Abstract
Detection of six species of lactic acid bacteria and six species of gram-positive catalase-positive cocci from low-acid fermented sausages (fuets and chorizos) was assessed by species-specific PCR. Without enrichment, Lactobacillus sakei and Lactobacillus curvatus were detected in 11.8% of the samples, and Lactobacillus plantarum and Staphylococcus xylosus were detected in 17.6%. Enriched samples allowed the detection of L. sakei and S. xylosus in all of the samples (100%) and of Enterococcus faecium in 11.8% of the sausages. The percentages of L. curvatus, L. plantarum, Staphylococcus carnosus, and Staphylococcus epidermidis varied depending on the sausage type. L. curvatus was detected in 80% of fuets and in 57% of chorizos. L. plantarum was found in 50% of fuets and 100% of chorizos. S. epidermidis was detected in only 11.8% of fuets, and S. carnosus was detected in only 5.9% of chorizos. Lactococcus lactis, Staphylococcus warneri, and Staphylococcus simulans were not detected in any sausage type. From a microbiological point of view, 70.6% of the samples could be considered of high quality, as they had low counts of Enterobacteriaceae and did not contain any of the food-borne pathogens assayed.
Low-acid fermented meat products (final pH, 5.3 to 6.2) are a group of traditional Mediterranean products with a great diversity within the different regions. The knowledge and control of their typical in-house microflora and the production processes are critical in terms of their microbiological quality and organoleptic characteristics. Food safety is a top priority for the European Communities, as indicated in the White Paper on Food Safety (19), and it is regulated by Commission of European Communities directive 93/43/CEE (22). The diversity of natural resources, traditions, competitiveness, and income levels of the agricultural sector are emphasized by the European Union in Agenda 2000 (18). The exploitation of the natural fermentative communities of slightly fermented sausages may diversify the market with different types of starter cultures that will be able to produce many typical regional slightly fermented sausages with their specific flavors.
Lactic acid bacteria (LAB) and coagulase-negative staphylococci (CNS) are the two main groups of bacteria that are considered technologically important in the fermentation and ripening of cured sausages. LAB are responsible for lactic acid production, for the “tangy” flavor of sausages, and for the small amounts of acetic acid, ethanol, acetoin, carbon dioxide, and pyruvic acid that are produced during fermentation, depending on the starter applied, the carbohydrate used, and the sources of meat proteins and additives (5, 26, 78). Staphylococcus and Kocuria are important for color stabilization, decomposition of peroxides (69; C. Barrière, M. C. Montel, and R. Talon, presented at the 44th International Congress of Meat Science and Technology, Barcelona, Spain, 1998), and aroma with their proteolytic and lipolytic activities (11, 12, 23, 47, 69, 72; C. Sajber, R. Karakas, and P. Mitic, presented at the 17th European Meeting of Meat Research Workers, Bristol, United Kingdom, 1971).
Among LAB, Lactobacillus sakei, Lactobacillus curvatus, and Lactobacillus plantarum are the species most widely described in acid-fermented meat products (35, 37, 48, 51, 62, 63, 66, 68, 79). However, in some slightly fermented sausages, such as Salame Felino, enterococcus and lactobacillus populations are very balanced (24). Within the group of gram-positive coagulase-negative cocci isolated from meat, CNS are mainly isolated from dry cured and ripened meats, whereas Kocuria spp. are dominant in freshly prepared sausages (16, 57, 74). Nevertheless, Kocuria varians and Kocuria kristinae are identified in 36 and 3% of dry sausages, respectively (27). Staphylococcus xylosus is frequently isolated as the main Staphylococcus species, but others have also been reported: Staphylococcus carnosus, Staphylococcus simulans, Staphylococcus saprophyticus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus warneri, and Staphylococcus equorum (16, 21, 27, 30, 38, 52, 70, 74, 80).
Phenotypic identification of fermentative microflora is time-consuming and often problematic due to ambiguous biochemical or physiological traits (36, 37, 73). In the last few years, the development of molecular typing methods has offered the possibility of accelerating a great deal of bacterial identification; moreover, direct sampling in complex matrices, such as foods, may avoid biases related to traditional methods.
Several molecular techniques have been applied for the identification of the main bacterial population isolated from meat products, such as sodium dodecyl sulfate-polyacrylamide gel electrophoresis of whole-cell proteins (64), hybridization with rRNA probes (49), restriction fragment length polymorphism analysis of the 16S rRNA gene (67), randomly amplified polymorphic DNA-PCR analysis (4, 7), PCR-temperature gradient gel electrophoresis (15), and species-specific PCR (83). Cocolin et al. (14) reported a PCR-denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during the fermentation of Italian sausages.
16S and 23S rRNA genes have been widely used for species identification and strain detection. However, in the case of closely related species, such as L. sakei and L. curvatus, 16S and 23S rRNA probes and primers cannot be used because of the high similarity of rRNA sequences. The variations in length and sequence of the 16S-23S rRNA intergenic spacer regions of the rRNA operon (rrn) have proved useful for identification of strains and species (6, 34, 41); the evolutionary rate of the region is 10 times greater than that of 16S rRNA, allowing one to distinguish closely related bacterial species (34).
Species-specific PCR is a rapid and reliable molecular technique for the characterization of bacterial communities without colony isolation; however, the sensitivity of PCR in foods can be reduced due to the complexity of the food matrix and the presence of many PCR inhibitors. Substances which have been proven to be PCR inhibitory, such as proteinases (53), heme compounds (1), chelating agents, and proteins (10, 59), may be present in fermented-sausage homogenates. Thus, many sample preparation methods, such as dilution, centrifugation, filtration, aqueous two-phase systems, adsorption methods, and DNA extraction, have been developed to overcome the effects of PCR-inhibitory substances (45).
The aim of this study was the assessment of the microbial hygienic quality of low-acid fermented sausages purchased at supermarkets and the identification of the main bacterial species of technological interest through development of species-specific PCR techniques, allowing the main species of LAB and CNS present in sausages to be identified directly from a food sample.
MATERIALS AND METHODS
Microbial and physicochemical analyses.
Seventeen ∼30-mm-diameter samples of commercial low-acid fermented sausages (pH 5.3 to 6.2), 10 fuets (cold ripened fermented sausages with black pepper) and 7 chorizos (cold ripened fermented sausages with red pepper), from different producers were analyzed in terms of hygienic quality and technologically important microflora (LAB and gram-positive catalase-positive cocci [GCC+]).
After removal of the casing, 10 g of each sample was homogenized in 90 ml of 0.1% peptone (Difco Laboratories, Detroit, Mich.) and 0.85% NaCl (Merck, Darsmstadt, Germany), pH 7.0, in a Stomacher Lab-Blender (model 400; Cooke Laboratories, Alexandria, Va.). After appropriate dilutions, the following determinations were carried out: LAB were enumerated by pour plating in MRS agar (Difco Laboratories) at 30°C for 72 h in anaerobiosis (Oxoid jars with AnaeroGen; Oxoid, Basingstoke, Hampshire, England), GCC+ were enumerated by spread plating in mannitol salt agar (Difco Laboratories) at 30°C for 2 days, enterococci were enumerated in poured kanamycin-esculin-azide agar (KAA) plates (Oxoid) at 37°C for 24 h, S. aureus was enumerated in poured Baird-Parker plates with rabbit plasma fibrinogen supplement (bioMerieux, Marcy l'Etoile, France) at 37°C for 48 h, sulfite-reducing clostridia were enumerated in SPS-agar tubes overlaid with paraffin (Merck) at 37°C for 24 h, Enterobacteriaceae were enumerated by pour plating in violet red bile glucose agar (Merck) at 30°C for 24 h, and Escherichia coli was enumerated by pour plating in Coli-ID medium (bioMerieux) at 37°C for 24 to 48 h. Microbiological counts were expressed as log CFU per gram.
The enumeration of Listeria monocytogenes was done in a 25-g sample by the most-probable-number (MPN) method. One milliliter of each of the three successive 10-fold dilutions was transferred into a three-test-tube set containing 10 ml of University of Vermont medium (UVM) I and incubated for 24 h at 30°C. A second step was carried out by transferring 1 ml from each tube to a new three-test-tube set containing 10 ml of UVM II and incubating the tubes at 30°C for 24 h, followed by spread plating in Palcam agar (Merck) for 48 h at 30°C. Typical colonies were confirmed by PCR (42) and API Listeria (reference no. 10300; bioMerieux). Salmonella was investigated in 25 g of the sample according to ISO 6579:1990(F) (39a).
