Abstract
Real-time PCR provides a means of detecting and quantifying DNA targets by monitoring PCR product accumulation during cycling as indicated by increased fluorescence. A number of different approaches can be used to generate the fluorescence signal. Three approaches—SYBR Green I (a double-stranded DNA intercalating dye), 5′-exonuclease (enzymatically released fluors), and hybridization probes (fluorescence resonance energy transfer)—were evaluated for use in a real-time PCR assay to detect Brucella abortus. The three assays utilized the same amplification primers to produce an identical amplicon. This amplicon spans a region of the B. abortus genome that includes portions of the alkB gene and the IS711 insertion element. All three assays were of comparable sensitivity, providing a linear assay over 7 orders of magnitude (from 7.5 ng down to 7.5 fg). However, the greatest specificity was achieved with the hybridization probe assay.
Members of the bacterial genus Brucella infect a wide range of animal hosts, including cows, goats, sheep, pigs, dogs, elk, bison, and even marine mammals. The genus Brucella comprises six species: B. abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae. It has been suggested from 16S rRNA gene sequence analysis and other biochemical characteristics (18, 29, 30) that the Brucella spp. constitute a monophyletic genus. Verger et al. (29, 30) proposed that the Brucella species be described as biovars of a single species, B. melitensis. However, Corbel (7) suggested that the existing nomenclature be retained for clarity. The extant literature associates particular species and biovars with brucellosis in different animal hosts: B. abortus with cows (also bison and elk), B. suis with pigs, B. ovis with sheep, B. melitensis with goats, and B. neotomae with desert wood rats. Brucellosis in bovine species can be associated specifically with B. abortus biovar 1, which accounts for 85% of reported cases (19). However, the ability of several Brucella species to infect organisms outside of their normal host range (e.g., B. melitensis and B. suis may infect cows, and B. melitensis, B. suis, B. abortus, and B. canis may infect humans) has been documented.
This pathogen has historically had a major economic impact on U.S. agriculture, especially the dairy industry, where it is estimated that over $3.5 billion has been spent to eradicate bovine brucellosis. It also represents a risk to humans, not only through the consumption of infected meat or milk (2) but also through the deliberate deployment of the organism as a biological weapon. B. suis was the first microorganism developed in the former U.S. biological weapons program, primarily because of the ease with which it can be spread through aerosol routes of infection (12). The effects of brucellosis in humans (also known as undulant fever) are generally nonlethal, but can be extremely debilitating.
Brucellosis remains a problem for U.S. agriculture today from the perspective of minimizing the potential for natural reservoirs of the disease to reinfect cattle, because brucellosis is nearly eradicated otherwise. As a consequence, wild bison and elk residing in areas such as Yellowstone National Park (YNP) are being studied to understand the relative risk wild animals pose to cattle ranging in the same region. This issue has generated intense interest after an Idaho cow was diagnosed with brucellosis in the spring of 2002. The long-term focus of the work reported here is to develop a rapid and sensitive real-time PCR assay for the specific detection of B. abortus in bison, cattle, and elk in the field.
Diagnostic methods for brucellosis currently rely on serological tests that detect antibodies against Brucella and cultivation. Blood or tissue cultures remain the standard for diagnosis, but since these microorganisms can be spread by aerosol routes of exposure, controlling the risk of infection in laboratory workers requires the use of additional biological containment measures, such as biosafety cabinets. Significant effort has been expended to develop DNA diagnostics for brucellosis that exploit the selectivity and sensitivity of PCR. Gene targets have included outer membrane proteins (3, 4, 8, 11, 13, 16), 16S rRNA gene sequences (15), 16S-23S spacer regions (23), housekeeping genes (8), erythritol utilization genes (6), and insertion sequences (6, 20, 22). These assays have been applied to bacterial isolates, as well as clinical specimens, with limits of detection down to 10 to 100 fg of bacterial genomic DNA and cell numbers ranging from 10 to 7,000 cells per ml of blood (13, 16, 17) or milk (16, 17, 23, 24, 26). Single primer sets or multiplexed assays (5, 6, 10, 26) with a cocktail of primers permit the detection of most of the known species or biovars of Brucella by conventional PCR. A variety of PCR assays have been developed that can differentiate Brucella species; however, they typically rely on post-PCR analysis, such as amplicon size (5), size discrimination combined with restriction fragment length polymorphism analysis (4, 8), or randomly amplified polymorphic DNA analysis (27). Most recently, real-time PCR assays for the detection of Brucella spp. have been developed (22).
