Abstract
Culture-dependent and culture-independent approaches were used to determine the relationship between the dehalogenase gene pool in bacteria enriched and isolated on 2,2-dichloropropionic acid (22DCPA) and the environmental metagene pool (the collective gene pool of both the culturable and uncultured microbes) from which they were isolated. The dehalogenases in the pure-cultures isolates, which were able to degrade 22DCPA, were similar to previously described group I and II dehalogenases. Significantly, the majority of the dehalogenases isolated from activated sludge by degenerate PCR with primers specific for α-halocarboxylic acid dehalogenases were not closely related to the dehalogenases in any isolate. Furthermore, the dehalogenases found in the pure cultures predominated in the enrichments but were a minor component of the community used to inoculate the batch cultures. Phylogenetic analysis of the dehalogenase sequences isolated by degenerate PCR showed that the diversity of the group II deh gene was greater than that of the group I deh gene. Direct plating of the activated sludge onto minimal media supplemented with 22DCPA resulted in biomass and DNA from which dehalogenases were amplified. Analysis of the sequences revealed that they were much more closely related to the sequences found in the community used to start the enrichments. However, no pure cultures were obtained with this isolation method, and thus no pure cultures were available for identification. In this study we examined the link between genes found in pure cultures with the metagene pool from which they were isolated. The results show that there is a large bias introduced by culturing, not just in the bacteria isolated but also the degradative genes that they contain. Moreover, our findings serve as a caveat for studies involving the culturing of pure cultures of bacteria and conclusions which are drawn from analysis of these organisms.
With the advent of culture-independent approaches for determining bacterial diversity, it has become apparent that only a small proportion of bacteria (between 0.0001 and 1%) can be cultured. At this time (as of January 2003), there are 1,137 genera containing 6,110 named species deposited in the German Culture Collection (Deutsche Sammlung von Mikroorganismen und Zellkulturen); however, the estimated number of species in 100 g of soil is between 13,000 (55, 56) and 500,000 (9). This disparity has far-reaching ramifications for microbial ecologists, microbiologists, biotechnologists, and environmental scientists. Furthermore, the bacteria which have been studied in detail are those which are easy to culture from the environment and thus are not necessarily environmentally relevant microorganisms (59). There is growing evidence (8, 59, 60) that the genes that code for the enzymes involved in processes in situ (i.e., environmentally relevant genes) are not found in cultured bacteria isolated as pure cultures that are able to perform the same task. The apparent differences between genes obtained by culturing and genes obtained by culture-independent approaches have been demonstrated for several groups of enzymes, including phenol hydroxylases (60), naphthalene-degrading enzymes (63), and 2,4-dichlorophenoxyacetate-degrading enzymes (8). Again, the differences are a point of concern since many situations require that we understand what genes are involved in an environmental process. Conclusions based on data which do not relate to in situ conditions but are generated due to biases introduced by the choice of the technique used can be incorrect.
Chlorinated compounds are probably the most important class of synthetic environmental pollutants, as illustrated by the European Union Dangerous Substances Directive (76/464/EEC) and by the U.S. Environmental Protection Agency's Register of Lists, which includes numerous halogenated compounds. Extensive research worldwide with pure-culture isolates, some containing more than one deh gene (12, 17, 22, 40, 46, 61), has shown that biodegradation of α-halocarboxylic acids (α-HAs) is always mediated by halocarboxylic acid dehalogenases. In a recent study, phylogenetic analysis (14) also showed that >95% of the α-HA dehalogenases that have been described fall into two distinct families (group I and II deh genes). The α-HAs dehalogenases have been shown to be a diverse group (14), even though the chemical similarity of the substrates which they dehalogenate is high. However, all the currently available sequences were derived from organisms that were isolated by enrichment on appropriate α-HAs. No known previous study has attempted to determine the relationship between the cultured and natural or metagene pools and the process being investigated (e.g., biodegradation). In this study the dehalogenase gene pool in bacteria isolated by enrichment on 2,2-dichloropropionic acid (22DCPA) was compared with the environmental metagene pool sampled by direct PCR amplification of the group I and II deh genes.
MATERIALS AND METHODS
Bacterial enrichment cultures and growth conditions.
Bacteria able to degrade 22DCPA were isolated from an enrichment culture by using Brunner's (3) minimal enrichment medium. The first enrichment culture (100 ml; primary enrichment) was inoculated with an aliquot (1%, vol/vol) of activated sludge from Cog Moor Sewage Treatment Works, Penarth, South Wales, United Kingdom, treating mixed domestic and industrial waste composed mainly of short-chain volatile fatty acids (isomers of butanoic and propionic acid), aromatic compounds (phenols), and long-chain fatty acids (hexadecanoic acid and octadecanoic acid). The culture was supplemented with 0.5 g of C liter−1 in the form of 22DCPA (equivalent to 22 mM) and 0.005% (wt/vol) yeast extract (YE) and incubated at 150 rpm and 30°C. Samples (2 ml) from the cultures were taken at regular intervals (every day) and used to determine chloride release (see below). Once biodegradation (>50% release of total available chloride, which occurred after 8 days) was observed, the enrichment was used to inoculate a fresh batch culture, termed the secondary enrichment. After dechlorination was measured, the culture was used to isolate pure cultures of 22DCPA-degrading bacteria. The bacteria were isolated by plating an aliquot (50 μl) of the secondary enrichment onto solidified Brunner's medium (1.5% [wt/vol] purified agar [LP028; Oxoid]) supplemented with 0.5 g of C liter−1 in the form of 22DCPA and 0.005% (wt/vol) YE and incubating the preparation at 30°C until single colonies were observed. Colonies were transferred with a sterile toothpick to a fresh flask containing Brunner's medium supplemented with 22DCPA and YE and incubated at 30°C and 150 rpm until dechlorination was observed. Further tests of purity were performed by serially diluting the culture on R2A medium (43), and cultures which were consistently able to degrade 22DCPA were stored as glycerol stocks at −70°C until they were used.
