Abstract
We developed an alternative nested-PCR-restriction fragment length polymorphism (RFLP) protocol for the detection of Cyclospora cayetanensis in environmental samples that obviates the need for microscopic examination. The RFLP method, with the restriction enzyme AluI, differentiates the amplified target sequence from C. cayetanensis from those that may cross-react. This new protocol was used to reexamine a subset (121 of 180) of surface water samples. Samples previously positive when the CYCF3E and CYCR4B primers (33) and RFLP with MnlI (20) were used were also PCR positive with the new primers; however, they were RFLP negative. We verified, by sequencing these amplicons, that while two were most likely other Cyclospora species, they were not C. cayetanensis. We can detect as few as one oocyst seeded into an autoclaved pellet flocculated from 10 liters of surface water. This new protocol should be of great use for environmental microbiologists and public health laboratories.
Cyclospora cayetanensis is a sporulating parasitic protozoan first characterized by Ortega et al. (31). It is a human pathogen that infects epithelial cells of the upper small intestine, usually the jejunum, and causes gastrointestinal distress and, in some cases, a low-grade fever (5). The United States and Canada experienced a number of large foodborne outbreaks in the 1990s (18), and in other developed countries, infection has been largely linked to international travel (37). In many developing countries, transmission has been associated with water (39), soil (4), and uncooked herbs and vegetables (32).
Microscopic detection methods take advantage of the oocyst's autofluorescence under UV light (31). Under a 365-nm dichromatic filter, C. cayetanensis oocysts autofluoresce dark blue, and under a 450- to 490-nm dichromatic filter, they autofluoresce mint green (31). While a trained microscopist can easily examine a few low-volume samples, examination of a large numbers of stool, food, and environmental samples is difficult and tedious.
Thus, molecular-analysis-based methods have been explored because of the potential for automation and high throughput. Ribosomal DNA is the most abundant constituent of nucleic acids within eukaryotes (45), and the 18S small-subunit ribosomal DNA (18S ssrDNA) contains highly conserved regions punctuated by sequences of hypervariability, making it an attractive region on which to focus. The first PCR method for clinical diagnosis, developed by Relman et al. (33), targeted this region. The outer primers, CYCF1E and CYCR2B, are Eukarya specific; the inner primers, CYCF3E and CYCR4B, were designed to specifically target C. cayetanensis. When it was noted that the inner primers would also amplify some Eimeria species, a restriction fragment length polymorphism (RFLP) method that uses the restriction enzyme MnlI was developed by Jinneman et al. (20) to distinguish C. cayetanensis from Eimeria spp. This nested-PCR-RFLP protocol is commonly used to detect this organism in stool (10, 30), foodstuffs (21, 26), and environmental waters (39; J. M. Shields and B. H. Olson, Abstr. Gen. Meet. Am. Soc. Microbiol., abstr. Q-92, 1999).
An important issue likely to be troublesome for environmental microbiologists is the presence of genetically similar microorganisms in environmental waters (Shields and Olson, Proc. Am. Water Works Assoc. 2000 Water Qual. Technol. Conf.) and soils. Of the 17 known Cyclospora species, sequence data are available for only 4: C. cayetanensis, C. cercopitheci, C. colobi, and C. papionis (11). These four species show a high degree of homology in the inner (CYCF3E and CYCR4B) PCR product (37). While there are differences in the sequences of this region, none of these occur within MnlI sites; therefore, these four Cyclospora species appear identical by PCR-RFLP analysis.
During a monitoring study of the Santa Ana River (SAR) in California, we detected a number of samples that were PCR (33)-RFLP (20) positive for C. cayetanensis but were unable to confirm these microscopically because of heavy debris and large concentrated volumes (5 to 8 ml of packed pellet for highly turbid waters). When other Cyclospora species sequences became available (11), we reexamined the 18S ssrDNA regions of the four available Cyclospora species sequences. We identified a hypervariable region and designed a nested-primer PCR-RFLP protocol that does not need microscopic confirmation. This protocol will be useful to microbiologists working with environmental samples and foodstuffs that come into contact with water and soil. Here, we describe the protocol and the results we obtained.
MATERIALS AND METHODS
Collection of environmental water samples.
Environmental water samples were collected in sterile 10-liter cubtainers (Fisher Scientific, Pittsburgh, Pa.). Samples were taken from various sites within the SAR watershed, i.e., SAR at Van Buren Bridge near Arlington, Calif. (SARVB), SAR at Prado Wetlands near Corona, Calif. (SARPW), SAR at Featherly Regional Park near Yorba Linda, Calif. (SARYL), and SAR at Pacific Coast Highway, Huntington Beach, Calif. (SARHB), and from the San Diego Creek (SDC) at Campus Drive, Irvine, Calif. The water used for the minimum detection study was collected from the SARYL site.
Flocculation.
The flocculation method used to concentrate the samples was that of Vesey et al. (43). Resultant pellets from environmental water samples were divided into three aliquots. One was stored in digestion buffer (6) and kept at −80°C for long-term storage. The other two were kept at 4°C, one in digestion buffer for genomic DNA extraction and the other in 2.5% potassium dichromate for microscopic analysis.
Genomic DNA extraction methods. (i) Digestion and extraction of genomic DNA from environmental samples.
The protocol described by daSilva et al. (6) for the detection of microsporidial species in stools was modified. The amounts of digestion buffer and silanized glass beads were adjusted on the basis of the size and nature of the pellet as follows. A 2:1 ratio of digestion buffer to packed pellet was generally used, although more buffer was added if the pellet was difficult to resuspend. An equal weight-to-volume ratio (milligrams per milliliter) of glass beads was used. In the case of packed pellets with a volume of less than 100 μl, a minimum of 300 μl of digestion buffer and 50 mg of glass beads was used.