Real-time PCR provides a means of detecting and quantifying DNA targets by monitoring PCR product accumulation, measured by increased fluorescence during cycling. A number of different approaches can be used to generate the fluorescence signal. In this work, three approaches: SYBR Green I (a double-stranded DNA [dsDNA] intercalating dye), 5′-exonuclease (enzymatically released fluors), and hybridization probes (fluorescence resonance energy transfer [FRET]) were evaluated for real-time PCR detection of B. abortus. Although the 5′-exonuclease assay (TaqMan) is dependent upon the annealing of a single probe to the target amplicon, in this article the term “hybridization probe assay” refers exclusively to the use of two adjacent probes in which fluorescence is generated by resonance energy transfer from a donor fluor on one probe to an acceptor fluor on the probe 2 bp away.
MATERIALS AND METHODS
Genomic DNA for PCR analysis.
The bacterial strains used to test the specificity of the primers and probes used in this study are listed in Table 1. Genomic DNA was used as the template for PCR cross-reactivity assessments. Samples provided by YNP were purified from methanol-killed strains obtained from the American Type Culture Collection (ATCC) or the National Animal Disease Center (NADC). Strains of the nearest relatives (ATCC and field isolates) were grown on ATCC-recommended media (http://www.atcc.org). Bordetella bronchiseptica, Ochrobactrum anthropi, Rhizobum leguminosarum, R. radiobacter, and R. rhizogenes strains were grown on nutrient agar; the R. radiobacter field isolate was grown on ATCC medium 1213 (King medium B), and Sinorhizobium meliloti was grown on ATCC medium 111 (Rhizobium X). Genomic DNA was subsequently isolated in our laboratory. All other genomic DNAs were supplied by the Armed Forces Institute of Pathology (AFIP).
TABLE 1.
Specificity of real-time PCR assays for the detection of B. abortus
| Template origin | Strain | Source | Result by:
|
||
|---|---|---|---|---|---|
| SYBR Green I assay | 5′-Exonuclease assay | Hybridization probe assay | |||
| Brucella abortus (standard curve reference strain)a | NADC 1144 | YNP | + | + | + |
| B. abortus | ATCC 4315 | YNP | + | + | + |
| B. abortus bv. 1 | ATCC 23448 | YNP | + | + | + |
| B. abortus bv. 2 | ATCC 23449 | YNP | + | + | + |
| B. abortus bv. 3 | ATCC 17385 | YNP | + | + | + |
| B. abortus bv. 4 | ATCC 23451 | YNP | + | + | + |
| B. abortus bv. 5 | ATCC 23452 | YNP | + | + | + |
| B. abortus bv. 9 | ATCC 23455 | YNP | + | + | + |
| B. abortusa | NADC 1145 | YNP | + | + | + |
| B. abortusa | NADC 1403 | YNP | + | + | + |
| B. abortusa | NADC 1409 | YNP | + | + | + |
| B. abortus | NADC 2308 | YNP | + | + | + |
| B. abortus | NADC RB51 | YNP | + | + | + |
| B. canis | ATCC 23365 | AFIP | + | + | ± |
| B. canis | ATCC 6133 | AFIP | + | + | − |
| B. melitensis | ATCC 16M | AFIP | + | − | − |
| B. neotomae | ATCC 23459 | AFIP | + | − | − |
| B. ovis | ATCC 25840 | AFIP | + | − | − |
| B. suis | ATCC 23444 | AFIP | + | − | − |
| Bordetella bronchiseptica | ATCC 10580 | ATCC | + | − | − |
| Ochrobactrum anthropi | ATCC 49237 | ATCC | + | − | − |
| Rhizobium leguminosarum | ATCC 14479 | ATCC | + | − | − |
| R. radiobacter | ATCC 15955 | ATCC | + | − | − |
| R. radiobacterb | ATCC 19358 | ATCC | + | − | − |
| R. radiobacterb | Field isolate | Our lab | + | − | − |
| R. rhizogenes | ATCC 15834 | ATCC | + | − | − |
| Sinorhizobium meliloti | ATCC 10310 | ATCC | + | − | − |
Bison isolate.
