. Development of a fungal biological control agent for potato cyst nematodes. PhD Thesis. University of Luton, Luton, United Kingdom.
- 16.Kerry, B. R. 2000. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant parasitic nematodes. Annu. Rev. Phytopathol. 38:423-441. [DOI] [PubMed] [Google Scholar]
- 17.Klimyuk, V. I., B. J. Carroll, C. M. Thomas, and J. D. G. Jones. 1993. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 3:493-494. [DOI] [PubMed] [Google Scholar]
- 18.Lovic, B. R., R. D. Martyn, and M. E. Miller. 1995. Sequence analysis of the ITS regions of RDNA in Monosporascus spp. to evaluate its potential for PCR mediated detection. Phytopathology 85:655-661. [Google Scholar]
- 19.Mitchell, D. J., M. E. Kannwischer-Mitchell, and D. W. Dickson. 1987. A semi-selective medium for isolation of Paecilomyces lilacinus from soil. J. Nematol. 19:255-256. [PMC free article] [PubMed] [Google Scholar]
- 20.Mulholland, V., L. Carde, K. J. O'Donnel, C. C. Fleming, and T. O. Powers. 1996. Use of the polymerase chain reaction to discriminate the potato cyst nematode at the species level, p. 247-252. In Proceedings of Diagnostics in Crop Production Conference. British Crop Protection Council, London, United Kingdom.
- 21.Orlando, C., P. P. Pinzani, and M. Pazzagli. 1998. Developments in quantitative PCR. Clin. Lab. Med. 36:255-269. [DOI] [PubMed] [Google Scholar]
- 22.Palm, M. E., W. Gams, and H. I. Nirenberg. 1995. Plectosporium, a new genus for Fusarium tabacinum, the anamorph of Plectosphaerella cucumerina. Mycologica 87:397-406. [Google Scholar]
- 23.Sikora, R. A., R. P. Schuster, and S. Kiewnick. 1994. Indexing biodiversity and antagonistic potential in agricultural soil of Madagaskar. Meded. Fac. Landbouwwet. Univ. Gent 59(2b):781-790. [Google Scholar]
- 24.Tsuchiya, D., and M. Taga. 2001. Application of fibre-FISH (fluorescence in situ hybridization) to filamentous fungi: visualization of the rRNA gene cluster of the ascomycete Cochliobolus heterostrophus. Microbiology 147:1183-1187. [DOI] [PubMed] [Google Scholar]
- 25.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innes, G. H. Gelfand, J. S. Sninsky, and T. J. White (ed.), PCR protocols. Academic Press, London, United Kingdom.
- 26.Winton, L. M., J. K. Stone, L. S. Watrud, amd E. M. Hansen. 2002. Simultaneous one-tube quantification of host and pathogen DNA with real-time polymerase chain reaction. Phytopathology 92:112-116. [DOI] [PubMed] [Google Scholar]
- 27.Yu, Q., and J. Coosemans. 1998. Fungi associated with cysts of Globodera rostochiensis, G. pallida, and Heterodera schachtii, and egg masses and females of Meloidogyne hapla in Belgium. Phytoprotection 79:63-69. [Google Scholar]