After pellet digestion and overnight incubation, the samples were centrifuged at 14,000 × g for 1 min to remove debris. Phenol-chloroform was used to remove proteins. The resultant aqueous layer was precipitated with ethanol-NH4 acetate and washed with cold 70% ethanol. Samples were resuspended in 100 μl of sterile double-distilled water (ddH2O). Chelex 100 was added at a concentration of 5%, and the samples were boiled for 2 to 3 min. After centrifugation, the supernatants were transferred to sterile microcentrifuge tubes.
The samples were further purified by passage through a Pharmacia ion-exchange spin column (Amersham Pharmacia Biotech and Science, Piscataway, N.J.) in accordance with the manufacturer's instructions. The samples were resuspended in 100 μl of sterile ddH2O and kept either at −20°C until PCR analysis or at −80°C for long-term storage.
(ii) Seeding of samples for minimum detection limit with primers CYCAO1, CYCAI2, and CYCAR1.
Pellets from flocculated water were divided into 300-mg fractions, 200 μl of phosphate-buffered saline was added to each, and the samples were autoclaved for 20 min to destroy the DNA of organisms present in the samples. The samples were stored in tightly sealed microcentrifuge tubes at 4°C until seeding and DNA extraction. Approximately 1,500 sporulated C. cayetanensis oocysts from Guatemala and Nepal (courtesy of Michael Arrowood of the Parasitic Diseases Division, Centers for Disease Control and Prevention, and Alan Lindquist of the Division of Parasitology, Environmental Protection Agency, respectively) were seeded into each autoclaved environmental sample. Unseeded autoclaved samples were also used to ensure that autoclaving had destroyed the genomic DNA.
(iii) Digestion and extraction of genomic DNA for minimum detection analysis of primers CYCAO1, CYCAI2, and CYCAR1.
The FastDNA kit for soils (Bio 101, Inc., Vista, Calif.) was used to extract and purify genomic DNA from the seeded sediment, as well as from autoclaved unseeded control samples. The manufacturer's instructions were followed, with certain modifications. After agitation in a bead beater, the crude DNA lysate was briefly centrifuged (∼6,000 × g for 30 s), the supernatant was removed, and more buffer and detergent were added to the pellet. This was repeated for a total of four cycles; the final centrifugation was at 14,000 × g for 5 min. The crude lysates were pooled, and crude DNA was precipitated overnight in 5 M ammonium acetate (0.1 volume) and ethanol (2.2 volumes) at −20°C. The crude DNA samples were then centrifuged (20 min at 14,000 × g), and the resultant pellet was washed with 70% cold ethanol, air dried, and then resuspended in 500 μl of phosphate buffer for further purification.
PCR methods. (i) PCR with primers CYCF1E, CYCR2B, CYCF3E, and CYCR4B.
The PCR primers and cycling protocol used were those described by Relman et al. (33), with the addition of HotStart (Perkin-Elmer, Boston, Mass.) and the inclusion of 100 μg of nonfat dry milk to prevent PCR inhibition during amplification of the outer fragment.
(ii) Design of new primers CYCAO1, CYCAI2, and CYCAR1.
Nested primers were designed with the 18S ssrDNA sequences from Cyclospora and Eimeria species found in the National Center for Biotechnology Information GenBank database. Sequences were aligned with CLUSTAL W version 1.8.1 (40). The sequences of the outer fragment primers were 5′-ATAACGAACGAGACCTTAGCCT (CYCAO1) and 5′-AAGGATGCAAAAGTCGTAACAC (CYCAR1). The inner fragment primers were CYCAI2 (5′-CAGGTCTGGGTAATCTTTTGAG-3′) (forward primer) and CYCAR1 (reverse primer).
(iii) Theoretical determination of new-primer (CYCAO1, CYCAI2, and CYCAR1) specificity.
The new primers were compared to those species identified during a BLAST inquiry (1). The 18S ssrDNA sequences of 31 species of Eimeria (11 complete and 20 partial), Isospora robini (complete), and the four Cyclospora species (complete) were examined. Tables 1, 2, and 3 list the species identified and examined (along with their GenBank accession numbers). These tables include the sequence where the inner primer may bind, the Tm (midpoint temperature in degrees Celsius) calculated as described by Rychlik and Rhoads (34) [Tm = 64.9 + (yG + zC − 16.4)/(wA + xT + yG + zC)], the resultant amplicon size in base pairs (when the complete 18S ssrDNA sequence is available), the predicted AluI fragments, and other pertinent information.
TABLE 1.
Species amplified with primers CYCAI2 and CYCAR1
| Organism(s) (reference) | Accession no(s). | Host species | Distribution | Tm (°C)a | Amplicon size (bp) | Alu I fragment sizes (bp) |
|---|---|---|---|---|---|---|
| Cyclosporaand Eimeria spp.b | ||||||
| C. cayetanensis (31) | U40261 | Human | Worldwide | 60.9 | 256 | 98, 88, 55, 15 |
| C. cercopitheci (strains 1 and 2) (11) | AF111184, AF111185 | Cerocopithecus aethiops (African green or vervet monkey) | Ethiopia | 60.9 | 257 | 104, 98, 55 (strains 1 and 2) |
| C. colobi (11) | AF111186 | Colobus guereza (colobus monkey) | Ethiopia | 60.9 | 256 | 104, 98, 55 |
| C. papionis (11) | AF111187 | Papio anubis (olive baboon) | Ethiopia | 60.9 | 257 | 159, 98 |
| E. necatrix (22) | U67119 | Chicken | Worldwide | 60.9 | 256 | 160, 96 |
| E. tenella (42) | U40264 | Chicken | Worldwide | 60.9 | 256 | 160, 96 |
| Isospora robini (27)c | AF080612 | Robin | United States | 55.5 | 255 | 103, 97, 55 |
Calculated as described by Rychlik and Rhoads (34).