Strains previously classified as Agrobacterium tumefaciens.
Instrumentation.
The field-portable Ruggedized Advanced Pathogen Identification Device (R.A.P.I.D.; Idaho Technology, Salt Lake City, Utah) was used for amplification and real-time quantification. This instrument holds up to 32 glass capillary reaction tubes in a rotating carousel. Air is the medium of heat transfer, facilitating rapid temperature transitions, resulting in shorter cycling times. Melt curves were generated by using the LightCycler Data Analysis software, version 3.1.106i, supplied with the R.A.P.I.D., with the digital filter enabled and a linear curve fit selected for quantification and a polynomial curve fit selected for melt curve analyses.
Primers and probes.
All assays utilized the same amplification primers, BAF and BAR (Table 2), to produce an identical 156-bp amplicon spanning a region of the B. abortus genome that includes portions of the alkB gene and the IS711 insertion element. These primers were designed using MacVector 7.0 (Accelrys, San Diego, Calif.). The theoretical specificity of the primers was determined by comparison to the GenBank database using the Basic Local Alignment Search Tool (BLAST) (1). The analysis of the primers indicated that they were specific for B. abortus. The sequence of the 5′-exonuclease probe, designed at the Armed Forces Institute of Pathology, is also given in Table 2. Hybridization probes for B. abortus (Table 2) were designed with MeltCalc (25). This software package was developed specifically for the design of hybridization probes and allows the user to set reagent concentrations, Tm values, and gaps between probes. In addition, it screens for interactions among the probes and primers so that noninteracting oligonucleotides can be selected.
TABLE 2.
Oligonucleotide sequences developed for assay of B. abortus
| Oligonucleotide | Position and orientation (B. abortus numbering; gb AF148682) | Sequence |
Tm (°C)a
|
Reference | ||
|---|---|---|---|---|---|---|
| TM Utility, version 1.3 | MeltCalc 2.06 Pro | R.A.P.I.D. Exp. | ||||
| BAF primer | Forward (1381-1399) | 5′-CCATTGAAGTCTGGCGAGC-3′ | 67.6 | 68.1 | 86.5 (amplicon) | This work |
| BAR primer | Reverse (1537-1516) | 5′-CGATGCGAGAAAACATTGACCG-3′ | 68.7 | 69.4 | This work | |
| TH 5′-exonuclease probe | Reverse (1492-1473) | 5′-FAM-GCATGCGCTATGATCTGGTTACGTT-(TAMRA)-3′ | 70.4 | 70.7 | Hadfield (unpublished) | |
| BAFR FAM hybridization probe | Reverse (1470-1489) | 5′-TGCGCTATGATCTGGTTACG-(FAM)-3′ | 64.9 | 64.6 | 63 (probe) | This work |
| BAFR CY5.5 Hybridization probe | Reverse (1467-1450) | 5′-(CY5.5)-AAATGCAGACACGCCCTA-(P)-3′ | 65.7 | 66.1 | This work | |
The Tm for the primer or probe was calculated by using Idaho Technology's TM Utility version 1.3 with optimized conditions for each reaction, MeltCalc 2.06 Pro, or R.A.P.I.D. For R.A.P.I.D., the values shown represent the average experimental value based on R.A.P.I.D. melt curve analysis with B. abortus NADC 1144 as the template.