Direct plating (8) was also used to isolate pure cultures of bacteria able to degrade 22DCPA. Serially diluted activated sludge (100 μl), diluted in Brunner's minimal medium, was spread onto minimal medium (Brunner's minimal medium solidified with 1.5% [wt/vol] Noble agar [Difco]) containing 22DCPA (final concentration, 22 mM) and 0.005% (wt/vol) YE. Plates were incubated at 20°C until colonies were observed. The colonies from the direct plating were further purified by plating them onto fresh minimal medium containing 22 mM 22DCPA and 0.005% (wt/vol) YE. To test for dechlorination of the 22DCPA, isolates were taken from the 22DCPA-containing plates, transferred to 10 ml of Brunner's minimal medium containing 22 mM 22DCPA, and incubated at 30°C. During this incubation the concentration of free chloride was measured coulometrically (see below). Once the colonies had been transferred to fresh medium, the remaining biomass was collected by pipetting 2 ml of 0.25× sterile Ringer's solution onto the plate surface. With the aid of a sterile swab the bacterial colonies were resuspended in the buffer, and 1 ml of the cell suspension was removed with a pipette and placed in a sterile microcentrifuge tube. The biomass was stored at −20°C until it was used.
Measurement of chloride release.
α-HA dehalogenation in liquid cultures was estimated by coulometric titration of free halide in samples with a Sherwood 926 chloride analyzer, as previously described (48).
DNA extraction, dehalogenase gene amplification, 16S rRNA gene amplification, and phylogenetic analysis.
DNA was extracted from bacterial biomass by the modified method of Ausubel et al. (1). The modification used has been described elsewhere (33).
PCR amplification, cloning (40 clones were screened for each library), and sequencing of the deh genes from either the pure cultures or DNA from enrichment cultures were performed by using primers described by Hill et al. (14). Briefly, the group I deh genes were amplified by using primers dehIfor1 (5′-ACGYTNSGSGTGCCNTGGGT) and dehIrev2 (5′-SGCMAKSRCNYKGWARTCACT), which resulted in amplification of a 504-bp product, referred to as the long group I deh product. A second primer set was used to amplify a short group I deh PCR product. The same forward primer, dehIfor1, was used; however, the reverse primer was dehIrev1 (5′-AWCARRTAYTTYGGATTRCCRTA), and this resulted in a PCR product that was 230 bp long. Group II deh genes were amplified by using dehIIfor1 (5′-TGGCGVCARMRDCARCTBGARTA) and dehIIrev1 (5′-TCSMADSBRTTBGASGANACRAA), which resulted in amplification of a 422-bp product. Phylogenetic analysis of the partial PCR products was performed as previously described (14). The percent coverage (C) of the clone libraries was determined as described by Good (13) by using the equation C = [1 − (n1/N)] × 100, where n1 is the number of clones that occurred only once and N is the total number of clones examined.
Amplification of the bacterial 16S rRNA genes from DNA samples was performed by using PCR primers and conditions described previously (32), and the results were analyzed as described by Hill and colleagues (14). For this analysis 50 clones were screened for each of the libraries created.
The partial dehalogenase gene sequences were compared with those in the EMBL database (2) by using Fasta3 (38, 39) at the European Bioinformatics Institute, Hinxton Hall, Cambridge, United Kingdom (http://www.ebi.ac.uk). In addition, the partial 16S rRNA gene sequences were analyzed using the Ribosomal Database Project with Similarity Rank (31) to identify most closely related database sequences. Sequence alignments were created using CLUSTAL W (54). The evolutionary distances for the nucleotide sequences encoding the dehalogenases were calculated by using the Kimura two-parameter algorithm (25), while the Poisson correction function in TREECON was used (57) to estimate the distances for the derived amino acid sequences of the dehalogenases. Phylogenetic trees for all the distance matrices were determined by the neighbor-joining method (45) using Treecon for Windows (57). Tree topologies were compared for trees constructed by the maximum-likelihood and maximum-parsimony methods using PHYLIP, version 3.5 (11), and by the neighbor-joining method. Bootstrap analyses (10) of up to 500 replicates were performed for all phylogenetic trees to estimate the reproducibility of the tree topologies. All the deh-derived amino acid sequences were checked to establish whether they contained residues which were described as essential by Kurihara et al. (28) and thus support the conclusion that they are potentially from active dehalogenases.