Inner forward primer, CYCAI2.
Inner forward primer, 5′-CAGGTCTAGGTAATCTTTTGAG. In the Isospora robini sequence, there is an A (underlined) rather than the C found in CYCA12.
(iv) PCR with primers CYCAO1, CYCAI2, and CYCAR1.
The PCR components, in a final volume of 50 μl, were 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 μg of nonfat dry milk, 200 μM each deoxynucleoside triphosphate, 2.0 mM MgCl2, 0.4 μM each primer, 1 U of AmpliTaq polymerase, and 5 μl of template. The second PCR (CYCAI2 and CYCAR1) consisted of 2.5 μl from the first reaction as the template and the same concentrations of PCR components but without the nonfat dry milk. The cycling protocol for both reactions was the same, i.e., 94°C for 5 min for initial denaturation; 45 cycles of 94°C for 30 s, 59.5°C for 30 s, and 72°C for 90 s; and a final extension of 72°C for 9 min.
(v) Cloning of positive-control plasmid and isolated PCR fragments.
The outer fragment, with primers CYCAO1 and CYCAR1 (444 bp), and PCR amplicons (310 and 257 bp) were cloned into 3,015-bp plasmids with p-GEM-T Easy-Vector (Promega, Madison, Wis.) in accordance with the manufacturer's instructions. Ligation reaction products were transformed with heat shock into competent Escherichia coli DH5α cells. Blue-and-white screening was done with Luria-Bertani agar medium supplemented with 100 μg of ampicillin per ml, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and 80 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. Plasmid DNA from individual white colonies was extracted (Qiagen spin kit; Qiagen) from overnight cultures and then screened by PCR and digestion with the appropriate restriction enzyme.
Analysis of PCR amplicons. (i) Visualization of PCR amplicons.
PCR products (10 μl) were visualized on 1.75% agarose gels supplemented with 0.5 μg of ethidium bromide per ml at constant current in Tris-borate-EDTA buffer. Three molecular size markers were used: 25- and 50-bp ladders (Promega) and DNA weight marker VIII (a mixture of pUCBM21 DNA cleaved with HpaII and pUCBM21 DNA cleaved with DraI and HindIII; Boehringer Mannheim, Indianapolis, Ind.).
(ii) Separation of fragments by denaturing high-performance liquid chromatography.
Because of the probability of multiple similar-size but dissimilar-sequence products, amplicons from PCR (primers CYCAI2 and CYCAR1) were separated by denaturing high-performance liquid chromatography (Wave System, Transgenomic, San Jose, Calif.). PCR products were separated by a 46 to 39% gradient of buffers A (0.1 M triethylammonium acetate [pH 7.0]) and B (0.1 M triethylammonium acetate [pH 7.0], 25% acetonitrile) at 60°C. The collected fractions were amplified and cloned for sequencing.
(iii) Sequencing and alignment of PCR products.
All sequencing was by Davis Sequencing (Davis, Calif.) with an ABI 377 automated sequencer with m13 (−21) as the forward sequencing primer for all cloned PCR fragments. Sequences were aligned with CLUSTAL W, version 1.8.1 (40). Accurate pairwise alignments were unweighted with IUB/Bestfit with a gap open penalty of 100 and a gap extra penalty of 10.
RFLP methods. (i) Confirmation of PCR products (CYCF3E and CYCR4B) by RFLP with MnlI.
The positive PCR products from the second reaction (10 μl) were digested as described by Jinneman et al. (20). Fragments were separated on a 4% NuSieve agarose gel (BioWhittaker Molecular Applications, Rockland, Maine) at constant current in Tris-borate-EDTA buffer for approximately 1.5 h. The agarose gels, containing ethidium bromide (0.5 μg/ml), were photographed under UV light with a gel documentation system (UVP ImageStore 5000; Ultra-Violet Products Ltd., Cambridge, United Kingdom).
(ii) Confirmation of PCR products (CYCAI2 and CYCAR1) by RFLP with AluI.
The positive PCR products from the second reaction (10 μl) were digested with 1 U of AluI (New England Biolabs) at 37°C for 2 h. Fragments were separated, visualized, and photographed as described above.
RESULTS
Environmental samples positive by PCR with primers CYCF3E and CYCR4B and RFLP with MnlI.
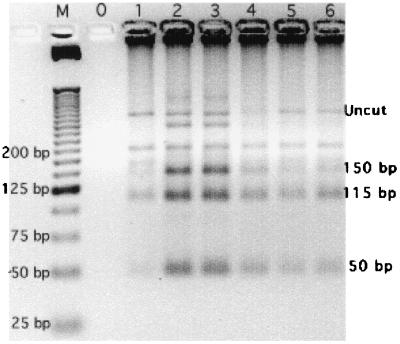
During our initial SAR monitoring study, we detected five samples that were PCR-RFLP positive for C. cayetanensis as described by Relman et al. (33) and Jinneman et al. (20). Figure 1 is the RFLP analysis of these samples. An additional sixth positive sample was detected in water from the SDC (Irvine, Calif.). Digestion of these PCR products (primers CYCF3E and CYCR4B) with the restriction enzyme MnlI resulted in three fragments of approximately 150, 115, and 50 bp (20). While the presence of multiple bands seen in the electrophoresis gel may be the result of incomplete digestion, PCR products from environmental samples are rarely homogeneous and thus may also indicate the presence of amplicons amplified from other, closely related organisms, such as Eimeria species, in the DNA extract.
FIG. 1.