Real-time PCR assay conditions.
Roche LightCycler kits, optimized for use with glass capillaries and containing a hot start polymerase, were used as the master mix base for all reactions. Each reaction mixture contained 5.0 mmol of MgCl2 and 500 nmol of each primer in a final volume of 20 μl. The LightCycler-FastStart DNA Master SYBR Green I kit was used for SYBR Green I assays. 5′-Exonuclease reaction chemistry consisted of the LightCycler-FastStart DNA Master Hybridization Probes kit with 100 nmol of the TH 5′-exonuclease probe (Table 2). The LightCycler-FastStart DNA Master Hybridization Probes kit was also used for the hybridization probe assay, which contained 300 nmol of BAFR-FAM probe and 500 nmol of BAFR-Cy5.5 probe (Table 2). Specific reaction conditions and cycling regimes for each of the three assay types are detailed in Table 3. Each template was tested at least three times to confirm the reproducibility of the assays.
TABLE 3.
Cycling regimes for SYBR Green I, 5′-exonuclease, and hybridization probe assays performed on the R.A.P.I.D. with Roche real-time PCR reagent kits
| Cycling regime | SYBR Green I
|
5′-Exonuclease
|
Hybridization probes
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of cycles | Analysis mode | Target temp (°C) | Incuba- tion time (s) | Transi- tion rate (°C/s) | Acquisi- tion mode | Gainsa | No. of cycles | Analysis mode | Target temp (°C) | Incuba- tion time (s) | Transi- tion rate (°C/s) | Acquisi- tion mode | Gainsa | No. of cycles | Analysis mode | Target temp (°C) | Incuba- tion time (s) | Transi- tion rate (°C/s) | Acquisi- tion mode | Gainsa | |
| Hot start (segment 1) | 1 | None | 95 | 600 | 20.0 | None | 1 | None | 95 | 600 | 20.0 | None | 1 | None | 95 | 600 | 20.0 | None | |||
| Amplification | 55 | Quantification | 55 | Quantification | 55 | Quantification | |||||||||||||||
| Segment 1 | 95 | 0 | 20.0 | None | 95 | 0 | 20.0 | None | 95 | 0 | 20.0 | None | |||||||||
| Segment 2 | 59 | 0 | 20.0 | None | 59 | 20 | 20.0 | Single | Ch 1 = 32 | 59 | 4 | 20.0 | Single | Ch 3 = 16 | |||||||
| Segment 3 | 72 | 12 | 20.0 | None | NAb | NA | NA | NA | 72 | 12 | 20.0 | None | |||||||||
| Segment 4 | 82 | 0 | 20.0 | Single | Ch 1 = 8 | NA | NA | NA | NA | NA | NA | NA | NA | ||||||||
| Melt | 1 | Melt curves | 1 | Melt curves | |||||||||||||||||
| Segment 1 | 95 | 60 | 20.0 | None | NA | NA | NA | NA | 95 | 60 | 20.0 | None | |||||||||
| Segment 2 | 60 | 120 | 5.0 | None | NA | NA | NA | NA | 45 | 120 | 5.0 | None | |||||||||
| Segment 3 | 94 | 0 | 0.2 | Step | Ch 1 = 8 | NA | NA | NA | NA | 75 | 0 | 0.2 | Step | Ch 3 = 16 | |||||||
Ch, channel.
NA, not applicable.
Standard curves and efficiency.
The standard curve was generated by the Fit Points analysis method included in the LightCycler Data Analysis software, version 3.1.106i, with the fit point number set at 2. Using this method, the log of the concentration of a dilution series of the standard or reference template DNA (B. abortus NADC strain 1144) was plotted versus the cycle number at which the fluorescent signal increased above background or threshold (Ct value). The slope of the standard curve generated for each detection approach was input into the following equation to determine reaction efficiency: efficiency = 10−(1/slope) (21).