The pH was determined by inserting into the sausages a model 52-32 electrode (Crison Instruments, S.A., Barcelona, Spain). Water activity (aw) was measured with an aw cryometer (awk-10; Nagy, Gäufelden, Germany). The fat, protein, and phosphates were analyzed by official Spanish methods for meat products (54); sodium chloride, nitrate, and nitrite contents were evaluated in the segmented continuous-flow Autoanalyzer II sampler (Technicon Ltd. Dublin, Ireland) by methods US-229-72A and 230-72A, as recommended by the manufacturer.
Reference strains.
The culture collection strains used as reference strains for evaluating the primers are listed in Table 1.
TABLE 1.
Reference strains and sources
| Species | Strain | Sourcea |
|---|---|---|
| L. sakei | CTC 494, CTC 232 | Spanish fermented sausages |
| L. curvatus | CTC 435, CTC 371 | Spanish fermented sausages |
| L. plantarum | CTC 305, CTC 300 | Spanish fermented sausages |
| E. faecium | CTC 492 | Spanish fermented sausages |
| CECT 410T | Equivalent to ATCC 19434, DSMZ 20477 | |
| Enterococcus faecalis | CECT 481 | Equivalent to ATCC 19433, DSMZ 20033 |
| L. lactis | LMG 2350 | Milk products |
| P. acidilactici | F | Meat products |
| Pediococcus pentosaceous | CTC 745 | Meat products |
| K. varians | CECT 230 | Equivalent to ATCC 15306, DSMZ 20033 |
| LMG 14231 | LMG Collection | |
| Kocuria roseus | CECT 52 | Equivalent to ATCC 186, DSMZ 20447 |
| Micrococcus luteus | CECT 245 | Equivalent to ATTC 10240, ATCC 10786, DSMZ 1790 |
| S. carnosus | DSMZ 20501 | DSMZ Collection |
| LTH 2102 | German meat products | |
| S. warneri | CECT 236 | Equivalent to ATCC 27836, DSMZ 20316 |
| CIT 863 | Meat products | |
| S. xylosus | CTC 3001 | Spanish fermented sausages |
| CTC 3050 | Spanish fermented sausages | |
| CECT 237 | Equivalent to ATCC 29971, DSMZ 20266 | |
| S. saprophyticus | CECT 235 | Equivalent to DSMZ 20229 |
| S. simulans | CECT 4538 | Equivalent to ATCC 27848, DSMZ 20322 |
| S. aureus | CTC 1008 | Spanish fermented sausages |
| S. epidermidis | CECT 231 | Equivalent to ATCC 1798, ATCC 12228, DSMZ 1798 |
CTC, IRTA Meat Technology Centre Collection (Monells, Spain); LMG, Laboratory of Microbial Gene Technology Collection, Agricultural University of Norway; DSMZ, German Collection of Microorganisms and Cell Culture; CECT, Spanish Collection of Microorganisms; CIT, INRA Collection (Theix, France); LTH, Food Technology Institute Collection, University of Hohemheim (Stuttgart, Germany).
LAB were grown anaerobically in MRS broth at 30°C for 24 h, and staphylococci and Kocuria spp. were grown in TSBYE broth (tryptic soy broth [Difco Laboratories] with 0.6% yeast extract) at 30°C for 24 h with agitation (140 rpm).
Biochemical identification of GCC+.
Randomly selected colonies with typical morphologies in the mannitol salt agar (MSA) plates were grown overnight in TSBYE broth at 30°C. The biochemical characterization was carried out by the API STAPH system (reference no. 20500; bioMerieux) following the manufacturer's recommendations. The identification was assessed by APILAB identification software (bioMerieux).
Bacterial DNA isolation and amplification of rrn operon regions. (i) DNA extraction from fermented meat products.
DNA extraction from the sausage homogenate, before and after enrichment, was done in duplicate using two different methodologies.
Two 10-g sausage portions were homogenized with 90 ml of MRS broth and 90 ml of mannitol salt broth in a Stomacher Lab-Blender for 1 min. Two 1.5-ml aliquots of each homogenate were placed in 1.5-ml tubes (Rubilabor S.L., Barcelona, Spain), centrifuged at 6,082 × g for 10 min, and frozen at −20°C. For LAB enrichment, the remaining MRS homogenate was grown in anaerobic jars for 24 h at 30°C, and the rest of the MSA homogenate was incubated aerobically (with shaking at 140 rpm) for 24 h at 30°C for GCC+ enrichment. Two 1.5-ml aliquots of each enriched culture were placed in 1.5-ml tubes, centrifuged at 8,000 rpm for 10 min, and frozen at −20°C. The frozen aliquots from enriched and unenriched samples were thawed, and DNA extraction proceeded in duplicate using two different isolation methods: the DNeasy tissue kit (catalog no. 69506; Qiagen, Hilden, Germany), using the manufacturer's recommended protocol for gram-positive bacteria, and the modified Anderson and McKay method (3). The lysis step was adapted in order to avoid biases between different species and to improve the final DNA yield. Lysozyme (5 μg/μl) and mutanolysine (0.05 U/μl) were used for LAB, and lysostaphine (0.03 μg/μl) plus lysozyme and mutanolysine were used for GCC+. The samples were incubated for 30 min at 37°C to ensure an efficient lysis. In the modified Anderson and McKay method for genomic DNA extraction, NaOH treatment was omitted and an RNase (1 μg/μl) treatment was carried out after the lysis step.
(ii) DNA extraction from pure cultures.
Control DNAs from pure cultures of different strains were extracted by the modified Anderson and McKay method (3) and with the DNeasy tissue kit.
To lyse LAB, a mixture of lysozyme (5 μg/μl) and mutanolysine (0.05 U/μl) was used. For GCC+, lysostaphine (0.03 μg/μl) was added to the mixture to ensure that there was no bias in the lysis of the different species assayed.
Primers.
The primers used in this study are listed in Table 2.
TABLE 2.
Primers used for PCR amplification, sequence, specificity, and location on the rRNA operon
| Species | Primer | R/Fa | Sequence (5′-3′) | Specificity | Position | Reference |
|---|---|---|---|---|---|---|
| L. sakei | Ls | R | ATGAAACTATTAAATTGGTAC | Species | 16S-23S spacer region | Berthier and Ehrlich (8) |
| 16S | F | GCTGGATCACCTCCTTTC | 16S RNA gene | 3′-end 16S rRNA | ||
| L. curvatus | Lc | R | TTGGTACTATTTAATTCTTAG | Species | 16S-23S spacer region | Berthier and Ehrlich (8) |
| 16S | F | GCTGGATCACCTCCTTTC | 16S RNA gene | 3′-end 16S rRNA | ||
| L. plantarum | Lpl | R | ATGAGGTATTCAACTTATG | Species | 16S-23S spacer region | Berthier and Ehrlich (8) |
| 16S | F | GCTGGATCACCTCCTTTC | 16S RNA gene | 3′-end 16S rRNA | ||
| E. faecium | Ef | R | CACACAATCGTAACATCCTA | Species | 23S rRNA | Frahm et al. (29) |
| 16S | F | GCTGGATCACCTCCTTTC | 16S RNA gene | 3′-end 16S rRNA | Berthier and Ehrlich (8) | |
| L. lactis | Lac | F | GCTGAAGGTTGGTACTTGTA | Species | 16S rRNA (V1) | Klijn et al. (43) |
| 23S | R | AGTGCCAAGGCATCCACC | 23S RNA gene | 5′-end 23S rRNA | Berthier and Ehrlich (8) | |
| P. acidilactici | Pda | F | CTGAATGAGATTTTAACACGAA | Species | 16S rRNA (V1) | Cai et al. (11) |
| 23S | R | AGTGCCAAGGCATCCACC | 23S RNA gene | 5′-end 23S rRNA | Berthier and Ehrlich (8) | |
| S. camosus | ScaI | F | GAACCGCATGGTTCTGCAA | Species | 16S rRNA | Gory et al. (33) |
| ScaII | R | CCGTCAAGGTGCGCATAGT | Species | 16S rRNA | This study | |
| S. warneri | Swa | F | TAGTGAAAGGCGGCTTTGCTG | Species | 16S rRNA | Gory et al. (33) |
| Sta2 | R | CCGTCAAGATGTGCACAGT | Genera | 16S rRNA | This study | |
| K. varians | Kva | R | CACGTTTACCTCCCCGGATC | Species | 23S rRNA (insertion region) | This study |
| Kva2 | F | GGTTTGTCGCGTCTTCTGTG | 16S RNA gene | 16S rRNA (V3-559-578pb) | This study | |
| S. xylosus | Sxyl FA | F | AAGTCGGTTGAAAACCCAAA | Strain | 16S-23S spacer region | This study |
| Sxyl FB | F | AAAATCGGCTGAAAACCTAAA | Strain | 16S-23S spacer region | This study | |
| Sxyl RA | R | CATTGACATATTGTCTTCAG | Strain | |||
| Sxyl RB | R | CATTGACATATTGTATTCAG | Strain | |||
| S. simulans | Si1 | F | ATTCGGAACAGTTTCGCAG | Species | 16S-23S spacer region | Forsman et al. (25) |
| Si2 | R | ATTGTGAGTAATCGTTTGCC | Species | 16S-23S spacer region | ||
| S. epidermidis | Epi1 | F | TCTACGAAGATGAGGGATA | Species | 16S-23S spacer region | Forsman et al. (25) |
| Epi2 | R | TTTCCACCATATTTTGAATTGT | Species | 16S-23S spacer region |
R, reverse; F, forward.