Nucleotide sequence accession numbers.
The partial sequences of the 16S rRNA genes determined in this study have been deposited in the EMBL database under accession numbers AJ514429 to AJ514456. The accession numbers for the group I and II dehalogenase sequences are AJ511295 to AJ511324.
RESULTS
Characterization of bacterial isolates able to degrade 22DCPA.
Ten pure cultures able to degrade 22DCPA were isolated from the secondary enrichment. All the isolates grew as yellow colonies on R2A medium (see above), and analysis of their partial 16S rRNA gene sequences showed that they all clustered close to sequences of members of the genus Xanthobacter and the isolates were most closely related to Xanthobacter autotrophicus strain 7c (>95% nucleotide identity) (Table 1) (41). Further analysis, based on restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA partial gene products (approximately 1,330 bp [data not shown]) (34) of the isolates, showed that there were two groups of isolates; six of the isolates gave one RFLP pattern (designated isolate AS1), and the remaining four isolates gave a second RFLP pattern (designated isolate AS2). An RFLP analysis of the group I and II deh partial gene products was conducted to determine the diversity of the 10 isolates. This analysis showed that isolates which were grouped as AS1 produced similar RFLP patterns for their group I and II deh genes. A similar result was obtained for AS2 (data not shown).
TABLE 1.
Phylogenetic affiliations of the 16S rRNA gene sequences obtained by PCR of DNA from activated sludge, primary and secondary enrichment cultures and the two pure cultures (isolates AS1 and AS2) able to degrade 22DCPA
| Clonea | Closest RDPII organismb | Phylogenetic affiliationc | Sab scored |
|---|---|---|---|
| ASG1 | Acidovorax defluvii | β-Proteobacteria, Burkholderiales | 0.956 |
| ASG2 | Prevotella albensis | Bacterioidales | 0.727 |
| ASG3 | Pseudomonas putida | γ-Proteobacteria, Pseudomonadales | 0.944 |
| ASG4 | Lactobacillus casei | Lactobacillales | 0.735 |
| ASG5 | Agrobacterium ferrugineum | α-Proteobacteria, Rhodobacteriales | 0.825 |
| ASG6 | Lactosphaera pasteurii | Lactobacillales | 0.963 |
| ASG7 | Aeromonas media | γ-Proteobacteria, Aeromonadales | 0.976 |
| ASG8 | Lactosphaera pasteurii | Lactobacillales | 0.951 |
| ASG9 | Aeromonas media | γ-Proteobacteria, Aeromonadales | 0.904 |
| ASG10 | Ruminococcus bromii | Clostridiales | 0.946 |
| ASG11 | Enterobacter agglomerans | γ-Proteobacteria, Enterobacteriales | 0.907 |
| AS1°1 | Paracoccus carotinifaciens | α-Proteobacteria, Rhodobacteriales | 0.674 |
| AS1°2 | Clone SJA-149 | Acidobacteriales | 0.593 |
| AS1°3 | Pseuodomonas putida | γ-Proteobacteria, Pseudomonadales | 0.917 |
| AS1°4 | Chryseobacterium indologenes | Flavobacteriales | 0.792 |
| AS1°5 | Caulobacter vibrioides | α-Proteobacteria, Caulobacterales | 0.937 |
| AS1°6 | Xanthobacter autotrophicus | α-Proteobacteria, Rhizobiales | 0.932 |
| AS1°7 | Caulobacter vibriobdes | α-Proteobacteria, Caulobacterales | 0.961 |
| AS1°8 | Sphingobacterium multivorum | Flavobacteriales | 0.887 |
| AS1°9 | Thiorhodovibrio sibirica | γ-Proteobacteria, Legionellales | 0.620 |
| AS1°10 | Chryseobacterium indologenes | Flavobacteriales | 0.817 |
| AS2°1 | Chryseobacterium indologenes | Flavobacteriales | 0.801 |
| AS2°2 | Burkholderia norimbergensis | β-Proteobacteria, Burkholderiales | 0.893 |
| AS2°3 | Achromobacter xylosidans | β-Proteobacteria, Burkholderiales | 0.903 |
| AS2°4 | Burkholderia norimbergensis | β-Proteobacteria, Burkholderiales | 0.902 |
| AS2°5 | Xanthobacter autotrophicus | α-Proteobacteria, Rhizobiales | 0.956 |
| Isolate AS1 | Xanthobacter autotrophicus | α-Proteobacteria, Rhizobiales | 0.942 |
| Isolate AS2 | Xanthobacter autotrophicus | α-Proteobacteria, Rhizobiales | 0.942 |
ASG, DNA from activated sludge; AS1°, DNA from primary enrichment culture; AS2°, DNA from secondary enrichment culture.
Determined using Sequence Match analysis at the Ribosomal Database Project II (RDPII) (31).
Phylogenetic groups as described in Bergey's Manual of Systematic Bacteriology, 2nd ed. (http://www.cme.msu.edu/bergeys/).
An Sab score of >0.7 corresponded to approximately >95% similarity (64).
