Abstract
Despite efforts to minimize fecal input into waterways, this kind of pollution continues to be a problem due to an inability to reliably identify nonpoint sources. Our objective was to find candidate source-specific Escherichia coli fingerprints as potential genotypic markers for raw sewage, horses, dogs, gulls, and cows. We evaluated 16S-23S rRNA intergenic spacer region (ISR)-PCR and repetitive extragenic palindromic (rep)-PCR analyses of E. coli isolates as tools to identify nonpoint fecal sources. The BOXA1R primer was used for rep-PCR analysis. A total of 267 E. coli isolates from different fecal sources were typed with both techniques. E. coli was found to be highly diverse. Only two candidate source-specific E. coli fingerprints, one for cow and one for raw sewage, were identified out of 87 ISR fingerprints. Similarly, there was only one candidate source-specific E. coli fingerprint for horse out of 59 BOX fingerprints. Jackknife analysis resulted in an average rate of correct classification (ARCC) of 83% for BOX-PCR analysis and 67% for ISR-PCR analysis for the five source categories of this study. When nonhuman sources were pooled so that each isolate was classified as animal or human derived (raw sewage), ARCCs of 82% for BOX-PCR analysis and 72% for ISR-PCR analysis were obtained. Critical factors affecting the utility of these methods, namely sample size and fingerprint stability, were also assessed. Chao1 estimation showed that generally 32 isolates per fecal source individual were sufficient to characterize the richness of the E. coli population of that source. The results of a fingerprint stability experiment indicated that BOX and ISR fingerprints were stable in natural waters at 4, 12, and 28°C for 150 days. In conclusion, 16S-23S rRNA ISR-PCR and rep-PCR analyses of E. coli isolates have the potential to identify nonpoint fecal sources. A fairly small number of isolates was needed to find candidate source-specific E. coli fingerprints that were stable under the simulated environmental conditions.
Fecal pollution is a serious environmental problem that affects many coastal and inland waters worldwide (1, 3, 4, 14). Point source discharges such as raw sewage, storm water, combined sewer overflows, effluents from wastewater treatment plants, and industrial sources are considered the major contributors to fecal pollution (22). Despite efforts to minimize fecal input from these sources into waterways in general and recreational water sites in particular, fecal contamination continues to be a problem, and policy makers have come to recognize the importance of nonpoint sources, such as agricultural runoff, dogs, horses, birds, and pleasure boats (32). These nonpoint source inputs are dispersed and sporadic, which makes their detection difficult. In order to develop public health management programs, identification of fecal sources is crucial.
A variety of methods, such as phage susceptibility (27) and multiple antibiotic resistance (MAR) profiles (21, 34), have been proposed to identify fecal sources in water. However, these techniques have some inherent uncertainties. The susceptibility of bacteria to specific phages undergoes periodic cycling due to the natural adaptation-counteradaptation cycle (49), and the relationship between phage and bacterial numbers is still not clear (12). MAR profiles can distinguish between human and nonhuman sources of Escherichia coli (26) and streptococci (23). However, antibiotic resistance genes are typically encoded on plasmids. These mobile genetic elements are often lost or obtained in response to altered environmental conditions, raising questions as to the stability of these markers (16) and the susceptibility of MAR profiles to changing environmental conditions (19).
Presently, DNA fingerprinting techniques such as ribotyping (7, 25, 39), pulsed-field gel electrophoresis (38), and PCR analysis of the 16S-23S rRNA intergenic spacer region (ISR) (6) and of repetitive extragenic palindromic (rep) sequences of E. coli isolates (15) are used to discriminate between human and nonhuman sources of fecal material. These methods depend on a library which consists of a collection of fingerprints of microorganisms from different potential fecal sources. The aim of these methods is to compare the fingerprints of environmental isolates to the library, which would indicate if the fecal pollution in the environment is derived from a particular host group represented in the library.
Rep-genotyping uses primers complementary to interspersed conserved repetitive DNA elements, present in multiple copies throughout the genome (44). Three families of repetitive elements have been identified: the repetitive extragenic palindromic (REP) sequences (42), the enterobacterial repetitive intergenic consensus (ERIC) sequences (29), and the BOX sequences (35). These repetitive elements are thought to be highly evolutionarily conserved because rep sites are essential protein-DNA interaction sites or because these sequences may propagate themselves as “selfish” DNA by gene conversion (44). Amplification of the distinct genomic regions located between these repetitive elements results in a distinctive strain pattern (45).
rRNA (rrn) operons are typically arranged in the order 16S-ISR-23S-ISR-5S and are present in multiple copies. For example, E. coli has seven rrn operons (10). These ISRs are under minimal selective pressure and often vary among strains, whereas the flanking rRNA genes are highly conserved. Amplification of the ISR can therefore be performed with universal primers targeted to conserved sites in the 16S and 23S rRNA genes (31). Thus, ISR analysis can be performed on all organisms and yet has the ability to discriminate between species and strains (2). The 16S-23S ISR is believed to be involved in the processing of precursor rRNA and in some organisms, such as E. coli, contains coding sequences for tRNA molecules (10, 46).
The objective of this study was to find candidate source-specific E. coli fingerprints as potential genotypic markers for raw sewage, horses, dogs, gulls, and cows. We evaluated 16S-23S rRNA ISR-PCR and rep-PCR analysis of E. coli isolates as tools to identify nonpoint fecal sources. The BOXA1R primer, based on the boxA sequence, was used in rep-PCR analysis (35). E. coli was selected because it has long been used as an indicator of human enteric pathogens in aquatic systems and is present in most warm-blooded animals (3, 22). Recent studies have shown that both ISR-PCR and rep-PCR analyses can conceptually differentiate between human and nonhuman fecal sources (6, 15). However, questions remain on whether these techniques can discriminate between nonhuman sources such as agricultural runoff or canine fecal material. Moreover, the stability of BOX and ISR fingerprints during the transition of E. coli from its primary habitat (the intestines of warm-blooded animals) to a secondary habitat (a water body) is unknown (19, 47). If these genotypic fingerprints are unstable, then this approach is likely of little use. Finally, the practicality of these methods is unresolved. For example, sample size requirements are critical to field implementation of these approaches since high costs are involved in the isolation and analysis of microorganisms (23).