Primers for species-specific PCR detection of S. carnosus, S. warneri, K. varians, and S. xylosus were developed. New primers based on the 16S rRNA sequences of S. carnosus and S. warneri (GenBank accession no. AB009934 and L37603, respectively) were coupled to the 16S rRNA probes previously reported by Gory et al. (33). For K. varians, the 23S rRNA insertion region (58) and 16S rRNA (76) were used to design a pair of species-specific PCR primers. For S. xylosus, previously reported 16S-23S spacer region primers (28) and newly designed primers of the intergenic region were tested for PCR amplification of type strains and fermented-sausage isolates.
Primers used for species-specific detection of L. sakei, L. curvatus, and L. plantarum were previously reported by Berthier and Ehrlich (8). For S. simulans and S. epidermidis, the primers used were those reported by Forsman et al. (28). The probes described for Enterococcus faecium (29), Lactococcus lactis (43), and Pediococcus acidilactici (11) were combined with nonspecific primers for 16S and 23S rRNA as described by Berthier and Ehrlich (8) for species-specific PCR amplification (Table 2).
The sequences of the species-specific primers were submitted to the BLAST search program (2) of the National Center for Biotechnology Information (Bethesda, Md.) (http://www.ncbi.nlm.nih.gov). The specificities of the primers were evaluated by PCR of the reference strain of each species. Cross-reactions among all the strains were tested (Table 1).
PCR amplification of rrn operon.
PCR was performed in a model 2400 DNA thermal cycler (Perkin-Elmer Corp. Applied Biosystems Division) as follows: initial denaturation at 94°C for 5 min, an appropriate number of cycles (Table 3) with denaturation at 94°C for 1 min, primer annealing (Table 3) for 1 min, and primer extension at 72°C for 1 min, followed by final extension at 72°C and cooling to 4°C for 7 min. A typical reaction mixture (25 μl) consisted of 20 mM Tris-HCl, pH 8.0, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate (Promega, Madison, Wis.), 0.5 μM each primer (Roche Molecular Biochemicals, Indianapolis, Ind.), 1 μl of genomic DNA, and 1 to 2 U of Taq DNA polymerase (Roche Molecular Biochemicals).
TABLE 3.
Optimized PCR protocol from each species tested
| Species | Primer pair | Cycles | Annealing temp (°C) | Size of amplified product (bp) |
|---|---|---|---|---|
| L. curvatus | Lc-16S | 20 | 50 | 305 |
| L. plantarum | Lpl-16S | 30 | 50 | 265 |
| L. sakei | Ls-16S | 20 | 45 | 290 |
| E. faecium | Ef-16S | 20 | 55 | 676/578 |
| L. lactis | Lac-23S | 20 | 59 | 2,060 |
| P. acidilactici | Pda-23S | 20 | 55 | 2,023/1,767 |
| S. carnosus | Sca1-Sca2 | 35 | 67 | 305 |
| K. varians | Kva-Kva2 | 35 | 67 | 3,362 |
| S. warneri | Swa-Sta2 | 20 | 72 | 289 |
| S. xylosus | Fa/Fb-Ra/Rb | 35 | 58 | 417/317/217 |
| S. epidermidis | Epi1-Epi2 | 30 | 55 | 240 |
| S. simulans | Sim1-Sim2 | 30 | 55 | 220 |
Previous assays with different strains of each species were tested at different matching temperatures to ensure experimental specificity (Table 3).
Gel electrophoresis.
Twenty-five microliters of the PCR product was electrophoresed at 100 V for 1 h on a 1.5% agarose gel (Roche Molecular Biochemicals) in 0.5× Tris-acetate-EDTA buffer stained with 0.1 μg of ethidium bromide (Sigma Chemical Co., St. Louis, Mo.)/ml. The smaller amount of ethidium bromide used ensured a high contrast between faint bands and the background. A 1-kb DNA ladder (Invitrogen, Merelbeke, Belgium) was used as a molecular size marker.
DNA sequence analysis.
16S-23S ribosomal DNA intergenic spacer regions from different S. xylosus strains (CTC 3001, CTC 3050, and DSMZ 20266) were amplified by the universal 16S-23S primer set as reported by Berthier and Ehrlich (8). The main band (∼350 bp) was cut from the electrophoresis gel and cleaned with a Gene Clean II kit (Bio 101, La Jolla, Calif.). Both strands were directly sequenced by a model 373A automated DNA sequencer (Applied Biosystems Inc.) according to the manufacturer's instructions. CLUSTAL W software (77) from the European Bioinformatics Institute (Wellcome Trust Genome Campus, Hinxton, United Kingdom) (http://www.ebi.ac.uk) was used for the analysis and comparison of sequences.
Statistical analyses.
Analyses of variance were performed using the nonparametric one-way ANOVA program from SAS software (SAS User's Guide: Statistics, SAS Institute Inc., 1988) in order to determine the significance of the differences between the results. The variables analyzed were microbial counts (LAB, GCC+, LAB/GCC+ ratio, enterococci, and Enterobacteriaceae) and physicochemical parameters (aw, pH, and nitrate, nitrite, protein, fat, and phosphate contents) for the two sausage types (fuet and chorizo) and species-specific PCR detection without enrichment.
The data were plotted as boxes and whiskers by the SAS software (SAS User's Guide: Statistics). This plot provided summary statistics for five numbers: minimum, maximum, median, 25th percentile, and 75th percentile.
In order to see the influence of physicochemical parameters on microflora, Pearson correlations from SAS software (SAS User's Guide: Statistics) between all microbial counts and the physicochemical parameters analyzed were done.
RESULTS
Physicochemical analysis of the products.
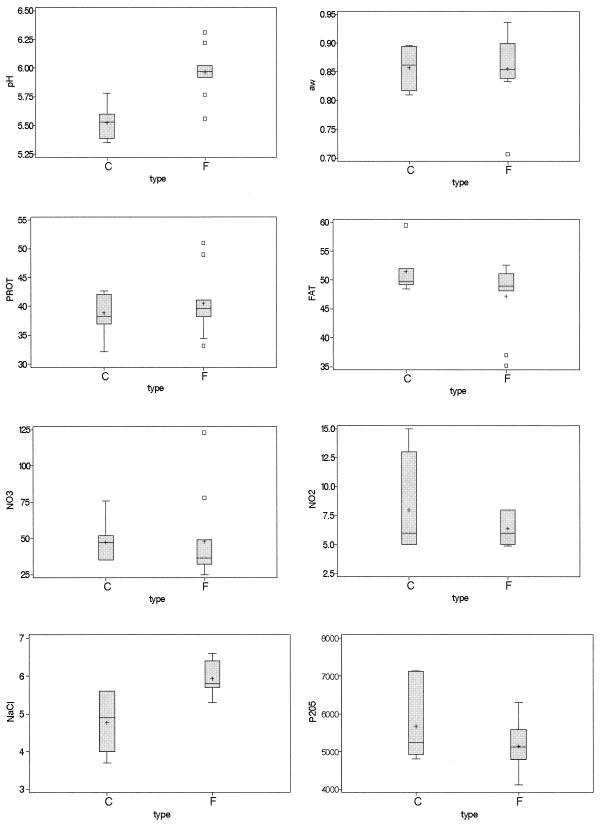
The physicochemical parameters of the samples are reported in Table 4 and are summarized in box-and-whisker plots for each variable and for the two types of sausages (fuet and chorizo) (Fig. 1).