Direct plating of the activated sludge onto minimal medium containing 22DCPA resulted in colonies which showed significant growth (colony size, >2 mm) after primary plating. However, these colonies resisted further purification by replating onto fresh medium containing 22DCPA and thus were not identified.
16S rRNA gene sequence analysis of activated sludge and enrichment cultures.
Analysis of partial 16S rRNA gene sequences was used to determine bacterial diversity in activated sludge and the primary and secondary enrichment communities. The levels of coverage for the activated sludge, primary enrichment, and secondary enrichment 16S rRNA gene sequence libraries were 23, 47, and 73%, respectively. The phylogenetic data generated from this analysis are summarized in Table 1. The sequences were found to be related to 16S rRNA gene sequences from members of both the gram-positive and gram-negative divisions. None of the sequences analyzed from the enrichment cultures were related to any gram-positive 16S rRNA gene sequences, and the majority of the sequences matched sequences from members of the phylum Proteobacteria. No common sequences were found in the three sample sources. The only sequences that matched 16S rRNA gene sequences from the pure cultures were AS1°6 and AS2°5; they were ≥95% similar at the nucleotide level to each other and to the isolate AS1 and AS2 16S rRNA gene sequences. These sequences matched sequences found in members of the α subclass of the Proteobacteria (α-Proteobacteria) and, more specifically in the genus Xanthobacter (41), which contains the haloalkane-degrading bacterium X. autotrophicus GJ10 (19).
Diversity of group I deh genes during biodegradation of 22DCPA.
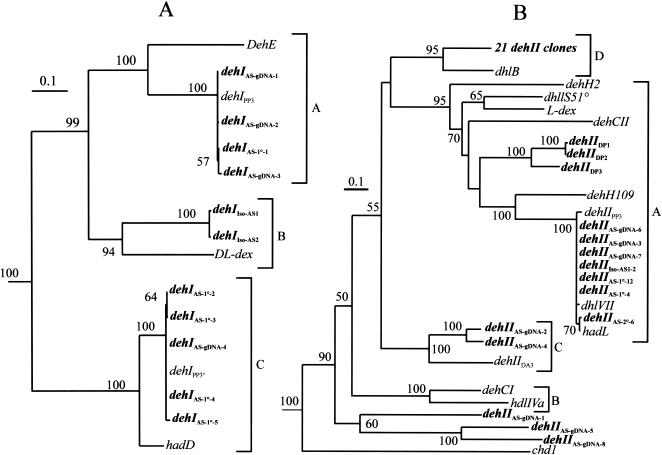
All of the sources of DNA except the direct plating biomass gave PCR products with both group I long and group I short deh PCR primer sets (see reference 14 for an explanation of primer sets; briefly, the short deh PCR product uses less-degenerate primers and is thus a more robust PCR product, but it provides less information); the results of the molecular analysis are summarized in Table 2. Due to the greater information content of the long group I deh PCR product, this product was used for the phylogenetic analysis (Fig. 1A). The partial gene products obtained all clustered in predefined group I deh subgroups (14). No group I deh sequences from the activated sludge, primary enrichment, or secondary enrichment clustered with the group I deh sequences of the pure-culture isolates. The group I deh PCR products obtained from the activated sludge and enrichment cultures clustered in two clades in the A and C subgroups (Fig. 1A). Again, the level of nucleotide identity for these clades was high, >94%. All group I deh clones from the secondary enrichment that were analyzed contained nonspecific PCR products that were not related to any known dehalogenase. The coverage values for these libraries were calculated and were found to be 50% for activated sludge DNA and 60% for primary enrichment DNA.
TABLE 2.
Summary of results of a molecular analysis of the samples from all biomass samples studied in this investigation
| Sample | PCR product
|
|||
|---|---|---|---|---|
| 16S rRNA gene | dehI (large)a | dehI (small)a | dehIIa | |
| Activated sludge | + | + | + | + |
| Primary enrichment | + | + | + | + |
| Secondary enrichment | + | ±b | − | + |
| Direct plating biomass | NAc | − | − | + |
| Isolates AS1 and AS2 | + | − | + | + |
Analysis in silico revealed statistically significant similarity to know dehalogenases in the EMBL database.
PCR product of the correct size was obtained; however, analysis in silico revealed no statistically significant similarity to dehalogenases in the EMBL database.
NA, not analyzed.
FIG. 1.