RFLP analysis with MnlI (20) of PCR (primers CYCF3E and CYCR4B 33)-positive samples. Lanes: M, 25-bp ladder; 0, sterile ddH2O control; 1, SDC sample collected on 21 April 1998; 2 and 3, SARHB samples collected on 5 June 1998; 4 and 5, SARVB samples collected on 28 August 1998; 6, SARYL sample collected on 28 September 1998. “Uncut” identifies undigested PCR products.
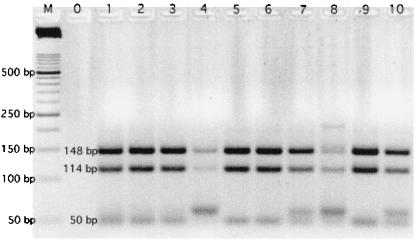
The PCR products from four of the positive environmental samples were cloned and screened by RFLP with MnlI (Fig. 2). While 4 (lanes 4, 7, 8, and 10) of the total of 10 isolates showed RFLP banding patterns not consistent with C. cayetanensis, 6 were consistent.
FIG. 2.
MnlI-digested cloned isolates from PCR (primers CYCF3E and CYCR4B 33)-RFLP (with MnlI 20)-positive samples (lanes 1 to 10). The isolates in lanes 1 to 4 were collected from the SARVB on 28 August 1998, and those in lanes 5 to 10 were collected from the SARYL on 28 September 1998. Lane M is a 50-bp ladder, and lane 0 is a PCR blank.
Four isolates (lanes 1, 2, 6, and 9 in Fig. 2) were sequenced and aligned (Fig. 3). While there are a few base pair differences among the environmental sample isolates and C. cayetanensis, the homology patterns are similar to those found among the available Cyclospora species sequences. In addition, as with Cyclospora species, none of the differences occur within the MnlI sites. Since we were unable to microscopically (under light and epifluorescence illumination) confirm these samples as C. cayetanensis, we reexamined the 18S ssrDNA region and identified a hypervariable area between bp 1486 and 1743. We developed a new nested-primer set and RFLP method.
FIG. 3.
CLUSTAL W (version 1.8.1) (40) multiple-sequence alignment of PCR (33)-RFLP (20)-positive environmental samples from the SAR. Shading indicates sequence differences, underlining indicates MnlI sites, and arrows indicate primer locations and directions. The numbers in sequence labels are the cloned isolate numbers, and the numbers in parentheses correspond to the lane numbers in Fig. 2.
Theoretical analysis for cross-reactivity of new primers.
The outer forward primer (CYCAO1) and the reverse primer (CYCAR1) were identified in all 16 species for which the entire 18S ssrDNA sequence was available (Tables 1 and 2). As shown in Table 1, the inner primer (CYCAI2) was found in all Cyclospora species, Eimeria necatrix, and Eimeria tenella and a very similar site was seen in Isospora robini. However, RFLP with AluI showed a distinctly different banding pattern (98-, 88-, 55-, and 15-bp fragments) for C. cayetanensis. Enzyme digestion of the two Eimeria species produces only two bands, while that of I. robini results in three. With two of the Cyclospora species, three bands are produced and digestion of C. papionis generates two. Table 2 lists those organisms that contain a site with high, although not exact, homology to primer CYCAI2. Since PCR stringency may be inadvertently decreased through errors during amplification or the presence of substances in the extract, we calculated Tm values (degrees Celsius) (34) at which mismatched primer annealing could occur. We determined that even if these species were amplified, AluI digestion would clearly distinguish them from C. cayetanensis.
TABLE 2.
Eimeria species that may be amplified with primers CYCAI2 and CYCAR1
| Organism (reference) | Accession no. | Host | Distribution | Inner forward primer (CYCAI2)a | Tm (°C)b | Amplicon size (bp) | Alu I fragment sizes (bp) |
|---|---|---|---|---|---|---|---|
| E. acervulina (42) | U67115 | Chicken | Worldwide | CGGGGCTGGGTAATCTTCTGAG | 69.4 | 254 | 156, 98 |
| E. bovis (23) | U77084 | Cattle | Worldwide | GAAGTCTAGGTAATCTTTTGAG | 52.4 | 253 | 156, 97 |
| E. brunetti (24) | U67116 | Chicken | Worldwide | CGGGGCTGGGTAATCTTGTGGG | 73.7 | 253 | 155, 98 |
| E. falciformis (35) | AF080614 | Mus (mouse) | NA,c Europe | CAGGTCTAGGTAATCTTTTGAG | 55.5 | 255 | 159, 96 |
| E. meleagrimitis (41) | AF041437 | Meleagris (turkey) | United States | GCAGTCTAGGTAATCTTTTGAG | 55.7 | 254 | 159, 95 |
| E. mitis (42) | U67118 | Chicken | Worldwide | TGGGCGTGGGTAATCTTGTGAG | 69.7 | 250 | 157, 94 |
| E. mivati (12) | U76748 | Chicken | NA, Europe | CGGGGCTGGGTAATCTTGTGAG | 70.2 | 254 | 158, 96 |
| E. nieschulzi (7) | U40263 | Rattus (rat) | Worldwide | CAGGTCTAGGTAATCTTTTGAG | 55.5 | 255 | 157, 98 |
| E. praecox (22) | U67120 | Chicken | Worldwide | TGGGGCTGGGTAATCTTGTGAG | 68.8 | 252 | 158, 94 |
Underlined letters are changes in sequence from CYCAI2.
Calculated as described by Rychlik and Rhoads (34).
NA, North America (includes Canada, the United States, and Mexico).