TABLE 4.
Physicochemical analysis of products
| Samplea | pH | aw | Protein (%)b | Fat (%) | Nitrate (ppm) | Nitrite (ppm) | NaCl (%) | Phosphate (ppm) |
|---|---|---|---|---|---|---|---|---|
| F1 | 5.97 | 0.707 | 39.8 | 48.1 | 37 | 8 | 6.5 | 6,305 |
| F5 | 5.97 | 0.833 | 38.2 | 49.6 | 49 | 6 | 5.8 | 5,105 |
| F6 | 5.92 | 0.781 | 41.1 | 48.5 | 29 | 6 | 6.0 | 5,429 |
| F8 | 6.31 | 0.710 | 38.4 | 49.5 | 78 | 5 | 5.7 | 4,795 |
| F9 | 5.97 | 0.875 | 33.2 | 52.6 | 36 | 8 | 6.4 | 4,120 |
| F11 | 6.02 | 0.739 | 39.4 | 52.2 | 25 | 7 | 5.3 | 5,612 |
| F12 | 5.77 | 0.840 | 40.7 | 48.2 | 123 | 6 | 5.5 | 5,587 |
| F15 | 5.94 | 0.781 | 51.0 | 35.2 | 32 | <5 | 5.7 | 5,119 |
| F16 | 6.22 | 0.887 | 49.0 | 37.0 | 34 | 5 | 5.8 | 5,134 |
| F20 | 5.56 | 0.936 | 34.4 | 51.1 | 38 | 8 | 6.6 | 4,210 |
| C2 | 5.78 | 0.826 | 37.0 | 51.8 | 35 | 5 | 3.7 | 5,237 |
| C3 | 5.39 | 0.879 | 37.7 | 52.0 | 36 | 5 | 4.0 | 5,125 |
| C7 | 5.45 | 0.817 | 38.3 | 49.6 | 76 | 5 | 5.6 | 4,805 |
| C10 | 5.53 | 0.852 | 32.2 | 59.5 | 47 | 7 | 4.6 | 4,918 |
| C13 | 5.35 | 0.894 | 42.0 | 49.2 | 51 | 13 | 5.0 | 7,120 |
| C14 | 5.60 | 0.896 | 42.7 | 49.7 | 52 | 15 | 4.9 | 7,131 |
| C17 | 5.58 | 0.810 | 42.1 | 48.5 | 35 | 6 | 5.6 | 5,415 |
F, fuet; C, chorizo.
%, grams per 100 g of dry product.
FIG. 1.
Box-and-whiskers plots for the physicochemical analysis of the products for the two types of sausages, chorizo (C) and fuet (F). FAT, fat content (percent); PROT, protein content (percent); NaCl, sodium chloride content (percent); NO2, nitrite content (parts per million); NO3, nitrate content (parts per million); P2O5, phosphate content (parts per million). The percentages represent grams per 100 g of dry product. Error bars indicate maximum and minimum values (n = 6), horizontal lines indicate median values, plus signs indicate means, and boxes indicate values between the 25th and 75th percentiles.
Fuet sausages had a significantly (P < 0.05) higher pH (5.94) than chorizo sausages (5.52). The fuet sodium chloride content (5.9%) was significantly higher (P < 0.05) than that in chorizo (4.8%). The aw and protein, fat, nitrate, and nitrite contents were not significantly different (P > 0.05) in the two types of products.
Microbial counts.
Seventeen low-acid fermented sausages (fuets and chorizos) from different producers were sampled for microbiological assays.
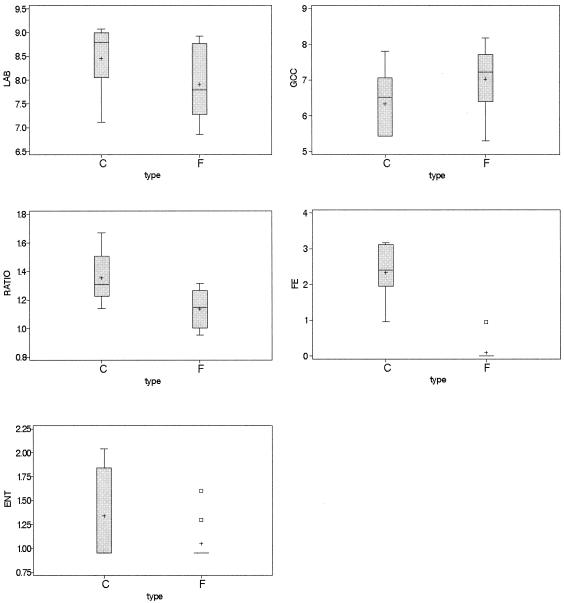
LAB and GCC+ constituted the main microflora (Table 5). The microbial counts for fuet and chorizo are summarized in Fig. 2. Counts of LAB were not significantly different (P > 0.05) in fuets and chorizos, with a mean of 8.13 log CFU/g. The counts of GCC+ also did not differ (P > 0.05) between fuets and chorizos, with a mean of 6.74 log CFU/g. The ratio between the counts of LAB and GCC+ was significantly different (P < 0.05) in the two groups of products. Fuets had a LAB/GCC+ ratio of 1.14, and chorizos had a ratio of 1.35. Enterococcus and Enterobacteriaceae counts were not significantly different (P > 0.05) in fuets and chorizos.
TABLE 5.
Microbial countsa
| Sample | LAB | GCC+ | FE | ENT | SA | LM | SRC | E. coli | SAL |
|---|---|---|---|---|---|---|---|---|---|
| F1 | 7.74 | 7.70 | 1.30 | <1 | <2 | <3 | <1 | <1 | Abs |
| F5 | 6.86 | 5.30 | 2.78 | <1 | <2 | <3 | <1 | <1 | Abs |
| F6 | 8.84 | 6.98 | 1.90 | <1 | <2 | <3 | <1 | <1 | Abs |
| F8 | 7.22 | 7.56 | 2.26 | <1 | <2 | <3 | <1 | <1 | Abs |
| F9 | 7.78 | 7.45 | 2.78 | <1 | <2 | 4 | <1 | <1 | Abs |
| F11 | 7.27 | 5.96 | 2.73 | <1 | <2 | <3 | <1 | <1 | Abs |
| F12 | 8.92 | 6.78 | 1.95 | 1.30 | <2 | <3 | <1 | <1 | Abs |
| F15 | 7.79 | 6.39 | 3.67 | <1 | <2 | <3 | <1 | <1 | Abs |
| F16 | 7.88 | 7.97 | 4.48 | 1.60 | <2 | <3 | <1 | <1 | Abs |
| F20 | 8.77 | 8.18 | <1 | <1 | <2 | <3 | <1 | <1 | Abs |
| C2 | 7.12 | 5.43 | 2.71 | <1 | <2 | <3 | <1 | <1 | Abs |
| C3 | 8.04 | 6.56 | <1 | <1 | <2 | <3 | <1 | <1 | Abs |
| C7 | 9.07 | 5.43 | 3.12 | <1 | <2 | <3 | <1 | <1 | Abs |
| C10 | 8.27 | 5.48 | 2.40 | 1.70 | <2 | 4 | 1 | <1 | Abs |
| C13 | 8.78 | 6.52 | 2.04 | 2.04 | <2 | <3 | <1 | <1 | Abs |
| C14 | 8.90 | 7.80 | 3.16 | 1.84 | 2 | 4 | 1 | 1 | Abs |
| C17 | 8.99 | 7.06 | 1.95 | <1 | <2 | <3 | <1 | <1 | Abs |
All microbial counts are expressed in log CFU per gram except for LM (expressed in MPN per gram) and SAL (expressed as absence [Abs] or presence in 25 g). FE, enterococci; ENT, Enterobacteriaceae; SA, S. aureus; LM, L. monocytogenes; SRC, sulfite-reducing clostridia; SAL, Salmonella spp.; C, chorizo; F, fuet.
FIG. 2.
Box-and-whiskers plots for the microbial counts of the products for the two types of sausages, chorizo (C) and fuet (F). LAB, counts of LAB; GCC, counts of GCC+; RATIO, LAB/GCC+ ratio; FE, counts of enterococci; ENT, counts of Enterobacteriaceae. All microbial counts are expressed as log CFU per gram. Error bars indicate maximum and minimum values (n = 6), horizontal lines indicate median values, plus signs indicate means, and boxes indicate values between the 25th and 75th percentiles.