Dendrograms illustrating the relationships among the α-HA dehalogenase gene sequences obtained from the enrichment cultures. (A) Dendrogram for group I dehalogenase genes (dehI type) from the enrichments and 22DCPA-degrading isolates. The sequences from pure-culture isolates are designated dehIIso-ASX, where X is the strain number of the pure culture. Group I deh sequences from the activated sludge are designated dehIAS-DNA-X, where X is the number of the clone from the library. The clones from the primary and secondary enrichments are designated dehIAS-1°-X and dehIAS-2°-X, respectively, where X is the clone number from the corresponding library. The DNA sequences were aligned by using CLUSTALW, and the tree was constructed with the neighbor-joining program from a similarity matrix of pairwise comparisons made using the Jukes-Cantor algorithm. Bootstrap values from 500 replicate trees are indicated at the nodes as percentages. The sequences of dehE (accession no. Y15517), DL-dex (accession no. U97030), hadD (accession no. M81841), and dehIIPP3° (silent deh gene) (accession no. AJ133461) were also included. (B) Dendrogram illustrating the relationships among the group II α-HA dehalogenase gene sequences obtained from the enrichment cultures. The designations are the same as those described above for the group I dehalogenases except that dehI is replaced by dehII. The tree was constructed like the tree for the group I dehalogenases. The sequences obtained from direct plating isolation are designated dehIIDPX, where X is the clone number from the clone library. For the dehII tree the accession numbers and/or references for the previously described α-HA genes used are as follows: dhlS5I°, reference 26 (not deposited in EMBL); dhlVII, accession no. X94147; cbbZp (2-phosphoglycolate phosphatase; outgroup [data not shown]), accession no. M68905; hadL, accession no. M81841; dehIIPP3, accession no. AJ133462; deh109, accession no. D17523; dehCI and dehCII, accession no. M62908 and M62909, respectively; dehH2, accession no. D90423; L-dex, accession no. S74078; hdlIVa, accession no. X66249; dhlB, accession no. M81691; chd1, accession no. AJ005843Z; and dehIIDA3, accession no. AJ133463. Since the dehII clones isolated from the primary and secondary enrichments and from isolates AS1 and AS2 were almost identical, these sequences are represented by one entry (21 dehII clones).
Both group I deh gene products from the two pure-culture isolates clustered together in a clade in the B subgroup and were ≥98% identical at the nucleotide level. Only the primer set that amplified the short group I deh product (14) gave a positive signal with the DNA from isolates AS1 and AS2. The phylogenetic affiliation of these sequences is shown in Fig. 1A. The sequences clustered in subgroup B along with the deh gene from Pseudomonas sp. strain 113, DL-dex (35). The DNA sequence identity with DL-dex was 68%.
Diversity of group II deh genes during biodegradation of 22DCPA.
All sources of biomass gave positive PCR products with the group II deh PCR primers (Table 2). The majority of the sequences (43 of 46 sequences) clustered in subgroups A, C, and D identified by Hill et al. (14), and there was very little sequence variation within each of these subgroups (Fig. 1B). Twenty-one sequences (none from the activated sludge biomass) clustered in subgroup D, which contained the group II deh gene dhlB from X.autotrophicus GJ10 (19). The levels of nucleotide identity among these sequences were not less than 88%. Of the 21 sequences analyzed, 11 were identical at the nucleotide level. These 11 sequences were derived from DNA extracted from primary and secondary enrichments and pure-culture isolates; none of these sequences were found in the activated sludge group II deh clone library. The members of a second, smaller group (seven sequences from all three sources of biomass and a pure-culture isolate), which formed a clade in subgroup A, were also very similar to each other and the group II deh from Pseudomonas putida PP3, and the levels of sequence similarity were >98%. Of the seven sequences in this clade three, which were derived from activated sludge DNA, were identical. They were also identical to a group II deh PCR product obtained from one of the isolates and two sequences obtained from the primary enrichment. Altogether, three group II deh sequences were obtained by PCR from the pure-culture isolates. Two of the group II deh sequences from isolates AS1 and AS2 were found to cluster most closely with dhlB (73% identical and 83% homologous at the amino acid level [58]) in subgroup D. The remaining sequence, which came from isolate AS1, was found to cluster with sequences in the A subgroup.
Also found in the A subgroup were the group II deh sequences from the direct plating biomass. These sequences formed their own clade, which was more closely related to the sequence encoding the dehalogenase from P. putida No. 109 (23) than to the sequence encoding any other dehalogenase. Within this clade the level of sequence identity was >97% between dehIIDP1 and dehIIDP2; dehIIDP3 was 80 and 82% identical to dehIIDP2 and dehIIDP1, respectively. The sequence similarity with the next-closest sequence to dehIIDP3, dehH109, was 61%. Sequences similar to these sequences were not found in any other source of biomass. The remaining sequences were quite dissimilar, and the levels of nucleotide identity among them varied from 36 to 83%. These sequences were also derived exclusively from the DNA extracted from activated sludge and not from any of the enrichments or pure cultures. Furthermore, several of these sequences were not closely related to any previously published sequences and hence may constitute new subgroups in the group II deh family.
Analysis in silico produced some unexpected results. BLAST searches of the EMBL database with our group II deh sequences returned statistically significant matches with sequences from microbial genome projects. On further inspection it became apparent that these sequences have been annotated as homologous to group II deh sequences. Many groups of workers have reported the presence of group II deh-like genes in prokaryotes that are not affiliated with the Proteobacteria, and these organisms included members of the gram-positive division (6, 52), the Thermus-Deinococcus group (62), the Chlamydiales (42), the Euryarchaeota (36), and the Crenarchaeota (47). However, analysis of amino acid alignments of the putative dehalogenases and dehalogenases characterized previously showed the tenuous nature of ascribing a gene function based solely on homology (the complete alignment of the putative dehalogenase sequences with previously characterized sequences is available at http://www.ucc.i.e./ucc/depts/microbio/papers/jrm-aem2003/dehalignmenttotal.pdf). Of the hundreds of haloacid-like dehalogenases reported for the genomes analyzed, only a handful may actually be genuine dehalogenases, while the remainder are more likely to be members of the HAD superfamily of hydrolases (27). An alignment of the putative group II deh sequences which show conservation of the nine amino acids essential for catalysis (14, 16, 28) and the seven residues which constitute the enzyme hydrophobic pocket (29) is provided as supplementary data available at http://www.ucc.i.e./ucc/depts/microbio/papers/jrm-aem2003/dehalignment.pdf. Interestingly, two of these sequences are from the archaea Sulfolobus solfataricus strain P2 (47) and Sulfolobus tokodaii strain 7 (21).