Table 3 lists those species containing a sequence similar to the CYCAI2 primer but whose complete 18S ssrDNA sequence was not available. It is impossible to predict that amplicons from these species will not contain AluI sites at the same locations as C. cayetanensis. Since RFLP patterns for the Eimeria species listed in Tables 1 and 2 resulted in no more than two bands (155 to 160 bp and 95 to 98 bp), it is unlikely that those listed in Table 3 will cause confusion in differentiation. However, further sequencing of these species, as well as other Cyclospora species, would be prudent.
TABLE 3.
Eimeria species that may be amplified with primers CYCAI2 and CYCAR1a
| Organism (reference) | Accession no. | Host | Distribution | Alul I fragment size (bp) |
|---|---|---|---|---|
| E. adeneodei (28) | AF324212 | Meleagris (turkey) | North America, Europe | 98 |
| E. antrozoi (36) | AF307876 | Antrozous (little brown bat) | North America | 98 |
| E. arizonensis (25) | AF307878 | Peromyscus (deer mouse) | Western North America | 98 |
| E. separata (3) | AF311643 | Rattus (rat) | Worldwide | 98 |
| E. pilarensis (36) | AF324215 | Chiroptera (bat) | SWb United States | No sites before 95 bp |
| E. catronensis (36) | AF324213 | Chiroptera (bat) | SW United States; northern Mexico | No sites before 104 bp |
| E. rioarribaensis (9) | AF307877 | Chiroptera (bat) | SW United States; northern Mexico | 98 |
| E. papillata (15) | AF311641 | Mus (mouse) | Not reported (likely SW United States; Mexico) | 98 |
| E. peromysci (25) | AF339492 | Peromyscus (deer mouse) | SW United States; northern Mexico | 96 |
| E. sevilletensis (25) | AF311644 | Onychomys (mouse) | SW United States | No sites before 101 bp |
| E. albigulae (25) | AF307880 | Neotoma (wood rat) and Chaetodipus (pocket mouse) | Western North America | No sites before 124 bp |
| E. chaetodipi (16) | AF339489 | Chaetodipus (pocket mouse) | Western United States | 98 |
| E. chobotari (14) | AF324214 | Dipodomys (kangaroo rat) | SW United States Mexico | 97 |
| E. dipodomysis (25) | AF339490 | Dipodomys (kangaroo rat) | SW United States | No sites before 103 bp |
| E. langebarteli (19) | AF311640 | Peromyscus (mouse) | SW United States, Mexico | 98 |
| E. leucopi (44) | AF339491 | Peromyscus (white-footed mouse) | SW United States | 98 |
| E. onychomysis (25) | AF307879 | Onychomys (grasshopper mouse) | SW United States | 95 |
| E. scholtysecki (13) | AF324216 | Dipodomys (kangaroo rat) and Chaetodipus (pocket mouse) | SW United States; Mexico | 98 |
| E. telekii (38) | AF246717 | Lemniscomys (striped grass mouse) | Kenya | 98 |
| E. tropidura (2) | AF324217 | Tropidurus delanonis (Hood Island lizard) | Galapagos Islands | 99 |
Inner forward primer, 5′ CAGGTCTAGGTAATCTTTTGAG; annealing temp, (Tm), 55.5°C. In the primer sequence, an A (underlined) replaces the C found in CYCA12. Only partial sequences are available. Tm was calculated as described by Rychlik and Rhoads (34).
SW, southwestern.
Minimum detection of oocysts and target DNA.
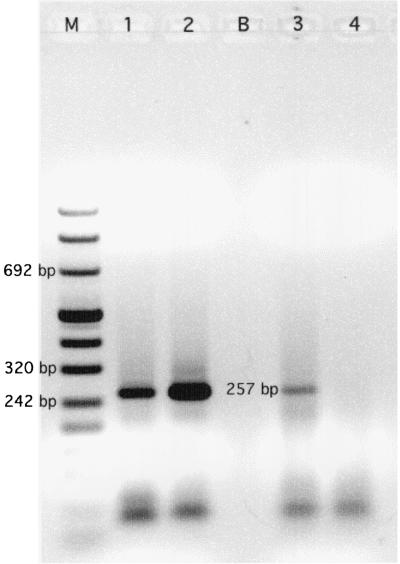
Our minimum detection limit, with primers CYCAO1, CYCAI2, and CYCAR1 assuming 100% recovery of DNA from the seeded environmental sample, was 0.75 oocyst (1.04 ng of genomic DNA as template) (Fig. 4). Since each oocyst contains four sporozoites and there are generally multiple copies of the 18S ssrDNA, detection of a fraction of an oocyst is reasonable. Our earlier work (Shields and Olson, Abstr. Gen. Meet. Am. Soc. Microbiol.) indicated a detection limit of one oocyst per liter of deionized water with the primers developed by Relman et al. (33). The first electrophoresis gel of the seeded environmental sample amplicons showed a considerable amount of smearing (data not shown), although a 257-bp band was visible; therefore, the second reaction was repeated with a 1:10 dilution of the first reaction product. No amplification was seen in unseeded environmental samples, and the interference assay, which used DNA extract inoculated with 260.5 pg of positive-control plasmid, showed little inhibition (Fig. 4).
FIG. 4.
Lanes: M, DNA molecular weight marker VIII; B, unused; 1 and 2, seeded environmental samples; 1, 0.75 oocyst (1.04 ng of DNA); 2, 7.5 oocysts (10.35 ng of DNA); 3, unseeded environmental sample inoculated with 260.5 pg of positive-control plasmid; 4, the same sample with no added positive-control plasmid.
Application of PCR with primers CYCAO1, CYCAI2, and CYCAR1 and RFLP with AluI to environmental samples.