MATERIALS AND METHODS
E. coli sources.
E. coli sources were chosen according to possible fecal sources responsible for pollution of the Belgian coast. Table 1 gives an overview of the fecal sources, the numbers of individuals sampled per fecal source, the number of isolates, and the date of collection. To obtain human-derived E. coli isolates, raw sewage samples were taken from the influent of a domestic wastewater treatment plant (Ossemeersen, Ghent, Belgium). The same plant was sampled on three different days. Dog fecal samples were collected at three different households (Ghent, Belgium), horse fecal samples were collected at three different maneges (Ghent, Belgium), cow fecal samples were taken from three different dairy cows on the same farm (Diksmuide, Belgium), and gull fecal samples were collected from three different gulls on the beach at Koksijde (Belgium). Larus argentatus and Larus ridibundus are the dominatant (>80%) gull species in this coastal area. Source samples were taken in a sterile container, stored at 4°C, and processed within 24 h after collection.
TABLE 1.
E. coli isolates used in this study
| Source | No. of isolates | No. of isolates
|
Sampling datesa | |
|---|---|---|---|---|
| BOX-PCR | ISR-PCR | |||
| Raw sewage | 3 | 49 | 48 | 10/6 (1)-3/18 (1)-4/29 (1) |
| Dog | 3 | 95 | 67 | 11/18 (1)-3/25 (2) |
| Cow | 3 | 65 | 65 | 11/3 (1)-2/18 (2) |
| Horse | 3 | 58 | 67 | 1/21 (1)-3/11 (2) |
| Gull | 3 | 66 | 82 | 1/13 (1)-2/3 (2) |
| Total no. analyzed | 333 | 329 | ||
The number of individuals sampled on each day is given in parentheses after the date. Sewage samples were obtained on three different days.
E. coli isolation.
A 10-fold dilution (n = 3) was plated on violet red bile agar (Oxoid Ltd. CM107, Hampshire, England) with the overlay method (33). After 24 h of incubation at 44.5°C, colonies were randomly picked and streaked onto MacConkey agar (Oxoid Ltd. CM7) and chromogenic E. coli/coliform agar (Oxoid Ltd. CM956). After 24 h of incubation at 37°C, colonies that turned red on MacConkey agar and blue on chromogenic E. coli/coliform agar were streaked onto plate count agar (Oxoid Ltd. CM325) and incubated at 37°C for 24 h. Streaking on plate count agar and incubation were repeated two more times. The pure cultures were then used to inoculate tryptone water (1%, wt/wt) (Oxoid Ltd. CM87) and EC broth containing 4-methylumbelliferyl-d-glucuronide (Oxoid Ltd. CM979) and incubated for 48 h at 37 and 44.5°C, respectively. Isolates that produced indole from tryptophan and were positive for gas production and fluorescence in EC broth containing 4-methylumbelliferyl-d-glucuronide were designated E. coli isolates and used for subsequent studies (33). Isolation efficiency was expressed as the ratio of the number of isolates with these characteristics over the total number of isolates randomly picked from violet red bile agar.
DNA extraction.
Half a plate of colonies grown on plate count agar was suspended in 500 μl of DNase- and RNase-free filter-sterilized water (Sigma-Aldrich Chemie, Steinheim, Germany). DNA was extracted according to the protocol from Hambly et al. (24). Fifty microliters of lysis buffer (1% [wt/wt] sodium dodecyl sulfate, 0.05 M EDTA, pH 8) was added to the cell suspension. After 15 min of incubation at room temperature, a 1:1 (vol/vol) phenol extraction was performed and followed by two 1:1 (vol/vol) phenol-chloroform extractions. Between each extraction step, the suspension was centrifuged for 10 min at 14,360 × g. DNA was precipitated by adding 10% (vol/vol) 3.3 M sodium acetate (pH 5.2) and 75% (vol/vol) isopropyl alcohol (100%), incubation for 1 h at −20°C, and centrifugation for 30 min at 14,360 × g. The pellet obtained was resuspended in 200 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). DNA concentrations of extractions of 10 isolates were measured with a spectrophotometer and found to be within 2 to 10 μg/μl.
Rep-PCR.
BOX-PCR fingerprints were obtained with the BOXA1R primer (5′-CTACGGCAAGGCGACGCTGACG- 3′) (45). PCR was performed as described by Rademaker and de Bruijn (40) with 2 μl of E. coli DNA in a volume of 25 μl. The PCR mixture contained 0.4 μM primer BOXA1R, 5 mM each deoxynucleoside triphosphate, 20 μl of 5× Gitschier buffer (83 mM NH4SO4, 335 mM Tris-HCl [pH 8.8], 55.5 mM MgCl2, 33.5 mM EDTA, and 150 mM β-mercaptoethanol in 200 ml of buffer), 8 U of Taq DNA polymerase (Promega, Madison, Wis.), 0.8 μl (20 mg/ml) of bovine serum albumin (Boehringer), 10 μl of dimethyl sulfoxide (Sigma-Aldrich Chemie), and DNase- and RNase-free filter-sterilized water (Sigma-Aldrich Chemie) to a final volume of 100 μl.