DISCUSSION
Cultivation of bacteria is often seen as a prerequisite for determining how their phenotypes contribute to the function of an ecosystem (37). However, the results of this study strongly support the conclusion (8, 59, 60, 63) that batch enrichment tends to result in bacteria which may not be environmentally relevant microorganisms. The results of this study directly relate the members of the culturable dehalogenase gene pool with the metagene pool from which they originated. By determining the intermediary dehalogenase gene pools in the primary and secondary enrichments and in pure cultures of bacteria, it was possible to investigate this relationship. There was a clear separation of the dehalogenase gene sequences isolated from the enrichments and pure cultures and the dehalogenase gene sequences found in the activated sludge. The exception to this observation was one group II deh sequence from isolate AS1, which did cluster with sequences from activated sludge. This result verified the advantage of using the culture-independent method to analyze the dehalogenase metagene pool, since traditional methods would have shown a much more limited diversity of dehalogenases.
The conclusion drawn was that isolates AS1 and AS2, while obviously able to degrade 22DCPA, were isolated because they were the faster growers and thus able to outcompete the other degraders for the nutrients present. The ability to degrade pollutants seems to be a common feature of Xanthobacter species (5, 18, 44, 49, 50, 53) and other genera (i.e., Pseudomonas) (51). The ability to isolate these genera may be an artifact of the innate abilities of the organisms not only to utilize the xenobiotic compound but more importantly to grow on laboratory media under laboratory conditions. However, the clear disparity between the cultured dehalogenase gene pool (dehalogenases in pure cultures of bacteria) and the uncultured dehalogenase gene pool is the key point for discussion. For both isolates the group I and II deh genes were related to genes encoding dehalogenases found in aerobically isolated pure cultures able to degrade α-HAs. The observation that no group I deh sequences similar to either isolate's group I deh sequences were found at any stage of the enrichment is not easily explained since group II deh sequences were observed in both situations. The explanation may lie in the efficiency of the degenerate PCR used to amplify the group I deh sequences from the biomass samples; thus, this needs to be investigated further. If bacteria which are easily cultured on pollutants as sole carbon sources have biodegradative genes which seem to be distantly related to the metagene pool from which they were isolated, conclusions related to environmental processes based on data from these organisms are tenuous. Very rarely do researchers attempt to determine whether the bacteria that they isolate which are able to degrade a test pollutant are actively involved in its biodegradation in situ. Dunbar et al. (8) showed that batch culturing resulted in isolation of the 2,4-dichlorophenoxyacetic acid-degrading genes tfdA and tfdB, which “were highly similar, if not identical” to each other and to genes of Alcaligenes eutrophus JMP134(pJP4) (7) but were different from genes isolated by direct plating. Other groups have obtained similar results with batch culturing. Watanabe et al. (60) found that batch culturing resulted in isolation of nondominant phenol-degrading bacteria from a consortium able degrade this pollutant. Using thermal gradient gel electrophoresis of phenol hydroxylase gene fragments, these workers were able to show that a dominant genotype in the activated sludge was lost during enrichment by batch culturing. In both studies the authors concluded that enrichment in batch cultures tended to select specific genotypes. This bias was also seen in the genes isolated in this study. For example, isolates AS1 and AS2 are related to X. autotrophicus GJ10, and they contained dhlB-like dehalogenase genes; thus, similar organisms isolated independently have similar genotypes.
To overcome the bias of batch culturing, alternative methods have been utilized, such as dilution to extinction or enrichment designed to promote or exclude particular taxa. One such method, direct plating of microbial biomass onto the test compound (4), was used successfully in this and other studies (8, 60) to isolate environmentally relevant genes. In the present investigation the dehalogenase genes amplified from the direct plating biomass were much more closely related to some of the dehalogenase genes in the activated sludge (Fig. 1B) than to genes isolated by enrichment in batch cultures. However, no pure cultures were successfully isolated by this method. Such stalling of bacterial cultures has been observed previously by Lilley et al. (30) and may occur because essential growth factors present in the aliquot used for the initial plating are absent in the laboratory media used for subsequent subculturing. The significant observation resulting from this method was that the biomass contained some of the dehalogenases, which were closely related to clones from the activated sludge. However, no group II deh genes with sequences similar to those found in the direct plating biomass were observed for any of the enrichments or the activated sludge. This observation may indicate that either the isolates were minor components of the activated sludge community or there is a bias associated with direct plating, which is different from that associated with batch culturing.