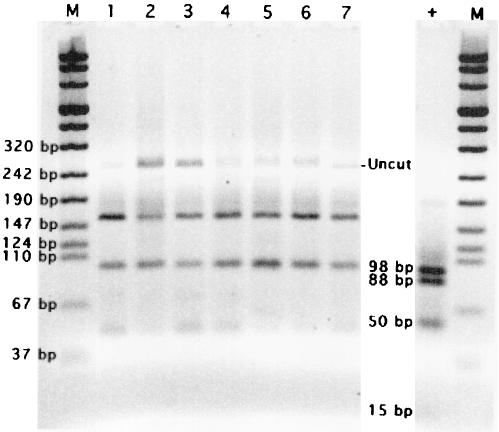
We applied our new PCR-RFLP protocol to the previously positive environmental samples. While these samples appeared positive with primers CYCF3E and CYCR4B and RFLP with MnlI (Fig. 1, 2, and 3) and remained PCR positive with primers CYCAO1, CYCAI2, and CYCAR1, RFLP analysis with AluI produced negative results (Fig. 5).
FIG. 5.
RFLP analysis with AluI of PCR (primers CYCAI2 and CYCAR1) amplicons from selected environmental samples. Lanes: M, DNA molecular weight marker VIII; +, C. cayetanensis oocysts with clear bands at 98, 88, and 50 bp and a faint band at 15 bp; 1, sample collected from the SDC on 21 April 1998; 2 to 5, individual water samples collected from the SARVB on 28 August 1998; 6 and 7, individual water samples collected from the SARYL on 28 September 1998. The term “Uncut” identifies amplicons that do not contain AluI sites.
To confirm these results, we isolated selected individual PCR amplicons and sequenced them. Figure 6 is an alignment of Cyclospora species and two of the isolates with sequences most closely related to C. cayetanensis. These amplicons were isolated from the samples in lanes 2 and 7 (Fig. 5), i.e., SARVB and SARYL, respectively. Both of these samples had previously been identified as C. cayetanensis with primers CYCF3E and CYCR4B (33) and RFLP with MnlI (20), and they correspond to those sequenced isolates shown in Fig. 2. In Fig. 6, each of these environmental isolate sequences contains one AluI site. C. colobi and C. cercopitheci contain two sites, and C. cayetanensis contains three. Thus, while they are likely Cyclospora species given the sequence similarity, they are clearly not C. cayetanensis.
FIG. 6.
CLUSTAL W (version 1.8.1) (40) multiple-sequence alignment of Cyclospora species and environmental samples positive by PCR with primers CYCAI2 and CYCAR1. SARVB B is from lane 2, and SARYL D is from lane 7, Fig. 5. Shading indicates sequence differences, underlining indicates AluI sites, and arrows indicate primer locations and directions.
DISCUSSION
Often, methods used to detect pathogens in environmental samples are modifications of those used for clinical samples. However, environmental samples present unique challenges to microbiologists and public health laboratories (Shields and Olson, Proc. Am. Water Works Assoc. 2000 Water Qual. Technol. Conf.), such as the presence of nonpathogenic but genetically similar organisms in sample matrices.
While the protocol using PCR with primers CYCF3E and CYCR4B (33) and RFLP with MnlI (20) is clearly able to identify C. cayetanensis oocysts in stool samples, it must be combined with microscopic confirmation to produce reliable results for environmental samples. Our newly designed primers and RFLP differentiate C. cayetanensis from Eimeria and other Cyclospora species, thus allowing researchers and public health laboratories to confidently identify C. cayetanensis in water and soil samples without microscopic confirmation, saving them time and resources.
Most of the cases of cyclosporiasis that have occurred in the United States and Canada have been linked to imported food (18); however, there have been sporadic reports of C. cayetanensis infection in the United States where no food source or history of international travel was implicated (17, 29, 46). Laboratory surveys of stool samples (collected during nonoutbreak periods), conducted in the United States and United Kingdom between 1992 and 1995, suggested that the prevalence in the general populations was <0.5% (18). Nevertheless, C. cayetanensis infection in developed countries has been viewed as linked either to travel or to foods imported from developing countries (37). However, a foodborne outbreak in southwestern Germany in December 2000 (8) suggested that C. cayetanensis may be present in the environmental waters and/or soils of European countries. The sources of this outbreak were epidemiologically traced to butterhead lettuce from southern France, mixed lettuce and leafy herbs from southern Italy, and chives from a greenhouse in Germany. While the implicated lettuce and herbs were no longer available for microscopic or molecular analysis, the relative risk associated with the consumption of these foods and illness was 5.0 (confidence interval = 1.4 < relative risk < 204 [P = 0.0045]).
The initial source of contamination of the German outbreak is still unknown. Possibilities include the soil, use of contaminated water for irrigation and pesticide dilution, and the poor sanitary facilities available to seasonal field workers (8). This outbreak clearly illustrates the potential for C. cayetanensis to become endemic in developed countries. As this potential increases, the need for methods, such as the one described in this paper, that both distinguish among Cyclospora species and differentiate Cyclospora from other coccidian genera becomes increasingly important as these organisms can be found in environmental waters used for irrigation and pesticide dilution.
Acknowledgments
We thank Michael Arrowood, Alexandre daSilva (Division of Parasitic Diseases, Centers for Disease Control and Prevention, Atlanta, Ga.), and Alan Lindquist (Parasitology Department, Environmental Protection Agency, Cincinnati, Ohio) for invaluable and inexhaustible advice and encouragement and the generous contribution of C. cayetanensis oocysts. We thank Menu Leddy and the Orange County Water District (Fountain Valley, Calif.) for the use of the Transgenomic WaveMachine, as well as their time and patience. We also thank David Relman of Stanford University for the positive-control plasmid. We are grateful to Arnold Demain (Drew University, Madison, N.J.) for proofreading the manuscript.