Each PCR was performed with a 9600 thermal cycler (Perkin-Elmer, Norwalk, Conn.). The PCR was initiated by incubating the reaction mixture at 95°C for 2 min, followed by 30 cycles of 3 s at 94°C, 30 s at 92°C, 1 min at 50°C, and 8 min at 65°C. The reaction was terminated with an extension step consisting of 8 min of incubation at 65°C. All PCR experiments contained a positive control (DNA of E. coli ATCC 25922) and a negative control (no DNA). DNA concentrations of PCR products of 10 isolates were measured with a spectrophotometer and found to be within 1.5 to 2 μg/μl. PCR samples (10 μl) were loaded on a 1.5% horizontal agarose gel (Gibco-BRL 15510-027; Invitrogen S.A., Merelbeke, Belgium) together with a 1-kb size ladder (0.5 μg/well; N.V. Invitrogen S.A.), and the positive control. All gels were run in 0.5× TAE (10 mM Tris, 5 mM acetate, 0.1 mM EDTA, pH 7.4) for 17.5 h at 4°C and 70 V, stained for 20 min in 0.5× TAE buffer containing 0.5 μg of ethidium bromide per ml, and immediately photographed on a UV transillumination table with a video camera module (Vilbert Lourmat, Marne-la-Vallée, France).
ISR-PCR and DGGE.
The two universal primers employed for amplification of the 16S-23S ISR were adapted from Jensen et al. (31). The forward primer, designated G1 (5′-GAAGTCGTAACAAAGG-3′), is located approximately 30 to 40 bp upstream of the spacer boundary. A GC-clamp (5′-CGCCCGCCGCGCCCCGCGCCGTCCCGCCGCCCCCCGCCCCC-3′) was added to the 5′ end of the G1 primer (36). The reverse primer, L1 (5′-CAAGGCATCCACCGT-3′), is located approximately 20 bp downstream of the spacer boundary.
PCR was carried out in a volume of 25 μl, which contained 1 μl of E. coli DNA. The PCR mixture contained 2 μM G1 and 0.2 μM L1, 200 μM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 μl of thermophilic DNA polymerase 10× reaction buffer (MgCl2 free), 2.5 U of Taq DNA polymerase (Promega), 0.25 μl (20 mg/ml) of bovine serum albumin (Boehringer), and DNase- and RNase-free filter-sterilized water (Sigma-Aldrich Chemie) to a final volume of 100 μl. The PCR was initiated with 3 min at 94°C, followed by 25 cycles of 1 min at 94°C, 2 min at 55°C, and 2 min at 72°C. The final cycle was followed by an additional 7 min at 72°C. All PCR experiments contained a positive control (DNA of E. coli ATCC 25922) and a negative control (no DNA).
Denaturing gradient gel electrophoresis (DGGE) (36) was performed with the Bio-Rad D Gene System (Bio-Rad, Hercules, Calif.). Samples (10 μl) were loaded onto 6% (wt/vol) polyacrylamide gels with a denaturing gradient ranging from 35 to 50% (where 100% denaturant contains 7 M urea and 40% formamide). Electrophoretic charge was applied in the same direction as the denaturants. All gels were run in 1× TAE buffer for 16 h at 60°C and 45 V, stained for 20 min in 200 ml of 1× TAE containing 16 μl of SYBR Green I nucleic acid gel stain (1:10,000 dilution; FMC BioProducts, Rockland, Maine), and immediately photographed as described above.
Fingerprint stability experiment.
The stability of the BOX and ISR E. coli fingerprints was assessed at three different temperatures (4, 12, and 28°C) in natural water. Two E. coli isolates from one dog (D1 and D2) and two E. coli isolates from one cow (C1 and C2) were grown overnight in nutrient broth (Oxoid Ltd. CM1). Then 50 μl of the overnight cultures was inoculated into 5 ml of autoclaved canal water (21 ± 1 mg of total organic carbon liter−1, 7.5 ± 0.1 mg of NO3− liter−1, <0.1 mg of NO2− liter−1) (Coupure, Ghent, Belgium), resulting in an initial concentration of approximately 106 CFU/ml. The tubes were shaken every 2 days to supply the cultures with oxygen. The fingerprint stability experiment ran for 150 days, after which isolates were recovered on MacConkey agar and transferred to plate count agar. The BOX and ISR fingerprints of five colonies per E. coli isolate and per temperature were compared to the fingerprint of the original isolate.
BOX and ISR fingerprint analysis.
Gel images and data were analyzed with Bionumerics software (version 1.5; Applied Maths, Kortrijk, Belgium). For the BOX fingerprints, band positions on each gel were normalized with the 1-kb size ladder as an external reference standard. Only DNA fragments ranging from 2,036 bp to 506 bp were used in the analysis (17). The bands were assigned to band classes, which resulted in transformation of a fingerprint into a binary table (0, absent in band class; 1, present in band class). The position tolerance settings for assigning band classes were calculated by tolerance and optimization analysis with the band-based Dice coefficient (13). The ISR fingerprints were analyzed similarly except for normalization. The ISR fingerprint of the E. coli ATCC 25922 strain was used as the external reference standard for normalization.
A matrix of similarities between the band class binary tables of the fingerprints was calculated based on the Dice coefficient (13). Dendrograms were created with the unweighted pair group method with arithmetic averages (UPGMA) linkage. To differentiate between different BOX and ISR fingerprints, a dendrogram was constructed based on Dice similarities between the binary tables of the E. coli ATCC 25922 strain, used as a positive control in all PCR runs and loaded on all gels. The similarity value of the cluster that contained all E. coli ATCC 25922 strains served as the similarity cutoff to identify distinctive BOX and ISR fingerprints.
Richness estimation.