The pH had a correlation of −0.64 with LAB counts in MRS (P < 0.05), −0.65 with enterococcus counts in KAA (P < 0.05), and −0.63 with the LAB/GCC+ ratio (P < 0.05). The sodium chloride content had correlations of 0.53 with GCC+ counts (P < 0.05) and −0.65 with enterococcus counts. Enterobacteriaceae counts had a correlation of 0.61 with the phosphate content (P < 0.05). Enterobacteriaceae counts could not be correlated with pH or aw (P > 0.05), but samples containing food-borne pathogens and higher counts of Enterobacteriaceae had aw values of ≥0.85.
From the point of view of hygiene, 4 MPN of L. monocytogenes/g were detected in three samples; one of them additionally contained 102 CFU of S. aureus cells/g, and another contained >102 CFU of Enterobacteriaceae/g. Salmonella was absent in all samples. E. coli and sulfite-reducing clostridium counts were <102 CFU/g in all of the samples (Table 5). Ninety-four percent of the samples met the Spanish hygienic microbiological standards (55), but only 70.6% of the samples had L. monocytogenes counts under 3 MPN/g and Enterobacteriaceae counts under 100 CFU/g.
Sequencing of the spacer region for identification of S. xylosus.
In order to specifically detect S. xylosus by PCR in meat samples, the previously reported primers (28) for the 16S-23S spacer regions of RNA operons were assayed with the reference strain CECT 237 (DSMZ 20266) and with two strains isolated from slightly fermented sausages (CTC 3001 and CTC 3050) and biochemically identified as S. xylosus. These primers were not able to amplify any of the S. xylosus strains tested.
Primers complementary to the conserved sequences near the 3′ end of the 16S and the 5′ end of the 23S rRNA genes were used to amplify the 16S-23S spacer from the above-mentioned strains for sequencing. Four different PCR products were detected for each strain. The predominant band (∼370 bp) was selected, cut from electrophoresis gels, and cleaned to sequence both strands directly. A comparison of the sequenced internal transcribed spacer (ITS) regions of the three strains mentioned above plus the previously reported S. xylosus ATCC 12162 is shown in Fig. 3. ATCC 12162 had a homology of 87% with strains CTC 3001 and CTC 3050, isolated from slightly fermented sausages. CTC 3001 and CTC 3050 had 100% homology with each other and 91% homology with the strain collection DSMZ 20266.
FIG. 3.
Intergenic sequence comparison of S. xylosus strains from different origins. 3001, CTC 3001; 3050, CTC 3050; DSMZ, DSMZ 20266; ATCC, ATCC 12162. Primers xylI and xylII from Forsman et al. (28) are shown in italics. The boldface letters indicate primers Fa/Fb and Ra/Rb from this study. Stars indicate conserved sequence positions; dashes indicate gaps (within sequences) or regions for which sequences are not available (outside sequences).
Two pairs of new primers were designed based on the polymorphisms observed in the ITS region in order to develop a dual PCR assay able to detect S. xylosus strains from different origins.
Species-specific PCR detection.
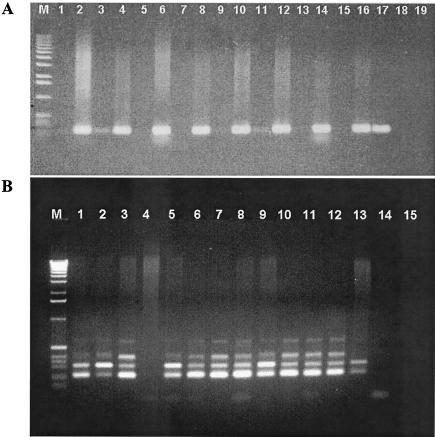
Six species of LAB (L. sakei, L. curvatus, L. plantarum, P. acidilactici, L. lactis, and E. faecium) and six GCC+ (K. varians, S. xylosus, S. carnosus, S. epidermidis, S. warneri, and S. simulans) were screened by species-specific PCR detection from the DNAs of low-acid fermented-sausage homogenates before and after enrichment. PCR amplification products for species-specific detection of L. sakei and S. xylosus from fermented sausages are shown in Fig. 4.
FIG. 4.
Agarose gel electrophoresis-PCR amplification products of L. sakei (A) and S. xylosus (B) from low-acid fermented sausages. (A) Lane M, 1-kb DNA ladder; lanes 1, 3, 5, 7, 9, 11, 13, and 15, genomic DNAs from nonenriched samples; lanes 2, 4, 6, 8, 10, 12, 14, and 16, genomic DNAs from enriched samples; lane 17, positive control (L. sakei CTC 494); lane 18, negative control (L. curvatus CTC 371); lane 19, negative control (no DNA). (B) Lane M, 1-kb DNA ladder; lane 1, positive control (S. xylosus CTC 3001); lane 2, positive control S. xylosus CTC 3050; lane 3, positive control (S. xylosus CECT 237); lane 4, negative control (S. carnosus DSMZ 20501); lanes 5 to 13, genomic DNAs from enriched samples; lane 14, negative control (S. epidermidis CECT 23); lane 15, negative control (no DNA).
Duplicates and a random selection of positive and negative controls in each assay were used to evaluate the reproducibility and specificity of the assay. No yield differences were observed in the PCR assay when Qiagen samples were compared with those from the modified Anderson and McKay DNA extraction method (3), and there seemed to be a balance between the higher yield of the modified Anderson and McKay protocol and the DNA cleaner preparations when the Qiagen kit was used. The yield of the PCR was optimized using 1 to 2 U of Taq polymerase per reaction (data not shown). Loading the entire PCR product in the electrophoresis gel was important in order to detect the faint bands. The sizes of the PCR products obtained after amplification of DNA from the different strains were those expected. No PCR products from other tested strains and species were detected.
Without enrichment and independently of the DNA extraction method used, L. sakei and L. curvatus were detected in 11.8% of the samples, and L. plantarum and S. xylosus were detected in 17.6%. No significant difference (P > 0.05) between microbial counts and physicochemical parameters was evident at time zero when considering the possibility of detection, or lack thereof, without enrichment (data not shown).
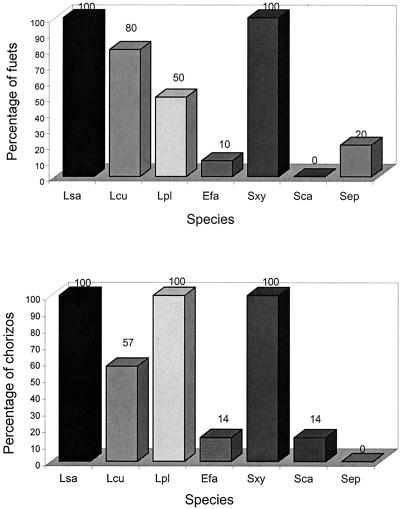
After enrichment for 24 h in MRS, L. sakei was detected in all of the samples, L. curvatus was detected in 71%, and L. plantarum was detected in 62%. E. faecium was found in 11.8% of the samples, while P. acidilactici and L. lactis were not detected. S. xylosus was detected in all samples assayed after enrichment (100%), S. carnosus was detected in 14% of chorizos, and S. epidermidis was detected in 20% of fuets. S. simulans and S. warneri were not detected in enriched samples (Fig. 5).
FIG. 5.
Percentages of sausages containing different species of LAB and GCC+ detected by species-specific PCR from enriched samples. Lsa, L. sakei; Lcu, L. curvatus; Lpl, L. plantarum; Efa, E. faecium; Lla, L. lactis; Pda, P. acidilactici; Sxy, S. xylosus; Sca, S. carnosus; Sep, S. epidermidis; Swa, S. warneri; Ssi, S. simulans; Kva, K. varians.
When the two types of products (fuet and chorizo) were compared, the species found in enriched samples were not exactly the same. L. sakei and S. xylosus were present at the same level (100%), but L. curvatus, L. plantarum, S. carnosus, and S. epidermidis varied in the two types of slightly fermented products. L. curvatus was detected in 80% of the fuets and in 57% of the chorizos. L. plantarum was detected in 50% of the fuets and in 100% of the chorizos. S. epidermidis was not detected in chorizos, while S. carnosus was not detected in fuets (Fig. 5).