This work was supported in part by grants from the California Center for Water and Wildlands and the National Water Research Institute (Fountain Valley, Calif.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aquino-Shuster, A. L., D. W. Duszynski, and H. L. Snell. 1990. Three new coccidia Apicomplexa from the Hood Island lizard, Tropidurus delanonis, from the Galapagos Archipelago Pacific Ocean. J. Parasitol. 76:313-318. [Google Scholar]
- 3.Becker, E. R., and P. R. Hall. 1931. Eimeria separata, a new species of coccidium from the Norway rat (Epimys norvegicus). Iowa State Coll. J. Sci. 6:131. [Google Scholar]
- 4.Bern, C., B. Hernandez, M. B. Lopez, M. J. Arrowood, M. A. de Mejia, A. M. de Merida, A. W. Hightower, L. Venczel, B. L. Herwaldt, and R. E. Klein. 1999. Epidemiologic studies of Cyclospora cayetanensis in Guatemala. Emerg. Infect. Dis. 5:766-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor B. A., J. Reidy, and R. Soave. 1999. Cyclosporiasis: clinical and histopathologic correlates. Clin. Infect. Dis. 28:1216-1222. [DOI] [PubMed]
- 6.DaSilva, A. J., F. J. Bornay-Llinares, C. del Aguila de la Puenta, H. Moura, J. M. Peralta, I. Sobottka, D. A. Schwartz, G. S. Visvesvara, S. B. Slemenda, and N. J. Pieniazek. 1997. Diagnosis of Enterocytozoon bieneusi (Microsporidia) infections by polymerase chain reaction in stool samples using primers based on the region coding for small-subunit ribosomal RNA. Arch. Pathol. Lab. Med. 121:874-879. [PubMed] [Google Scholar]
- 7.Dieben, C. P. A. 1924. Over der morphologic en biologic van het rattencoccidium Eimeria nieschuki n. sp., en zijne verspreiding in Nederland. Ph.D. thesis. Veeartsenijk. Hoogeschool, Utrecht, The Netherlands.
- 8.Döller, P. C., K. Dietrich, N. Filipp, S. Brockmann, C. Dreweck, R. Vonthein, C. Wagner-Wiening, and A. Wiedenmann. 2002. Cyclosporiasis outbreak in Germany associated with the consumption of salad. Emerg. Infect. Dis. 8:992-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duszynski, D. W., D. T. Scott, J. Aragon, A. Leach, and T. Perry. 1999. Six new Eimeria species from vespertilionid bats of North America. J. Parasitol. 85:496-503. [PubMed] [Google Scholar]
- 10.Eberhard, M. L., N. J. Pieniazek, and M. J. Arrowood. 1997. Laboratory diagnosis of Cyclospora infections. Arch. Pathol. Lab. Med. 121:792-797. [PubMed] [Google Scholar]
- 11.Eberhard, M. L., A. J. DaSilva, B. G. Lilley, and N. J. Pieniazek. 1999. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp. n., C. colobi sp. n., and C. papionic sp. n. Emerg. Infect. Dis. 5:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edger, S. A., and C. T. Seibold. 1964. A new coccidium chickens, Eimeria mivati sp. n. (Protozoa: Eimeriidae) with details of its life history. J. Parasitol. 50:193-204. [PubMed] [Google Scholar]
- 13.Ernst, J. V., M. J. Frydendall, and D. M. Hammond. 1967. Eimeria scholtysecki, a sp. from Ord's kangaroo rat (Dipodomys ordii). J. Protozool. 14:181-182. [DOI] [PubMed] [Google Scholar]
- 14.Ernst, J. V., E. C. Oaks, and J. R. Sampson. 1970. Eimeria reedi sp. n. and E. chobotari sp. n. (Protozoa: Eimeriidae) from heteromyid rodents. J. Protozool. 17:453-455. [Google Scholar]
- 15.Ernst, J. V., B. Chobotar, and D. M. Hammond. 1971. The oocysts of Eimeria vermiformis sp. n. and E. papillata sp. n. (Protozoa: Eimeriidae) from the mouse Mus musculus. J. Protozool. 18:221-223. [DOI] [PubMed] [Google Scholar]
- 16.Ford, P. L., D. W. Duszynski, and C. T. McAllister. 1990. Coccidia (Apicomplexa) from heteromyid rodents in the southwestern United States, Baja California, and northern Mexico with three new species from Chaetodipus hispidus. J. Parasitol. 76:325-331. [PubMed] [Google Scholar]
- 17.Hale, D., W. Aldeen, and K. Carroll. 1994. Diarrhea associated with cyanobacteria-like bodies in an immunocompetent host: an unusual epidemiological source. JAMA 271:144-145. [PubMed] [Google Scholar]
- 18.Herwaldt B. L. 2000. Cyclospora cayetanensis: a review, focusing on the outbreaks of cyclosporiasis in the 1990s. Clin. Infect. Dis. 31:1040-1057. [DOI] [PubMed] [Google Scholar]
- 19.Ivens, V., F. J. Kruidenier, and N. D. Levine. 1959. Further studies of Eimeria (Protozoa: Eimeriidae) from Mexican rodents. Trans. III Acad. Sci. 51:53-57. [Google Scholar]
- 20.Jinneman K. C., J. H. Wetherington, A. M. Adams, J. M. Johnson, B. J. Tenge, N.-L. Dang, and W. E. Hill. 1996. Differentiation of Cyclospora sp. and Eimeria spp. by using the polymerase chain reaction amplification products and restriction fragment length polymorphisms. Food and Drug Administration Laboratory Information Bulletin LIB no. 4044. [Online.] http://vm.cfsan.fda.gov/∼mow/kjcs19c.html.