RESULTS
Sensitivity and specificity.
All three assays demonstrated similar sensitivity when using genomic B. abortus from NADC strain 1144 as a template, with a linear assay over 7 orders of magnitude (from 7.5 ng down to 7.5 fg). Figure 1 shows the amplification curves generated for each of the three detection approaches. Based on the whole genome sequence (size and GC content) of B. melitensis (9), we estimate that the lower limit of detection corresponds to detection of two genome copies of B. abortus. Multiple copies of IS711 are known to be distributed in the genomes of the Brucella, but for the purposes of this calculation, it was assumed that only one copy was inserted into the alkB locus (14).
FIG. 1.
Amplification curves for SYBR Green I (A), 5′-exonuclease (B), and hybridization probe (C) assays. Dilutions of B. abortus genomic DNA (NADC strain 1144) were used as a template for each reaction. A 10-fold dilution series (from 7.5 ng down to 7.5 fg) was analyzed in triplicate. LightCycler Data Analysis software, version 3.1.1.06i, was used to generate curves by plotting cycle number (x axis) against fluorescence, read in channel 1 for SYBR Green I and 5′-exonuclease assays and channel 3 for the hybridization probe assay. For clarity, only one curve per concentration is depicted, with concentrations decreasing from left to right.
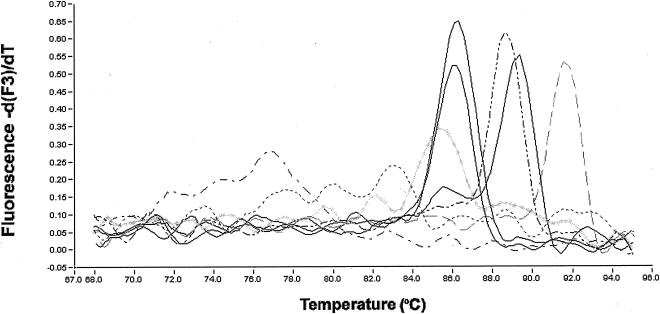
In Table 1, real-time PCR assays are arranged such that overall specificity increases from left to right. The cross-reactivity for each genomic DNA examined is also indicated. All genomic DNAs tested generated a fluorescent signal when SYBR Green I was used as the detection approach. Representative melt curves for amplicons generated in the SYBR Green I assay are shown in Fig. 2, with peak Tms indicated in Table 4. Seven B. abortus biovar 1 strains, as well as samples of biovars 2 and 4, generated coincident peak Tms that could be clearly distinguished from all other Brucella and closely related species, with the exception of R. rhizogenes. Melting of products for three biovars of B. abortus (biovars 3, 5, and 9) produced two peaks, one of which was close to that of the other B. abortus samples and another peak with a Tm approximately 3°C higher. Agarose gel electrophoretic analysis of the amplicons provided further evidence of the lack of specificity of the SYBR Green I assay, because heterogeneous products were observed, except for those generated from authentic B. abortus templates (data not shown). The 5′-exonuclease assay demonstrated improved specificity compared to the SYBR Green I assay, since it did not detect any of the closely related genera. However, both B. canis templates generated fluorescence signals. The hybridization probe assay was the most specific, with only one of the two B. canis strains tested giving positive amplification results and only when present at concentrations greater than 10 pg (indicated in Table 1 by “±”). Melt curve analysis of the hybridization probe assay products was unable to discriminate between B. abortus and B. canis. In each case, a single peak with a Tm of 63°C was observed (Table 2) (data not shown).
FIG. 2.
Representative SYBR Green I amplicon melt curves analyzed with LightCycler Data Analysis software, version 3.1.1.06i, with the digital filter enabled and polynomial curve fitting. Shown are results for the no-template control (— - —), the distantly related strain B. bronchiseptica ATCC 10580 (------), B. abortus NADC 1144 (——), B. abortus biovar 1 ATCC 23448 (coincident with NADC 1144) (——), B. abortus biovar 3 ATCC 17385 (doublet peaks with markedly higher Tm) (——), O. anthropi (— .. —), R. leguminosarum (— — —), and R. rhizogenes ATCC 15834 ( ).