The relative prevalence of S. xylosus as a representative of GCC+ in the species-specific PCR assays performed directly on food samples was corroborated by comparing the biochemical profiles of 65 randomly selected colonies, using the API STAPH system (bioMerieux) and the species-specific PCR protocol, after isolation of chromosomal DNAs from different colonies isolated from several sausage samples. The API STAPH galleries allowed 4 strains of S. carnosus and 46 strains of S. xylosus with different biochemical patterns to be identified (99% identification). Nineteen strains showed differences in biochemical identities for assessing colonies as specific species by the API test. Species-specific PCR of pure cultures allowed these 19 species to be identified as S. xylosus.
DISCUSSION
Microbial counts.
A few sporadic studies have been carried out on traditional low-acid fermented meat products, showing that hygienic shortcomings can cause up to 25% of product loss, with serious economic consequences, and that they may undermine consumer confidence in traditional products (25, 44). In this study, 29.4% of the low-acid fermented sausages analyzed had L. monocytogenes counts over 3 MPN/g and Enterobacteriaceae counts over 100 CFU/g. Most of the traditional low-acid dry-fermented sausages rely on natural contamination by environmental microflora for fermentation. The environmental microflora is composed not only of LAB and GCC+ for the fermentation and flavor of sausages, but also of spoilage and pathogenic microorganisms that contaminate raw meat. Since food safety is one of the primary concerns of European policy (22) and consumers, the study of the microbial (hygienic and technological) quality of these types of products is crucial in order to help traditional manufacturers produce safe and standardized products.
In this study, Salmonella was not detected in any of the products analyzed. Salmonella has been shown to survive during pepperoni manufacture (39). S. aureus, considered the third most common pathogen causing food-borne outbreaks after Salmonella and Campylobacter, was isolated in 5.9% of the sausage samples. The growth of S. aureus and the presence of enterotoxin in fermented sausages, particularly in Genoa- and Italian-type dry salami, have caused several outbreaks of food poisoning (75).
L. monocytogenes, a food-borne pathogen that poses a risk for immunodeficient persons and that has been involved in sporadic food poisoning, was isolated in 17.6% of the traditional low-acid fermented sausages. L. monocytogenes is a ubiquitous microorganism that is often isolated in manufacturing plants as an in-house bacterium (13, 81). Salvat et al. (61) reported that as many as 68% of environmental samples in a curing plant were positive for L. monocytogenes and that after cleaning, 17% of the samples remained positive. Recent European investigations reported 12 to 16% Listeria-positive isolations in industrial fermented meat products (17).
The ratio between LAB and GCC+ was highly balanced in the slightly fermented sausages analyzed, and a correlation between the LAB/GCC+ ratio and pH could be established. It has been reported that sausages with a predominance of LAB are tangier and less flavorful (46, 50, 56), while sausages with higher numbers of GCC+ contain more aromatic compounds (25).
Species-specific PCR detection of LAB and GCC+.
All strains of L. sakei tested were detected by a unique homogeneous band of the ITS region, thus confirming their uniformity as reported by Berthier and Ehrlich (8). The presence of two different amplified bands when E. faecium was detected by the species-specific PCR protocol applied in this study may reflect the presence of several rrn operons in these species. Sechi and Daneo-Moore (71) described the characterization of rrn operons of two different lengths in Enterococcushirae, one with tRNA (Ala) and one without it in the ITS region.
Polymorphisms in the ITS regions of different S. xylosus strains from different origins have been detected by sequencing. PCR products amplified with the species-specific primer pair gave the same four-band pattern in the three strains tested, although the different numbers of the amplified bands allowed a different number of rRNA operons in the various strains to be hypothesized (40). The development of a dual PCR assay to sort out these polymorphisms allowed S. xylosus to be detected in all the slightly fermented sausages assayed, and moreover, the different strains could be distinguished. Identification of S. xylosus by the dual PCR assay matched perfectly with the results of biochemical methods, and even when phenotypic identification was difficult due to exceptions in some biochemical traits that are officially considered genus or species specific, PCR could help identification to species level, as previously reported by Watts and Yancey (82).
By species-specific PCR, LAB could be detected in 29.4% of the nonenriched samples and GCC+ could be detected in only 17.6% of the samples without enrichment. The low detection level in nonenriched samples could not be statistically related to microbial counts or to the measured physicochemical variables (the type of product [chorizo or fuet] and the fat, protein, phosphate, and sodium chloride contents of the product) (P > 0.05). It has been reported that substances such as heme compounds may copurify with DNA and act as inhibitors (1) and that nonspecific DNA (0.4 mg in 100 μl of reaction mixture) could completely inhibit the reaction (59). Some PCR inhibitors from the food matrix were not obviously sorted out by the DNA extraction procedures used. Further studies to investigate new DNA extraction protocols combining different filters and prefilters, the introduction of bovine serum albumin in the PCR mixture, and the use of different DNA polymerases to avoid PCR inhibition are in progress.
From enriched samples, L. sakei and S. xylosus were detected in all the sausages by species-specific PCR, and they can be considered the predominant species in slightly fermented sausages. L. sakei has been reported as the main species in different fermented products all over southern Europe (20, 37, 51, 62, 63, 65, 79). The predominance of S. xylosus over other staphylococci and the lack of K. varians in the low-acid fermented sausages by the end of the process are in agreement with the results of several authors for different types of Mediterranean sausage (16, 21, 30, 38, 52, 60, 70, 74, 80). L. plantarum was detected in 100% of chorizos and in only 50% of fuets. Some technological differences, such as sugar content, processing temperature, and naturally selected microflora, may define the final pH of the fermented sausages. L. plantarum is more aciduric and produces higher amounts of d-lactic acid than L. sakei and L. curvatus (9, 31). S. carnosus and S. epidermidis, described as seldom isolated from fermented sausages (16, 21, 30, 38, 52, 60, 70, 74, 80), were identified in this study in 5.9 and 11.8% of the low-acid sausages, respectively. S. simulans and S. warneri were not detected in this study. The presence of K. varians in 36% and K. kristinae in 3% of dry sausages from other countries of Central Europe was probably due to climatic differences and diverse production practices (27).
Enterococci were not quantitatively important compared to lactobacilli, in contrast to the results of Dellapina et al. (24) for the slightly fermented Felino salami. E. faecium was detected in only 11.8% of the sausages through species-specific PCR. The role of enterococci in fermented meat products has not been thoroughly studied, and their presence is highly controversial. Some authors consider them to be technologically unacceptable, reflecting a certain level of contamination and a poor curing process. Other authors consider E. faecium strains to be potential starter cultures, as has been reported for dairy products, silage, and probiotics. However, the latest findings on biogenic amine production and antibiotic resistance determinants and their potential transfer to pathogenic gram-positive species point to the need for wide phenotypic and genotypic characterization of each strain used (32).
The identification of LAB and GCC+ in low-acid sausages by species-specific PCR from enriched samples may offer the possibility of reducing the labor time and enhancing the identification of bacterial composition. Moreover, molecular methods can complement biochemical species identification of isolated colonies.
Acknowledgments
This research was funded by the Spanish Inter-Ministerial Commission of Science and Technology (CICYT ALI99-0308). We thank the Ministry of Science and Technology for the scholarship for Belen Martin.
We thank Y. Beltran for technical assistance, A. Valero for the physicochemical analyses, and P. Gou for support of the statistical analyses.
REFERENCES
- 1.Akane, A., K. Matsubara, H. Nakamura, S. Takahashi, and K. Kimura. 1994. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from blood stains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 39:362-372. [PubMed] [Google Scholar]
- 2.Altschul, S. F., A. A. Hadden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrighetto, C., L. Zampese, and A. Lombardi. 2001. RAPD-PCR characterization of lactobacilli isolated from artisanal meat plants and traditional fermented sausages of Veneto region (Italy). Lett. Appl. Microbiol. 33:26-30. [DOI] [PubMed] [Google Scholar]
- 5.Bacus, J. N. 1986. Fermented meat and poultry products, p. 123-164. In A. M. D. Pearson (ed.), Advances in meat and poultry microbiology. Macmillan, London, United Kingdom.