The richness of the E. coli population in each fecal source type was estimated by the Chao1 index (9) with the freeware program EstimateS (version 6.0; R. K. Colwell, http://viceroy.eeb.uconn.edu/estimates). For each fecal source, a dendrogram was created with UPGMA linkage based on the matrix of similarities between the binary tables of the E. coli isolates of one sample. Clusters with a similarity coefficient lower than the similarity cutoff, were considered distinctive BOX and ISR fingerprints. The amount of E. coli isolates present in the distinctive BOX and ISR fingerprints served as input for the nonparametric richness estimation.
Selection of candidate source-specific E. coli fingerprints.
To search for candidate source-specific E. coli fingerprints, a dendrogram was created with UPGMA linkage based on the matrix of similarities between the binary tables of the 267 E. coli isolates from the five fecal sources. Clusters with a similarity coefficient lower than the similarity cutoff were considered distinctive BOX and ISR fingerprints. Distinctive BOX and ISR fingerprints that contained E. coli isolates from multiple samples of one fecal source type were selected as candidate source-specific E. coli fingerprints for the technique used.
Internal stability of BOX and ISR library.
Discriminant analysis was used to verify the internal stability of each fecal source, i.e., to calculate how many of the 267 isolates would be assigned to the correct fecal source. This was done by jackknife analysis (maximum similarity) on the Dice similarity data of the BOX and ISR fingerprints individually and for the combination of both fingerprints of each E. coli isolate. The average rate of correct classification (ARCC) for the library was obtained by averaging the correct classification percentages for all sources (48).
RESULTS
E. coli isolation.
The isolation procedure had a high efficiency for selection of E. coli isolates. Isolation from fecal material resulted in 100% isolation efficiency for horse and gull, 97% for cow, and 95% for dog samples. However, isolation from raw sewage samples resulted in a lower isolation efficiency of 67%.
BOX and ISR fingerprints.
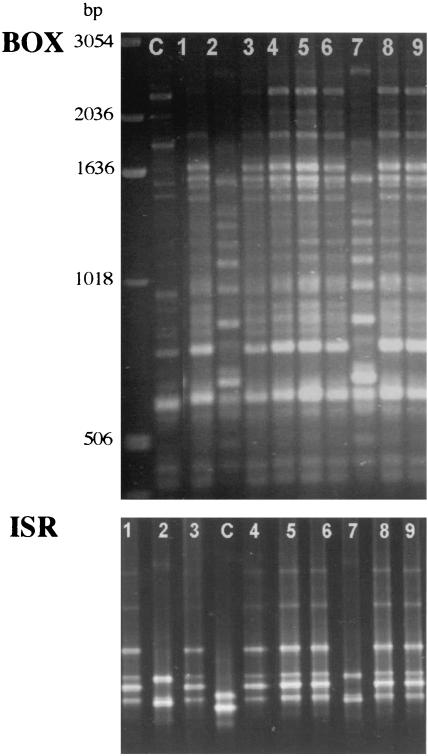
Figure 1 shows typical fingerprints of E. coli isolates from a gull sample generated by rep-PCR with primer BOXA1R and by ISR-PCR. The BOX fingerprints were relatively complex and generally consisted of 15 to 25 bands. The ISR fingerprints were less complex and consisted of two to six bands.
FIG. 1.
BOX and ISR fingerprints of gull E. coli isolates (lanes 1 to 9) and of E. coli ATCC 25922 (lane C). The first lane contains a 1-kb molecular size ladder for normalization of the BOX fingerprints.
Similarity cutoff.
E. coli ATCC 25922 strain was used as the positive control in all PCR runs and loaded on all gels. The similarity value of the cluster that contained all these E. coli ATCC 25922 strains served as a similarity cutoff to identify distinctive BOX and ISR fingerprints. The similarity cutoff was 49% for BOX-PCR and 68% for ISR-PCR. Clusters with a similarity coefficient lower than the cutoff value were considered distinctive BOX and ISR fingerprints. The choice of the similarity cutoff has an influence on the number of distinctive BOX and ISR fingerprints. In general, a lower similarity cutoff yields fewer fingerprints and a higher percentage of sharing among sources than a high similarity cutoff. The similarity cutoff chosen here can be considered very stringent because its low value results in the grouping of many isolates with undifferentiated fingerprints that are shared between many sources.
Selection of candidate source-specific E. coli fingerprints.
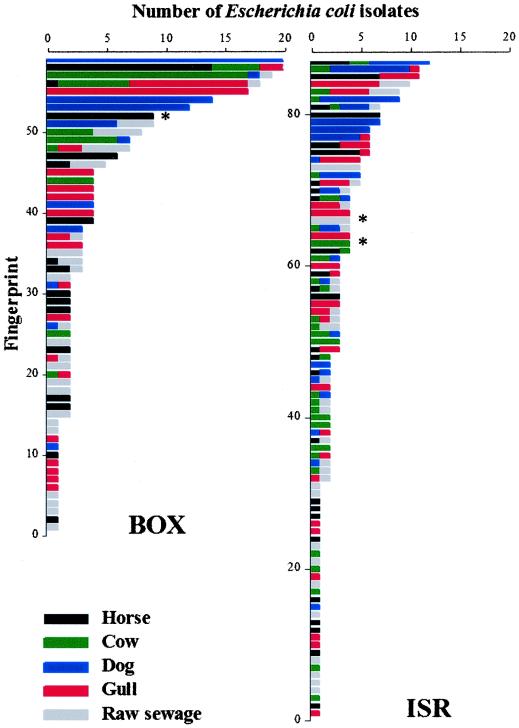
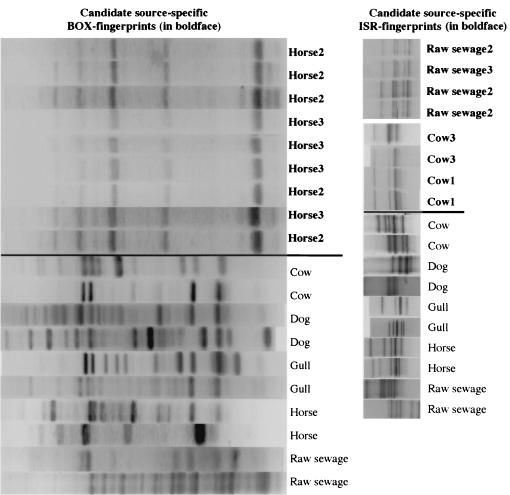
Figure 2 gives the distribution of the 267 E. coli isolates from the different fecal sources between the distinctive BOX and ISR fingerprints. A high diversity in fingerprints was encountered: 267 isolates yielded 59 distinctive BOX fingerprints and 87 distinctive ISR fingerprints. One horse-specific E. coli fingerprint was found with BOX-PCR, and one cow- and raw sewage-specific E. coli fingerprint was obtained with ISR-PCR (Fig. 3) Most of the candidate source-specific E. coli fingerprints of the different isolates were identical, although we do not have sequence information to confirm this. These candidate source-specific E. coli fingerprints were present in only two of the three source samples. Neither the gull nor the individual dogs had any BOX and ISR fingerprints in common. In each individual gull and dog, multiple E. coli isolates had identical BOX and ISR fingerprints.