However, even these sequences were not very closely related to several dehalogenases that were found in the activated sludge. Clones dehIIAS-gDNA-1, dehIIAS-gDNA-5, and dehIIAS-gDNA-8 were not affiliated with any of the subgroups described previously (14). While the approaches used here do not reveal whether these genes are actively expressed and thus translated into a dehalogenase, they do show that the catabolic genes involved in the biodegradation of chlorinated pollutants are much more diverse than investigations based on pure-culture studies have led us to believe. The derived amino acids from these partial gene sequences were analyzed and did show conservation of the 16 amino acids essential for catalysis (14, 16, 28) and the hydrophobic pocket of the enzymes (29), thus leading us to conclude that they may be active genes. Reverse transcription-PCR is thus a logical approach to determine whether these genes are actively being expressed or are silent. However, when any PCR-based method is used, the limitations and biases due to DNA extraction, primer design, and PCR efficiency need to considered when conclusions are drawn from the results.
In addition to exclusion of environmentally relevant microorganisms, our results showed that environmentally relevant genes are excluded through enrichment and isolation of dehalogenase-containing bacteria. Silent dehalogenase genes, a feature reported for several dehalogenases, are also overlooked unless culture-independent approaches are utilized in conjunction with the traditional culturing methods. These conclusions are a worrying development if one is trying to describe the contribution that the degradative genes make to environmentally relevant processes. The extent to which we are missing environmentally relevant genes is not fully understood; however, the fact that we are missing them is now unequivocal.
With the results of this study we are now in a stronger position to state that the diversity and predominance of the group II deh genes seem to be much greater than the limited diversity and distribution of the group I deh genes. Group II deh genes have long been recognized to be part of a superfamily of hydrolase-encoding genes (14, 27); however, group I deh genes do not appear to be related to any homologous genes. It may be that group I deh genes evolved more recently than group II genes; however, it should be noted that α-HA-degrading bacterial isolates often contain more than one dehalogenase encoded by genes belonging to both families (61). The selection may be associated with the fact that DehI-type dehalogenases extend stereospecificity to d enantiomers and thus provide a survival advantage to the host (i.e., the ability to use more of the chlorinated compound as a growth substrate or more complete or faster degradation of the pollutant). Thus, the group I deh genes may have evolved in response to human activities.
The predominance of Proteobacteria in enrichments and isolates is again consistent with previous studies (Table 3), but it was most probably due to the inherent bias of cultivation. Inspection of the 16S rRNA gene sequences from the enrichment process showed that proteobacteria dominated the primary and secondary enrichments, and no detectable gram-positive 16S rRNA gene sequences were found. Furthermore, isolates AS1 and AS2 were not predominant organisms in the activated sludge. The levels of coverage of the 16S rRNA gene libraries were 23, 47, and 73% for the activated sludge, the primary enrichment, and the secondary enrichment, respectively. Even with the low coverage of the activated sludge population, gram-positive 16S rRNA gene sequences accounted for a significant fraction of the sequences analyzed. Our analysis in silico led us to conclude that although group II deh genes are found mainly in the Proteobacteria, they are also present in other prokaryotes, including Archaea. In addition, there have been reports of gram-positive bacteria that are able to degrade α-HAs, but none of these organisms have been deposited in recognized culture collections or are currently available for study (15, 20, 24). This analysis highlights how culturing selected against the gram-positive bacteria, which may be the reason that no gram-positive α-HA degraders have been reported since 1964. The results further strengthen the conclusion that culturing biases skew our view of the microbial world and thus influence the conclusions that we draw.
TABLE 3.
Phylogenetic affiliations of α-HA dehalogenase hosts isolated in previous studies
| Dehalogenase genea | Proteobacterial group | Host genus | Accession no. or reference |
|---|---|---|---|
| dhlS51° | α | Agrobacterium | 26 |
| dehIIDA3 | α | Bradyrhizobium | AJ133463 |
| dhlB | α | Xanthobacter | M81691 |
| dehII°PII | β | Burkholderia | AJ133468 |
| dehIIK13 | β | Burkholderia | AJ133466 |
| dehIII11 | β | Burkholderia | AJ133465 |
| dehIIG02 | β | Burkholderia | AJ133464 |
| hdIIVα | β | Burkholderia | X66249 |
| dehCII | γ | Pseudomonas | M62908 |
| dehH2 | γ | Moraxella | D90423 |
| L-dex | γ | Pseudomonas | S74078 |
| dehH109 | γ | Pseudomonas | D17523 |
| hadL | γ | Pseudomonas | M81841 |
| dehIIK55 | γ | Pseudomonas | AJ133467 |
| dhIVII | γ | Pseudomonas | X94147 |
| dehIIPP3 | γ | Pseudomonas | AJ133462 |
| dehCI | γ | Pseudomonas | M62908 |
Designation of the gene as reported previously. A degree symbol indicates that the gene is described as silent.