- 6.Barry, T., G. Colleran, M. Glennon, L. Dunican, and F. Gannon. 1991. The 16S/23S ribosomal spacer region as a target for DNA probes to identify Eubacteria. PCR Methods Appl. 1:51-56. [DOI] [PubMed] [Google Scholar]
- 7.Berthier, F., and S. D. Ehrlich. 1999. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int. J. Syst. Bacteriol. 49:997-1007. [DOI] [PubMed] [Google Scholar]
- 8.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. [DOI] [PubMed] [Google Scholar]
- 9.Bucharles, C., J. P. Girard, J. Sirami, and S. Pascal. 1984. Characteristics of a dry sausage showing excessive acidity. Sci. Aliments 4:137-143. [Google Scholar]
- 10.Buffone, G. J., G. J. Demmler, C. M. Schimbor, and J. Greer. 1991. Improved amplification of cytomegalovirus DNA from urine after purification of DNA with glass beads. Clin. Chem. 37:1945-1949. [PubMed] [Google Scholar]
- 11.Cai, Y., S. Kumai, M. Ogawa, Y. Benno, and T. Nakase. 1999. Characterization and identification of Pediococcus species isolated from forage crops and their application for silage preparation. Appl. Environ. Microbiol. 65:2901-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantoni, C., M. R. Molnar, P. Renon, and G. Gioletti. 1967. Lipolytic micrococci in pork fat. J. Appl. Bacteriol. 30:190-196. [DOI] [PubMed] [Google Scholar]
- 13.Chasseignaux, E. 1999. Ecologie de Listeria monocytogenes dans les ateliers de transformation de viandes des volailles et de porcs. Université Claude Bernard, Lyon, France.
- 14.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocolin, L., M. Manzano, and G. Comi. 2000. Development of a rapid method for the identification of Lactobacilus spp. isolated from naturally fermented Italian sausages using a polymerase chain reaction-temperature gradient gel electrophoresis. Lett. Appl. Microbiol. 30:126-129. [DOI] [PubMed] [Google Scholar]
- 16.Comi, G., B. Citterio, M. Manzano, and C. Cantoni. 1992. Evaluation and characterization of Micrococcaceae strains in Italian dry fermented sausages. Fleischwirtschaft 72:1679-1683. [Google Scholar]
- 17.Commission de l'Agence Française de Sécurité Sanitaire des Aliments. 2000. Étude des risques liés a Listeria monocytogenes. Commission de l'Agence Française de Sécurité Sanitaire des Aliments, Maisons-Alfort, France.
- 18.Commission of European Communities. 1999. Europe's agenda 2000. Priority Publications Programme XD5 final version 31.8. Commission of European Communities, Brussels, Belgium.
- 19.Commission of European Communities. 2000. White paper on food safety. Commission of European Communities, Brussels, Belgium.
- 20.Coppola, R., B. Giagnacovo, M. Iorizzo, and L. Grazia. 1998. Characterization of lactobacilli involved in the ripening of sopressata molisana, a typical southern Italy fermented sausage. Food Microbiol. 15:347-353. [Google Scholar]
- 21.Coppola, R., M. Iorizzo, A. Sorrentino, E. Sorrentino, and L. Grazia. 1996. Resistenza al congelamento di lattobacilli mesofili isolati da insaccati e paste acide. Ind. Aliment. 35:349-351, 356.
- 22.Council of the European Communities. 1993. Directive 93/43/CEE, relative to the hygiene of the food products. OJEC L175:1-11.
- 23.Debevere, J. M., J. P. Voets, F. De Schryver, and A. Huyghebaert. 1976. Lipolytic activity of Micrococcus sp. isolated from a starter culture in pork fat. Lebensmittel Wiss. Technol. 9:160-162. [Google Scholar]
- 24.Dellapina, G., D. Blanco, E. Pancini, S. Barbuti, and M. Campanini. 1994. Microbiological evolution in Italian Felino, Milan and Hungarian-style salami. Ind. Conserv. 69:85-90. [Google Scholar]
- 25.Demeyer, D., M. Raemaekers, A. Rizzo, A. Holck, A. De Smedt, B. Ten Brink, B. Hagen, C. Montel, E. Zanardi, E. Murbrekk, F. Leroy, F. Vandendriessche, K. Lorentsen, K. Venema, L. Sunesen, L. H. Stahnke, L. De Vuyst, R. Talon, R. Chizzolini, and S. Eerola. 2000. Control of bioflavour and safety in fermented sausages: first results of a European project. Food Res. Int. 33:171-180. [Google Scholar]
- 26.Demeyer, D. I. 1982. Stoichiometry of dry sausage fermentation. Antonie Leeuwenhoek 48:414-416. [Google Scholar]
- 27.Fischer, U., and K. H. Schleifer. 1980. Vorkommen von staphylokokken un mikrokokken in Rohwurst. Fleischwirtschaft 60:1046-1049. [Google Scholar]
- 28.Forsman, P., A. Tilsala-Timisjarvi, and T. Alatossava. 1997. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 143:3491-3500. [DOI] [PubMed] [Google Scholar]
- 29.Frahm, E., I. Heiber, S. Hoffmann, C. Koob, H. Meier, W. Ludwig, R. Amann, K. H. Schleifer, and U. Obst. 1998. Application of 23S rDNA-targeted oligonucleotide probes specific for enterococci to water hygiene control. Syst. Appl. Microbiol. 21:450-453. [DOI] [PubMed] [Google Scholar]
- 30.García-Varona, M., E. M. Santos, I. Jaime, and J. Rovira. 2000. Characterisation of Micrococcaceae isolated from different varieties of chorizo. Int. J. Food Microbiol. 54:189-195. [DOI] [PubMed] [Google Scholar]
- 31.Garriga, M., M. Hugas, P. Gou, M. T. Aymerich, J. Arnau, and J. M. Montfort. 1996. Technological and sensorial evaluation of Lactobacillus strains as starter cultures in fermented sausages. Int. J. Food Microbiol. 32:173-183. [DOI] [PubMed] [Google Scholar]
- 32.Giraffa, G. 2002. Enterococci from foods. FEMS Microbiol. Rev. 26:163-171. [DOI] [PubMed] [Google Scholar]
- 33.Gory, L., L. Millet, J. J. Godon, and M. C. Montel. 1999. Identification of Staphylococcus carnosus and Staphylococcus warneri isolated from meat by fluorescent in situ hybridization with 16S rRNA-targeted oligonucleotide probes. Syst. Appl. Microbiol. 22:225-228. [DOI] [PubMed] [Google Scholar]
- 34.Gürtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 141:3-16. [DOI] [PubMed] [Google Scholar]
- 35.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165-174. [Google Scholar]
- 36.Hastings, J. W., and W. H. Holzapfel. 1987. Conventional taxonomy of lactobacilli surviving radurization of meat. J. Appl. Bacteriol. 62:209-216. [DOI] [PubMed] [Google Scholar]
- 37.Hugas, M., M. Garriga, T. Aymerich, and J. M. Monfort. 1993. Biochemical characterization of lactobacilli from dry fermented sausages. Int. J. Food Microbiol. 18:107-113. [DOI] [PubMed] [Google Scholar]
- 38.Hugas, M., and M. Roca. 1997. Selección de cepas autóctonas de Staphylococcus spp. como cultivos iniciadores en embutidos cárnicos. Eurocarne 54:45-47. [Google Scholar]
- 39.Ihnot, A. M., A. M. Roering, R. K. Wierzba, N. G. Faith, and J. B. Luchansky. 1998. Behavior of Salmonella typhimurium DT104 during the manufacture and storage of pepperoni. Int. J. Food Microbiol. 40:117-121. [DOI] [PubMed] [Google Scholar]
- 39a.International Organization for Standardization. 2002. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp. ISO 6579:2002. International Organization for Standardization, Geneva, Switzerland.
- 40.Jensen, M. A., and N. Straus. 1993. Effect of PCR-conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 3:186-194. [DOI] [PubMed] [Google Scholar]
- 41.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein, P. G., and V. K. Juneja. 1997. Sensitive detection of viable Listeria monocytogenes by reverse transcription-PCR. Appl. Environ. Microbiol. 63:4441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1991. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 57:3390-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagrange, L., and J. Lelièvre. 1995. Propos sur la production fermière ou problématique de la production fermière, p. 67-72. In L. Lagrange (ed.), Différenciation et qualité des produits alimentaires. ENITA, Clermont-Ferrand, France.