FIG. 2.
Distribution of the 267 E. coli isolates from different fecal sources between distinctive BOX and ISR fingerprints. The asterisks (*) indicate the candidate source-specific E. coli BOX and ISR fingerprints.
FIG. 3.
Candidate source-specific E. coli BOX fingerprint for horse and the candidate source-specific E. coli ISR fingerprint for both raw sewage and cow, as opposed to randomly chosen non-source-specific BOX and ISR fingerprints of all fecal source types. The candidate source-specific fingerprints are situated above the line. The sample number of the candidate source-specific E. coli fingerprints is indicated in boldface.
Internal stability of BOX and ISR library.
Discriminant analysis was done with jackknife analysis with maximum similarity instead of average similarity in order to avoid averaging out unique fingerprints. The average rate of correct classification (ARCC) for the five source categories was 83% for BOX-PCR analysis, 67% for ISR-PCR analysis, and 84% for BOX- plus ISR-PCR analysis. ISR-PCR was almost as useful as BOX-PCR for correctly classifying dog and gull isolates. The high classification rates obtained with ISR-PCR and BOX-PCR analysis were 87% and 89% for gull and 83% and 94% for dog, respectively (Table 2). However, the ability to correctly classify the isolates of the horse, cow, and raw sewage samples was substantially higher with BOX-PCR. The use of BOX plus ISR fingerprint data of each E. coli isolate in comparison with the ISR fingerprint data of each E. coli isolate resulted in a major improvement in correctly classifying the isolates. For example, the correct classification rate increased to 31% for the horse samples. The improvement was minor when the BOX plus ISR fingerprint data were used in comparison with only the BOX fingerprint data.
TABLE 2.
Assignment of E. coli isolates to fecal source groups by jackknife analysis (maximum similarities) for BOX-PCR, ISR-PCR, and the combination of BOX-plus ISR-PCRa
| Method | Source | % of E. coli isolates in assigned group
|
||||
|---|---|---|---|---|---|---|
| Raw sewage | Horse | Gull | Cow | Dog | ||
| BOX-PCR | Raw sewage | 68 | 2 | 3 | 9 | 1 |
| Horse | 5 | 85 | 3 | 2 | 2 | |
| Gull | 7 | 4 | 89 | 5 | 0 | |
| Cow | 16 | 4 | 2 | 80 | 3 | |
| Dog | 5 | 6 | 3 | 4 | 94 | |
| ISR-PCR | Raw sewage | 47 | 13 | 0 | 11 | 2 |
| Horse | 9 | 68 | 2 | 16 | 1 | |
| Gull | 7 | 6 | 87 | 4 | 1 | |
| Cow | 26 | 11 | 8 | 51 | 13 | |
| Dog | 10 | 2 | 3 | 18 | 83 | |
| BOX-plus ISR-PCR | Raw sewage | 65 | 0 | 3 | 7 | 0 |
| Horse | 0 | 91 | 3 | 2 | 2 | |
| Gull | 7 | 4 | 89 | 2 | 0 | |
| Cow | 23 | 2 | 2 | 82 | 3 | |
| Dog | 5 | 4 | 3 | 7 | 95 | |
Values in boldface indicate the percentage of isolates correctly assigned to fecal source groups.
When nonhuman sources were pooled so that each isolate was classified as animal or human derived (raw sewage) the correct classification rate with BOX-PCR was 67% for human-derived and 96% for animal samples. When ISR-PCR was used, the correct classification rate was 49% for human-derived and 94% for animal samples. This resulted in an ARCC of 82% for BOX-PCR analysis and 72% for ISR-PCR analysis with pooled data.
Richness estimation.