TABLE 4.
Melt curve analysis of SYBR Green I amplification products
| Template origin | Avg amplicon Tm (°C)a |
|---|---|
| B. abortus NADC 1144, 1145, 1403, 1409, 2308, RB51, bv. 1, 2, 4 | 86.4 |
| B. abortus bv. 3, 5, 9 | 85.8, 89.5 |
| B. canis, B. melitensis, B. neotomae, B. ovis, B. suis | 89.6 |
| R. rhizogenes ATCC 15955, R. rhizogenes (field isolate), S. meliloti ATCC 10310, O. anthropi ATCC 49237 | 89.2 |
| R. radiobacter ATCC 19358, R. rhizogenes ATCC 15834 | 85.6 |
| R. leguminosarum ATCC 14479 | 91.6 |
| B. bronchiseptica ATCC 10580 | 80.5 |
Tms listed are for each prominent peak in the melt curve analysis.
Reaction efficiency.
The equations for the linear regression line for the standard curve generated by each detection approach and its corresponding R2 value are as follows: SYBR Green I, y = −4.29x + 21.42, R2 = 0.9953; 5′-exonuclease, y = −3.31x + 21.14, R2 = 0.9985; hybridization probe, y = −3.59x + 22.54, R2 = 0.9956. Based on the slopes of these lines, the levels of efficiency were 1.71 for the SYBR Green I assay, 2.00 for the 5′-exonuclease assay, and 1.90 for the hybridization probe assay.
DISCUSSION
A perfect amplification, in which each target sequence is replicated every cycle, would have an efficiency of 2.00. Conversely, a reaction in which no amplification occurred would have an efficiency of 1.00. The efficiency for each of these real-time assays using B. abortus genomic DNA as a template ranged from 1.71 to 2.00, indicating that in all cases, at least moderate amplification efficiency was achieved. In the case of the SYBR Green I assay, the lower efficiency may be explained by the presence of the intercalating dye, which can be inhibitory to PCR (32). The reduced efficiency of the hybridization probe assay relative to the 5′-exonuclease assay may be a function of the necessity for four oligonucleotides to anneal in order for amplification and detection to occur.
The fact that all templates examined generated a fluorescent signal in the SYBR Green I assay (Table 1 and Table 4) emphasizes the need for experimental confirmation of the in silico analysis that indicated primers were specific for B. abortus. Furthermore, although SYBR Green I assays have the advantage of not requiring probe design, the lack of additional discrimination that can be gained by incorporating one or more probes may result in false positives. This may be overcome in part by conducting a melt analysis and/or gel analysis, but then the benefits of real-time PCR are lost.
Using hybridization probes, the only nontarget organism found to cross-react was the organism most closely related to B. abortus, B. canis (28; P. Keim, personal communication), and only when present at concentrations greater than 10 pg (indicated in Table 1 by “±”). Cross-reactivity at high concentrations of template DNA was also seen in the development of a real-time PCR assay for detection of legionellae (31).
There are two primary advantages for the use of hybridization probes in real-time PCR. First, the requirement that two additional probes must bind, as compared to one probe in 5′-exonuclease assays, can result in increased specificity. This specificity may be particularly important in cases in which very little sequence divergence between closely related targets exists, as is the case in Brucella. Second, detection via hybridization probes is not dependent on a hydrolysis reaction, and thus, a melt analysis can be conducted. Thus, not only does use of hybridization probes require complementarity of two primers and two probes with a target sequence, but it also permits discrimination based on the probe Tm.