- 45.Lantz, P. G., B. Hahn-Hägerdal, and P. Radström. 1994. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci. Technol. 5:384-389. [Google Scholar]
- 46.Metaxopoulos, J., J. Samelis, and M. Papadelli. 2001. Technological and microbiological evaluation of traditional processes as modified for the industrial manufacturing of dry fermented sausage in Greece. Ital. J. Food Sci. 1:3-18. [Google Scholar]
- 47.Miralles, M. C., J. Flores, and G. Perez-Martinez. 1996. Biochemical tests for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 13:227-236. [Google Scholar]
- 48.Montel, M. C., R. Talon, J. Fournaud, and C. Champomier. 1991. A simplified key for identifying homofermentative Lactobacillus and Carnobacterium spp. from meat. J. Appl. Bacteriol. 70:469-472. [DOI] [PubMed] [Google Scholar]
- 49.Nissen, H., and R. H. Dainty. 1995. Comparison of the use of rRNA probes and conventional methods in identifying strains of Lactobacillus sake and L. curvatus isolated from meat. Int. J. Food Microbiol. 25:311-315. [DOI] [PubMed] [Google Scholar]
- 50.Nurmi, E. 1966. Effect of bacterial inoculation on characteristics and microbial flora of dry sausage. Acta Agralia Fennica 108:1-77. [Google Scholar]
- 51.Parente, E., S. Griego, and M. A. Crudele. 2001. Phenotypic diversity of lactic acid bacteria isolated from fermented sausages produced in Basilicata (Southern Italy). J. Appl. Microbiol. 90:943-952. [DOI] [PubMed] [Google Scholar]
- 52.Pirone, G., and E. Manganelli. 1990. Caratterizzazione di Micrococcaceae isolate da salami tipo Napoli. Ind. Conserv. 65:220-223. [Google Scholar]
- 53.Powell, H. A., C. M. Gooding, S. D. Garrett, B. M. Lund, and R. A. McKee. 1994. Proteinase inhibition of the detection of Listeria monocytogenes in milk using the polymerase chain reaction. Lett. Appl. Microbiol. 18:59-61. [Google Scholar]
- 54.Presidencia del Gobierno Español. 1979. Métodos de análisis de productos cárnicos. Bol. Of. Estado 207:2022. [Google Scholar]
- 55.Presidencia del Gobierno Español. 1977. Normas microbiológicas para chorizo, salchichón y lomo embuchado. Bol. Of. Estado Esp.
- 56.Reuter, G., H. J. Langner, and H. J. Sinell. 1968. Entwicklung der Mikroflora in schenell-reifender deutscher Rohwurst und analoge quantitative Aminosäureanalyse bei einer Salami. Fleischwirtschaft 48:170-176. [Google Scholar]
- 57.Rodríguez, M., F. Núñez, J. J. Córdoba, C. Sanabria, E. Bermúdez, and M. A. Asensio. 1994. Characterization of Staphylococcus spp. and Micrococcus spp. isolated from Iberian ham throughout the ripening process. Int. J. Food Microbiol. 24:329-335. [DOI] [PubMed] [Google Scholar]
- 58.Roller, C., W. Ludwig, and K. H. Schleifer. 1992. Gram-positive bacteria with a high DNA G*C content are characterized by a common insertion within their 23S rRNA genes. J. Gen. Microbiol. 138:1167-1175. [DOI] [PubMed] [Google Scholar]
- 59.Rossen, L., P. Norskov, K. Holmstrom, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 60.Rossi, F., R. Tofalo, S. Torriani, and G. Suzzi. 2001. Identification by 16S-23S rDNA intergenic region amplification, genotypic and phenotypic clustering of Staphylococcus xylosus strains from dry sausages. J. Appl. Microbiol. 90:365-371. [DOI] [PubMed] [Google Scholar]
- 61.Salvat, G., M. T. Toquin, Y. Michel, and P. Colin. 1995. Control of Listeria monocytogenes in the delicatessen industries: the lessons of a listeriosis outbreak in France. Int. J. Food Microbiol. 25:75-81. [DOI] [PubMed] [Google Scholar]
- 62.Samelis, J., F. Maurogenakis, and J. Metaxopoulos. 1994. Characterisation of lactic acid bacteria isolated from naturally fermented Greek dry salami. Int. J. Food Microbiol. 23:179-196. [DOI] [PubMed] [Google Scholar]
- 63.Samelis, J., J. Metaxopoulos, M. Vlassi, and A. Pappa. 1998. Stability and safety of traditional Greek salami—a microbiological ecology study. Int. J. Food Microbiol. 44:69-82. [DOI] [PubMed] [Google Scholar]
- 64.Samelis, J., E. Tsakelidou, J. Metaxopoulos, and G. Kalantzopoulos. 1995. Differentiation of Lactobacillus sake and Lact. curvatus isolated from naturally fermented Greek dry salami by SDS-PAGE of whole-cell proteins. J. Appl. Bacteriol. 78:157-163. [DOI] [PubMed] [Google Scholar]
- 65.Santos, E. M., C. González-Fernández, I. Jaime, and J. Rovira. 1998. Comparative study of lactic acid bacteria house flora isolated in different varieties of “chorizo.” Int. J. Food Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 66.Santos, J. A., T. Lopez-Diaz, M. C. Garcia-Fernandez, M. L. Garcia-Lopez, and A. Otero. 1996. Effect of a lactic starter culture on the growth and protease activity of Aeromonas hydrophila. J. Appl. Bacteriol. 80:13-18. [Google Scholar]
- 67.Sanz, Y., M. Hernández, M. A. Ferrus, and J. Hernández. 1998. Characterization of Lactobacillus sake isolates from dry-cured sausages by restriction fragment length polymorphism analysis of the 16S rRNA gene. J. Appl. Microbiol. 64:600-606. [DOI] [PubMed] [Google Scholar]
- 68.Schillinger, U., and F. K. Lücke. 1987. Identification of Lactobacilli from meat and meat products. Food Microbiol. 4:199-208. [Google Scholar]
- 69.Schleifer, K. H. 1986. Gram positive cocci, p. 999-1003. In P. H. A. Sneath, N. S. Mair, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 70.Seager, M. S., J. G. Banks, C. Blackburn, and R. G. Board. 1986. A taxonomic study of Staphylococcus spp. isolated from fermented sausages. J. Food Sci. 51:295-297. [Google Scholar]
- 71.Sechi, L. A., and L. Daneo-Moore. 1993. Characterization of intergenic spacers in two rrn operons of Enterococcus hirae ATCC 9790. J. Bacteriol. 175:3213-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selgas, D., B. Sanz, and J. A. Ordóñez. 1989. Actual identity of six micrococcal strains selected as potential starter for dry fermented sausages. Microbiologia SEM 5:53-55. [PubMed] [Google Scholar]
- 73.Sierra, M., M. E. Gonzales Fandos, M. C. García, M. L. García, and B. Moreno. 1995. Numerical taxonomy of an “atypical” population of Gram-positive cocci isolated from freshly dressed lamb carcasses. Int. J. Food Microbiol. 24:363-373. [DOI] [PubMed] [Google Scholar]
- 74.Simonetti, P., and C. Cantoni. 1983. Coagulase negative staphylococci for dry sausage ripening. Ind. Aliment. 22:262-264. [Google Scholar]
- 75.Smith, J. L., and S. A. Palumbo. 1980. Inhibition of aerobic and anaerobic growth of Staphylococcus aureus. Int. J. Food Safety 4:221-233. [Google Scholar]
- 76.Stackebrandt, E., C. Koch, O. Gvozdiak, and P. Schumann. 1995. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Bacteriol. 45:682-692. [DOI] [PubMed] [Google Scholar]
- 77.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thornill, P. J., and T. M. Cogan. 1984. Use of gas-liquid chromatography to determine the end-products of growth of lactic acid bacteria. Appl. Environ. Microbiol. 47:1250-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Torriani, S., F. Dellaglio, and M. Palummeri. 1990. Characterization of lactobacilli isolated from Italian salami. Ann. Microbiol. 40:225-233. [Google Scholar]
- 80.Torriani, S., R. Di Bucchianico, F. Pattarini, G. Zabeo, and F. Dellaglio. 1994. Presenza e characterizzazione biotecnologica di batteri lattici e Micrococcaceae negli insaccati abruzzesi. Ind. Conserv. 69:3-9. [Google Scholar]
- 81.van den Elzen, A. M., and J. M. Snijders. 1993. Critical points in meat production lines regarding the introduction of Listeria monocytogenes. Vet. Q. 15:143-145. [DOI] [PubMed] [Google Scholar]
- 82.Watts, J. L., and R. J. J. Yancey. 1994. Identification of veterinary pathogens by use of commercial identification systems and new trends in antimicrobial susceptibility testing of veterinary pathogens. Clin. Microbiol. Rev. 7:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yost, C. K., and F. M. Nattress. 2000. The use of multiplex PCR reactions to characterize populations of lactic acid bacteria associated with meat spoilage. Lett. Appl. Microbiol. 31:129-133. [DOI] [PubMed] [Google Scholar]