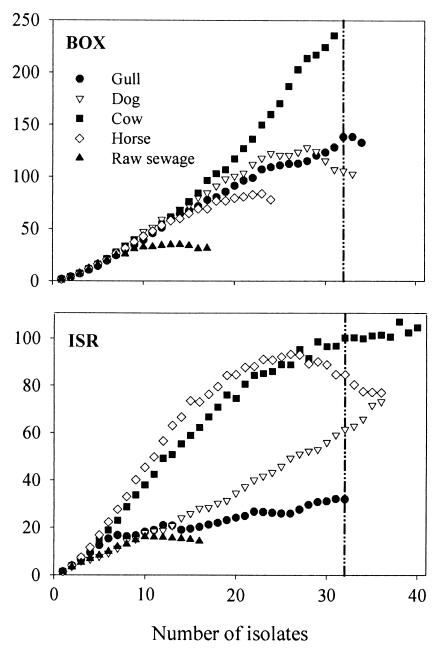
Figure 4 gives the Chao1 richness estimation curves for both fingerprinting techniques, based on one sample of each fecal source. In total, 139 isolates were used in the BOX-PCR analysis and 158 isolates in the ISR-PCR analysis. Nonparametric richness estimators have been developed to estimate species richness from samples of macroorganisms. Recently, interest is emerging in applying these tools to microorganisms (28). The operational taxonomic units for the nonparametric richness estimation for this study were defined not as species but as the number of distinctive BOX and ISR fingerprints. The Chao estimators (8, 9) compare the proportion of fingerprints that have been observed before, i.e., in another sampling, to those that were observed only once. In a very rich community, the probability that a fingerprint will be observed more than once is low, and most fingerprints are only represented by one individual in a sample. However, in a depauperate community, the probability that a fingerprint will be observed more than once is higher, and many fingerprints are observed multiple times in a sample (28). In this study, the Chao1 estimator was used to determine the number of E. coli isolates needed to reliably estimate the richness of the E. coli population in the different fecal sources. Since the number of samples per fecal source was limited to three individuals, the actual E. coli richness cannot be conclusively assessed from this study.
FIG. 4.
Chao1 estimated richness based on one sample per fecal source. Isolates were differentiated by BOX-PCR (n = 139) and ISR-PCR (n = 158). Each value represents the mean for 50 randomizations of sample order. The vertical line indicates the mean value of the number of isolates used per fecal source.
The Chao1 estimator for horse, gull, and raw sewage leveled off after 22, 32, and 15 isolates, respectively, with BOX-PCR and after 25, 32, and 15 isolates, respectively, with ISR-PCR, suggesting that after these points, the Chao1 estimate is relatively independent of sample size. Thus, this number of isolates is a representative sample of all the fingerprint types present in the E. coli population of the fecal source, which implies that further isolation would not result in greater richness. There was almost no difference in the number of isolates between BOX-PCR and ISR-PCR for horse, gull, and raw sewage samples. However, the number of isolates for cow and dog differed according to the fingerprint technique used. When BOX-PCR was used, the Chao1 estimate for dog was already independent of sample size after 29 isolates. However, for cow no plateau was observed after 31 isolates. ISR-PCR resulted in a Chao1 estimate independent of sample size at 32 isolates for cow, whereas for dog the Chao1 estimator continued to rise after 36 isolates.
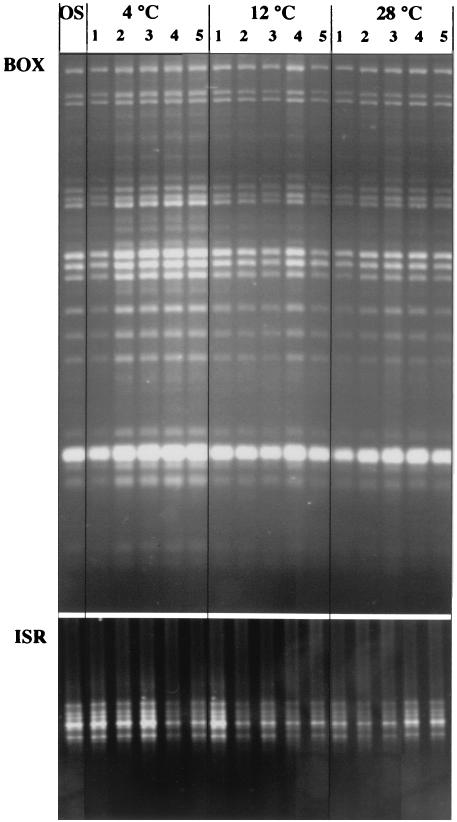
Fingerprint stability experiment.
All four isolates incubated at all three temperatures except C2 at 4°C yielded colonies after plating on MacConkey agar. The BOX and ISR fingerprints of five colonies per E. coli isolate and per temperature were compared to the fingerprint of the original isolate. Figure 5 shows the BOX and ISR fingerprints of C1 of the five colonies typed per temperature. No change in BOX and ISR fingerprint was observed for this isolate at the different incubation temperatures after prolonged incubation. The same result was found for all the other isolates.
FIG. 5.
BOX- and ISR-PCR fingerprints of a cow E. coli isolate (C1) after 150 days of incubation in natural water at 4, 12, and 28°C. The BOX and ISR fingerprints of five colonies (lanes 1 to 5) per E. coli isolate and per temperature were compared to the original isolate fingerprint (lane OS).
DISCUSSION
Candidate source-specific E. coli fingerprints were found for cow, horse, and raw sewage samples. Raw sewage and cows are widely known to pose a public health risk because raw sewage carries high levels of human pathogens (20) and ruminants are considered the main carrier of E. coli O157:H7 (43). Thus, we obtained candidate source-specific E. coli fingerprints for the most important fecal sources in terms of human health risk assessment.
As noted by others (11, 47), the E. coli isolates were highly diverse, with only two candidate source-specific E. coli fingerprints for cow and raw sewage, identified out of the 87 ISR fingerprints present in the 267 isolates typed. Similarly, there was only one candidate source-specific E. coli fingerprint for horse out of a total of 59 BOX fingerprints. Most of the candidate source-specific E. coli fingerprints in the different isolates were identical, although we do not have sequence information to confirm this. The use of the similarity cutoff to select candidate source-specific E. coli fingerprints was rather stringent and based on the reproducibility of DNA extraction, PCR, and electrophoresis of a standard E. coli strain. This approach limits the number of candidate source-specific E. coli fingerprints because a lower similarity cutoff results in fewer candidate source-specific E. coli fingerprints, whereas a higher similarity cutoff can increase the number of candidate source-specific E. coli fingerprints. For example, when the similarity cutoff for ISR-PCR analysis was increased to 80%, one additional candidate source-specific E. coli fingerprint for horse was found.