Redkar et al. (22) also developed a hybridization probe-based real-time PCR assay for the detection of B. abortus that used primers derived from the IS711 insertion element and the species-specific chromosomal locus, alkB. The reported sensitivity of this assay was 16 to 50 genome copies with methanol-killed cells as a template. However, in our laboratory, no fluorescence signal could be detected by this assay with B. abortus genomic DNA as a template. Analysis of the primer and probe sequences provided predicted an amplicon size of 173 bp that completely contained the amplicon described in this work (156 bp). In theory, a smaller amplicon is preferred for real-time PCR assays. Differences in the amplification primers and probe sequences compared with published alkB sequences (mismatches were identified by BLAST) or in the locations of these primers and probes relative to those described here may also account for the inability to reproduce the previous work. Our assay worked reliably and had a lower limit of detection (7.5 fg).
Redkar et al. (22) also reported the successful design of hybridization probe assays for the specific detection of B. melitensis and B. suis with only slight modification in oligonucleotide sequences from those for B. abortus, suggesting the feasibility of using hybridization probes to selectively detect very closely related species. We made no attempt to reproduce these assays.
All three of the detection approaches examined successfully detected the B. abortus amplicon generated with primers BAF and BAR (Table1); however, only the hybridization probe assay afforded species-level discrimination. Given the high degree of similarity between Brucella spp. and their nearest neighbors and the results presented here, we anticipate that the use of the hybridization probe assay will permit reliable assessment of B. abortus-infected animals in the field and will serve as a useful tool for protecting the public and agriculture against bioterrorism.
Acknowledgments
This work was supported with funding provided by the U.S. Department of Energy, Office of Intelligence, to the Idaho National Engineering and Environmental Laboratory, operated by Bechtel BWXT Idaho, LLC, under contract DE-AC07099-ID13727.
We thank Robert Lindstrom, Yellowstone National Park, for providing cells and DNA originating from the ATCC and NADC.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Brucellosis. [Online.] World Health Organization, Geneva, Switzerland. www.who.int/inf-fs/en/fact173.html
- 3.Bailey, G. G., J. B. Krahn, B. S. Drasar, and N. G. Stoker. 1992. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J. Trop. Med. Hyg. 95:271-275. [PubMed] [Google Scholar]
- 4.Barrera-Saldaña, H. A., A. M. Sifuentes-Rincón, and A. Revol-de Mendoza. 1996. Diagnosticó de brucelosis por la reacción en cadena de la polimerasa. Gac. Med. Mex. 132:300-302. [Google Scholar]
- 5.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker, B. J., and S. M. Halling. 1995. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J. Clin. Microbiol. 33:1640-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbel, M. J. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Costa, M., J. P. Guillou, B. Garin-Bastuji, M. Thiébaud, and G. Dubray. 1996. Specificity of six gene sequences for the detection of the genus Brucella by DNA amplification. J. Appl. Bacteriol. 81:267-275. [DOI] [PubMed] [Google Scholar]
- 9.DelVecchio, V., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, L. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J.-J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewalt, D. R., and B. J. Bricker. 2000. Validation of the abbreviated Brucella AMOS PCR as a rapid screening method for differentiation of Brucella abortus field strain isolates and the vaccine strains, 19 and RB51. J. Clin. Microbiol. 38:3085-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fekete, A., J. A. Bantle, S. M. Halling, and M. R. Sanborn. 1990. Preliminary development of a diagnostic test for Brucella using polymerase chain reaction. J. Appl. Bacteriol. 69:216-227. [DOI] [PubMed] [Google Scholar]