- 21.Jinneman, K. C., J. H. Wetherington, W. E. Hill, A. M. Adams, J. M. Johnson, B. J. Tenge, N.-L. Dang, R. L. Manger, and M. M. Wekell. 1998. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. oocysts directly from raspberries. J. Food. Prot. 61:1497-1503. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, W. T. 1930. Coccidiosis of the chicken. Oregon Agric. Exp. State Bull. 314:16.
- 23.Kheysin, Y. M. 1972. Life cycles of coccidia of domestic animals, p. 198. University Park Press, Baltimore, Md.
- 24.Levine, P. P. 1942. A new coccidium pathogenic for chickens, Eimeria brunetti n. sp. Cornell Vet. 32:430-439. [Google Scholar]
- 25.Levine, N. D., V. Ivens, and F. J. Kruidenier. 1957. New species of Eimeria from Arizona rodents. J. Protozool. 4:80-88. [Google Scholar]
- 26.Lopez, A. S., D. R. Dodson, M. J. Arrowood, P. A. Orlandi, Jr., A. J. Da Silva, J. W. Bier, S. D. Hanauer, R. L. Kuster, S. Oltman, M. S. Baldwin, K. Y. Won, E. M. Nace, M. L. Eberhard, and B. L. Herwaldt. 2001. Outbreak of cyclosporiasis associated with basil in Missouri in 1999. Clin. Infect. Dis. 32:1010-1017. [DOI] [PubMed] [Google Scholar]
- 27.McQuistion, T. E., and B. B. Holmes. 1988. Isospora robini, new species of coccidian parasite (Apicomplexa: Eimeriidae) from the American robin (Turdus migratorius). Proc. Helminthol. Soc. Wash. 55:324-325. [Google Scholar]
- 28.Moore, E. N., and J. A. Brown. 1951. A new coccidium pathogenic for turkeys, Eimeria adenoeides n. sp. (Protozoa: Eimeriidae). Cornell Vet. 41:124-135. [PubMed] [Google Scholar]
- 29.Ooi, W. W., S. K. Zimmerman, and C. A. Needham. 1995. Cyclospora species as a gastrointestinal pathogen in immunocompetent hosts. J. Clin. Microbiol. 33:1267-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlandi, P., A. and K. A. Lampel. 2000. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J. Clin. Microbiol. 38:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortega, Y. R., C. R. Sterling, R. H. Gilman, V. A. Cama, and F. Diaz. 1993. Cyclospora species—a new protozoan pathogen of humans. N. Engl. J. Med. 328:1308-1312. [DOI] [PubMed] [Google Scholar]
- 32.Ortega, Y. R., C. R. Roxas, R. H. Gilman, N. J. Miller, L. Cabrera, C. Taquiri, and C. R. Sterling. 1997. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region in Peru. Am. J. Trop. Hyg. 57:683-686. [DOI] [PubMed] [Google Scholar]
- 33.Relman, D. A., T. M. Schmidt, A. Gajadhar, M. Sogin, J. Cross, K. Yoder, O. Sethabutr, and P. Echeverria. 1996. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 173:440-445. [DOI] [PubMed] [Google Scholar]
- 34.Rychlik, W., and R. E. Rhoads. 1989. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 17:8543-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider, A. C. U. 1875. Note sur la psorospermie oviforme du pouipe. Arch. Zool. Exp. Gen. 4:xi-xiv.
- 36.Scott, D. T., and D. W. Duszynski. 1997. Eimeria from bats of the world: two new species from Myotis spp. (Chiroptera: Vespertilionidae). J. Parasitol. 83:495-501. [PubMed] [Google Scholar]
- 37.Shields, J. M., and B. H. Olson. 2003. Cyclospora cayetanensis: a review of an emerging pathogenic coccidian. Int. J. Parasitol. 33(4):371-391. [DOI] [PubMed] [Google Scholar]
- 38.Slapeta, J. R., D. Modry, J. Votypka, M. Jirku, M. Obornik, J. Lukes, and B. Koudela. 2001. Eimeria telekii n. sp. (Apicomplexa: Coccidia) from Lemniscomys striatus (Rodentia: Muridae): morphology, pathology and phylogeny. Parasitology 122:133-143. [DOI] [PubMed] [Google Scholar]
- 39.Sturbaum, G. D., Y. R. Ortega, R. H. Gilman, C. R. Sterling, L. Cabrera, and D. A. Klein. 1998. Detection of Cyclospora cayetanensis in wastewater. Appl. Environ. Microbiol. 64:2284-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyzzer, E. E. 1927. Species and strains of coccidia in poultry. J. Parasitol. 13:215. [Google Scholar]
- 42.Tyzzer, E. E. 1929. Coccidiosis in gallinaceous birds. Am. J. Hyg. 10:269-383. [Google Scholar]
- 43.Vesey, G., J. S. Slade, M. Byrne, K. Shepherd, and C. R. Fricker. 1993. A new method for the concentration of Cryptosporidium oocysts from water. J. Appl. Bacteriol. 75:82-86. [DOI] [PubMed] [Google Scholar]
- 44.Von Zeilen, B. W. 1961. Eimeria leucopi n. sp., a coccidium from the deer mouse Peromyscus leucopus. J. Protozool. 8:134-138. [Google Scholar]
- 45.Waters, A. P., and T. F. McCutchan. 1990. Ribosomal RNA: nature's own polymerase-amplified target for diagnosis. Parasitol. Today 6(2):56-59. [DOI] [PubMed] [Google Scholar]
- 46.Wurtz, R. 1994. Cyclospora: a newly identified intestinal pathogen of humans. Clin. Infect. Dis. 18:620-623. [DOI] [PubMed] [Google Scholar]