The observed high diversity suggests that more samples per fecal source type need to be analyzed to determine if the candidate source-specific E. coli fingerprints are indeed source specific. Instead of isolating E. coli from additional fecal samples, primers can be designed for the candidate source-specific E. coli fingerprints. Multiple fecal samples can then be analyzed with a multiplex PCR to determine the source specificity of these candidate source-specific E. coli fingerprints.
Questions also remain on whether the use of these candidate source-specific E. coli fingerprints is geographically limited and why the candidate source-specific E. coli fingerprints were present in only two of the three source samples. The latter could be explained if these candidate source-specific E. coli fingerprints are intrinsic to some source individuals or by temporal diversity of E. coli (18). Follow-up studies are needed to assess the temporal diversity of these source-specific E. coli fingerprints. It is possible that the candidate source-specific E. coli fingerprints are only present during a certain period of the year, which would limit their applicability as potential genotypic markers.
Another approach to evaluating if 16S-23S rRNA ISR-PCR and rep-PCR analyses have the potential to identify fecal sources is by means of jackknife analysis. Despite the limited number of candidate source-specific E. coli fingerprints found, we were able to create a fingerprint library with an average rate of correct classification (ARCC) for the five source categories of 83% for BOX-PCR analysis, 67% for ISR-PCR analysis, and 84% for BOX- plus ISR-PCR analysis. According to Harwood et al. (26), policy makers find correct classification rates of 60 to 70% very useful. Sometimes water quality managers are primarily interested in discriminating between animal and human contamination and consider determining the major source(s) of animal contamination a secondary objective. When the data were pooled into a human-derived (raw sewage) and an animal group, the ARCC value remained similar for BOX-PCR analysis (82%) but increased from 67 to 72% for ISR-PCR analysis.
BOX-PCR analysis resulted in excellent classification rates for all fecal sources except raw sewage, which still had a relatively good classification rate of 68%. ISR-PCR analysis resulted in low classification rates for raw sewage and cow and high classification rates for horse, gull, and dog. Raw sewage streams can contain small amounts of other fecal sources due to storm water runoff in the sewer system (30). This explains why, for both techniques, most raw sewage fingerprints were incorrectly classified during jackknife analysis. This inherent mixed character can be circumvented with human feces as the source for human fecal contamination instead of raw sewage (7, 15). Nevertheless, we used raw sewage because it is one of the most important human fecal contamination sources at the Belgian coast.
The high classification rates for gull and dog obtained with BOX- and ISR-PCR analysis should be interpreted with caution. During jackknife analysis each fingerprint present in the library is individually taken out of its fecal source group and reassigned to a fecal source group by comparing maximum similarity of the fingerprint with all the other fingerprints remaining in the library. It was found that the different source individuals for gull and dog did not have BOX and ISR fingerprints in common and each source individual contained multiple identical BOX and ISR fingerprints. Therefore it is more likely that gull or dog fingerprints are correctly classified since identical fingerprints are still present in the gull and dog fecal group. Jackknife analysis does not correct for such information. This demonstrates the necessity to use an external validation data set of E. coli BOX and ISR fingerprints out of an additional sample for each fecal source before this fingerprint library can be applied in the field.
The Chao1 estimator indicated that enough isolates were analyzed to characterize the richness of the E. coli population with BOX- and ISR-PCR for horse, gull, and raw sewage. For these sources, 32 isolates were sufficient to obtain a richness estimate independent of sample size. This number includes a safety margin for horse and raw sewage, since the Chao1 estimator leveled off before 32 isolates. However, for dog and cow the number of isolates differed according to the technique used. Additional BOX fingerprints for cow and ISR fingerprints for dog would be obtained when more isolates would be analyzed. In contrast to our approach, some studies obtained only one to three E. coli per individual, and multiple individuals were sampled to obtain a larger number of E. coli isolates (7, 15). With this approach, the composition of the E. coli population within one individual of a fecal source remains unknown, and isolates which are thought to be specific for one source might be common E. coli clones between several sources. In this study, generally 32 isolates of a fecal source were sufficient to characterize the richness of the E. coli population from that source. It is not clear how many individuals need to be sampled to characterize the richness of the E. coli population between different individuals of a fecal source.
The BOX and ISR fingerprints were stable in natural water over a 150-day period at all incubation temperatures. Since the water temperature at the Belgian coast varies from 4 to 20°C during the year, it was decided to incubate the E. coli isolates at 4, 12, and 28°C. The isolates were incubated for 150 days under low-nutrient conditions because Naas et al. (37) found a large amount of genetic variation in E. coli isolates under conditions of starvation, which further increased with the length of time spent in the stationary phase. Our results do not correspond with the findings of Wittam (47), who used multilocus enzyme electrophoresis. The results of the multilocus enzyme electrophoresis analysis indicated substantial changes in E. coli population composition during transition from the host to the external environment. Given the dependence of multilocus enzyme electrophoresis on loci and operating conditions (41), it is difficult to compare his results to our fingerprint results. Therefore, the stability of genotypic markers in E. coli during transition from the host to the external environment may be dependent on the microbial source tracking method used.
In conclusion, 16S-23S rRNA ISR-PCR and rep-PCR analyses of E. coli isolates have the potential to identify nonpoint sources of fecal pollution. A fairly small number of isolates was needed to find candidate source-specific E. coli fingerprints that were stable under the simulated environmental conditions. In view of automation, direct detection of source-specific genotypic markers in water samples without isolation holds promise. However, to make this approach widely applicable, the possible temporal diversity, geographical limitations, and survival of these source-specific genotypic markers in the environment need to be assessed.
Acknowledgments
This research was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT; grant number 001386).