- 12.Franz, D. R., P. B. Jahrling, A. M. Friedlander, D. J. McClain, D. L. Hoover, W. R. Byrne, J. A. Pavlin, G. W. Christopher, and E. M. Eitzen, Jr. 1999. Clinical recognition and management of patients exposed to biological warfare agents, p. 37-79. In J. Lederberg (ed.), Biological weapons: limiting the threat. Belfer Center for Science and International Affairs, Cambridge, Mass. [DOI] [PubMed]
- 13.Guarino, A., L. Serpe, G. Fusco, A. Scaramuzzo, and P. Gallo. 2000. Detection of Brucella species in buffalo whole blood by gene-specific PCR. Vet. Rec. 147:634-636. [DOI] [PubMed] [Google Scholar]
- 14.Halling, S. M., and E. S. Zehr. 1990. Polymorphism in Brucella spp. due to highly repeated DNA. J. Bacteriol. 172:6637-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman, L., and H. De Ridder. 1992. Identification of Brucella spp. by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:2099-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leal-Klevezas, D. S., I. O. Martínez-Vázques, A. López-Merino, and J. P. Martínez-Soriano. 1995. Single-step PCR for detection of Brucella spp. from blood and milk of infected animals. J. Clin. Microbiol. 33:3087-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leal-Klevezas, D. S., I. O. Martínez-Vázques, J. Garcia-Cantú, A. López-Merino, and J. P. Martínez-Soriano. 2000. Use of polymerase chain reaction to detect Brucella abortus biovar 1 in infected goats. Vet. Microbiol. 75:91-97. [DOI] [PubMed] [Google Scholar]
- 18.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicoletti, P. 1980. The epidemiology of bovine brucellosis. Adv. Vet. Sci. Comp. Med. 24:69-98. [PubMed] [Google Scholar]
- 20.Ouahrani-Bettache, S., M. P. Soubrier, and J. P. Liautard. 1996. IS6501-anchored PCR for the detection and identification of Brucella species and strains. J. Appl. Bacteriol. 81:154-160. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen, R. 2001. Quantification on the LightCycler, p. 21-34. In S. Meuer, C. Wittwer, and K.-I. Nakagawara (ed.), Rapid cycle real-time PCR. Springer, New York, N.Y.
- 22.Redkar, R., S. Rose, B. J. Bricker, and V. DelVecchio. 2001. Real-time detection of Brucella abortus, Brucella melitensis and Brucella suis. Mol. Cell. Probes 15:43-52. [DOI] [PubMed] [Google Scholar]
- 23.Rijpens, N. P., G. Jannes, M. Van Asbroeck, R. Rossau, and L. M. F. Herman. 1996. Direct detection of Brucella spp. in raw milk by PCR and reverse hybridization with 16S-23S rRNA spacer probes. Appl. Environ. Microbiol. 62:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero, C., and I. Lopez-Goni. 1999. Improved method for purification of bacterial DNA from bovine milk for detection of Brucella spp. by PCR. Appl. Environ. Microbiol. 65:3735-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schütz, E., and N. von Ahsen. 1999. Spreadsheet software for thermodynamic melting point prediction of oligonucleotide hybridization with and without mismatches. BioTechniques 27:1218-1224. [DOI] [PubMed] [Google Scholar]
- 26.Sreevatsan, S., J. B. Bookout, F. Ringpis, V. S. Perumaalla, T. A. Ficht, L. G. Adams, S. D. Hagius, P. H. Elzer, B. J. Bricker, G. K. Kumar, M. Rajasekhar, S. Isloor, and R. R. Barathur. 2000. A multiplex approach to molecular detection of Brucella abortus and/or Mycobacterium bovis infection in cattle. J. Clin. Microbiol. 38:2602-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tcherneva, E., N. P. Rijpens, B. Jersek, and L. M. Herman. 2000. Differentiation of Brucella species by random amplified polymorphic DNA analysis. J. Appl. Microbiol. 88:69-80. [DOI] [PubMed] [Google Scholar]
- 28.Velasco, J., C. Romero, I. Lopez-Goñi, J. Leiva, R. Diaz, and I. Moriyon. 1998. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 48:759-768. [DOI] [PubMed] [Google Scholar]
- 29.Verger, J.-M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35:292-295. [Google Scholar]
- 30.Verger, J.-M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1987. Taxonomy of the genus Brucella. Ann. Inst. Pasteur/Microbiol. 138:235-238. [DOI] [PubMed] [Google Scholar]
- 31.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittwer, C. R., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-139. [DOI] [PubMed] [Google Scholar]