We thank Astrid Van Bossche for technical assistance and Wim De Windt, Nico Boon, Liesbeth De Vos, Sofie Dobbelaere, Lutgart Raskin, Kurt Sys, and Kristof Verthé for critically reading the manuscript.
REFERENCES
- 1.Anderson, S. A., S. J. Turner, and G. D. Lewis. 1997. Enterococci in the New Zealand environment: implications for water quality monitoring. Water Sci. Technol. 35:325-331. [Google Scholar]
- 2.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1998. Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evolution. 47:62-72. [DOI] [PubMed] [Google Scholar]
- 3.Baker, K. H., and D. S. Herson. 1999. Detection and occurrence of indicator organisms and pathogens. Water Environ. Res. 71:530-551. [Google Scholar]
- 4.Baudart, J., J. Grabulos, J. P. Barusseau, and P. Lebaron. 2000. Salmonella spp. and fecal coliform loads in coastal waters from a point vs. nonpoint source of pollution. J. Environ. Qual. 29:241-250. [Google Scholar]
- 5.Bennet, A. F., and R. E. Lenski. 1993. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution 47:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Buchan, A., M. Alber, and R. E. Hodson. 2001. Strain-specific differentiation of environmental Escherichia coli isolates via denaturing gradient gel electrophoresis (DGGE) analysis of the 16S-23S intergenic spacer region. FEMS Microbiol. Ecol. 35:313-321. [DOI] [PubMed] [Google Scholar]
- 7.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 9.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 10.Condon, C., J. Philips, Z. Fu, C. Squires, and C. L. Squires. 1992. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 11:4175-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornuet, J., S. Piry, G. Luikart, A. Estoup, and M. Solignac. 1999. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 14.Dionisio, L. P. C., M. Joao, V. S. Ferreiro, M. L. Fidalgo, M. E. Garcia, and J. J. Borrego. 2000. Occurrence of Salmonella spp in estuarine and coastal waters of Portugal. Antonie von Leeuwenhoek Int. J. Genet. 78:99-106. [DOI] [PubMed] [Google Scholar]
- 15.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freter, R. 1983. Factors affecting conjugal plasmid transfer in natural communities, p. 105-114. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. Academic Press, New York, N.Y.
- 17.Fromin, N., W. Achouak, J. M. Thiery, and T. Heulin. 2001. The genotypic diversity of Pseudomonas brassicacearum populations isolated from roots of Arabidopsis thaliana: influence of plant genotype. Fems Microbiol. Ecol. 37:21-29. [Google Scholar]
- 18.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 20.Gostin, L. O., Z. Lazzarini, V. S. Neslund, and M. T. Osterholm. 2000. Water quality laws and waterborne diseases: Cryptosporidium and other emerging pathogens. Am. J. Public Health 90:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graves, A. K., C. Hagedorn, A. Teetor, M. Mahal, A. M. Booth, and R. B. Reneau. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31:1300-1308. [DOI] [PubMed] [Google Scholar]
- 22.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. BioScience 51:817-825. [Google Scholar]
- 23.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartel, P. G., J. D. Summer, J. H. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed]
- 26.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havelaar, A. H., W. M. Pot-Hogeboom, K. Furuse, R. Pot, and M. P. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 28.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulton, C. S. J., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genome of Escherichia coli, Salmonella typhimurium and other enteric bacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 30.Jagals, P., W. O. K. Grabow, and J. C. Devilliers. 1995. Evaluation of indicators for assessment of human and animal fecal pollution of surface runoff. Water Sci. Technol. 31:235-241. [Google Scholar]
- 31.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2002. A biomarker for the identification of cattle fecal pollution in water with the LTIIa toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biot. 59:97-104. [DOI] [PubMed] [Google Scholar]
- 33.Krieg, N. R., and J. G. Holt. 1984. Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 34.Livermore, D. M., M. Warner, L. M. C. Hall, V. I. Enne, S. J. Projan, P. M. Dunman, S. L. Wooster, and G. Harrison. 2001. Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from west Wales. Environ. Microbiol. 3:658-661. [DOI] [PubMed] [Google Scholar]
- 35.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, R. Hakenbeck, D. A. Morrison, G. J. Boulnois, and J.-P Claverys. 1992. A highly conserved repeated DNA element in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muyzer, G., E. C. de Wall, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198-207. [DOI] [PubMed] [Google Scholar]
- 38.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Phenotypic and genotypic characterization of human and nonhuman Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 39.Parveen, S., M. Portier, K. Robinson, L. Edmiston, M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rademaker, J. L. W., and F. J. de Bruijn. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis, p. 151-171. In G. Caetano-Anollés and P. M. Gresshoff (ed.), DNA markers: protocols, application, and overviews. J. Wiley and Sons, New York, N.Y.
- 41.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern, M. J., G. F.-L. Ames, N. H. Smith, E. C. Robinson, and C. F. Higgins. 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37:1015-1026. [DOI] [PubMed] [Google Scholar]
- 43.Vernozy-Rozand, C., M. P. Montet, and S. Ray-Gueniot. 2002. E. coli O157:H7 in water: a public health problem. Rev. Med. Vet. Toulouse 153:235-242. [Google Scholar]
- 44.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA-sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria with repetitive sequence-based polymerase chain reaction. Methods Cell Biol. 5:25-40. [Google Scholar]
- 46.Whiley, R. A., B. Duke, J. M. Hardie, and L. M. C. Hall. 1995. Heterogeneity among 16S-23S rRNA intergenic spacers of species within the Streptococcus milleri group. Microbiology 141:1461-1467. [DOI] [PubMed] [Google Scholar]
- 47.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie van Leeuwenhoek J. Microbiol. 55:23-32. [DOI] [PubMed] [Google Scholar]
- 48.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]