The biodegradation of halogenated carboxylic acids has received a great deal of attention due the hazardous nature of the xenobiotic compounds. Many of the pure cultures able to degrade α-HAs have been isolated by enrichment by using batch culturing, and in light of the results of this study and other studies in which the bias introduced by batch culturing was investigated, whether the genes isolated previously are the genes that perform the task in situ must be considered. In this study we did not fully address whether the genes that were shown to be present in the activated sludge and were not found in the pure cultures were active (i.e., transcribed and translated into dehalogenases). However, the weight of evidence points to the conclusion that by batch culturing we indeed miss the environmentally relevant genes involved in biodegradation of α-HAs. Moreover, the results presented here may be interpreted as showing that the gene pool that exists in the unculturable community is not represented in the culturable fraction. The ramifications of this and other studies are that researchers are determining contributions which bacteria make to processes based on data derived from organisms that are not environmentally relevant microorganisms. This issue warrants further investigation to determine the true relationship between the cultured gene pool and the metagene pool from which it arose.
Acknowledgments
We express our gratitude to the Wellcome Trust for supporting this research.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology, p. 2.4. Greene Publishing Associates Wiley Interscience, New York, N.Y.
- 2.Baker, W., A. van den Broek, E. Camon, P. Hingamp, P. Sterk, G. Stoesser, and M. A. Tuli. 2000. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 28:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunner, W., D. Staub, and T. Leisinger. 1980. Bacterial degradation of dichloromethane. Appl. Environ. Microbiol. 40:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buyuksonmez, F., T. F. Hess, R. L. Crawford, and R. J. Watts. 1998. Toxic effects of modified Fenton reactions on Xanthobacter flavus FB71. Appl. Environ. Microbiol. 64:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykhuizen, D. E. 1998. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Leeuwenhoek 73:25-33. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics University of Washington, Seattle.
- 12.Goldman, P., G. W. Milne, and D. B. Keister. 1968. Carbon-halogen bond cleavage. 3. Studies on bacterial halidohrolases. J. Biol. Chem. 243:428-434. [PubMed] [Google Scholar]
- 13.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 14.Hill, K. E., J. R. Marchesi, and A. J. Weightman. 1999. Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. J. Bacteriol. 181:2535-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch, P., and M. Alexander. 1960. Microbial decomposition of halogenated propionic and acetic acids. Can. J. Microbiol. 6:241-249. [DOI] [PubMed] [Google Scholar]
- 16.Hisano, T., Y. Hata, T. Fujii, J. Q. Liu, T. Kurihara, N. Esaki, and K. Soda. 1996. Crystallization and preliminary X-ray crystallographic studies of l-2-haloacid dehalogenase from Pseudomonas sp. YL. Proteins 24:520-522. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, D. B., J. E. Oppentocht, and G. J. Poelarends. 2001. Microbial dehalogenation. Curr. Opin. Biotechnol. 12:254-258. [DOI] [PubMed] [Google Scholar]
- 18.Janssen, D. B., F. Pries, J. R. van der Ploeg, B. Kazemier, P. Terpstra, and B. Witholt. 1989. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J. Bacteriol. 171:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssen, D. B., A. Scheper, L. Dijkhuizen, and B. Witholt. 1985. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl. Environ. Microbiol. 49:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman, D. D. 1964. Microbial degradation of 2,2-dichloropropionic acid in five soils. Can. J. Microbiol. 10:843-852. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, H., N. Tone, and K. Tonomura. 1981. Purification and properties of haloacetate halidohydrolase specified by plasmid from Moraxella sp. strain B. Agric. Biol. Chem. 45:35-42. [Google Scholar]
- 23.Kawasaki, H., T. Toyama, T. Maeda, H. Nishino, and K. Tonomura. 1994. Cloning and sequence analysis of a plasmid-encoded 2-haloacid dehalogenase gene from Pseudomonas putida No. 109. Biosci. Biotechnol. Biochem. 58:160-163. [DOI] [PubMed] [Google Scholar]
- 24.Kearney, P. C., D. D. Kaufman, and M. L. Beall. 1964. Enzymatic dehalogenation of 2,2-dichloropropionate. Biochem. Biophys. Res. Commun. 14:29-33. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Kohler, R., A. Brokamp, R. Schwarze, R. H. Reiting, and F. R. Schmidt. 1998. Characteristics and DNA-sequence of a cryptic haloalkanoic acid dehalogenase from Agrobacterium tumefaciens RS5. Curr. Microbiol. 36:96-101. [DOI] [PubMed] [Google Scholar]
- 27.Koonin, E. V., and R. L. Tatusov. 1994. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 244:125-132. [DOI] [PubMed] [Google Scholar]
- 28.Kurihara, T., J. Q. Liu, V. Nardi-Dei, H. Koshikawa, N. Esaki, and K. Soda. 1995. Comprehensive site-directed mutagenesis of l-2-halo acid dehalogenase to probe catalytic amino acid residues. J. Biochem. 117:1317-1322. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y. F., Y. Hata, T. Fujii, T. Hisano, M. Nishihara, T. Kurihara, and N. Esaki. 1998. Crystal structures of reaction intermediates of l-2-haloacid dehalogenase and implications for the reaction mechanism. J. Biol. Chem. 273:15035-15044. [DOI] [PubMed] [Google Scholar]
- 30.Lilley, A. K., J. C. Fry, M. J. Bailey, and M. J. Day. 1996. Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris). FEMS Microbiol. Ecol. 21:231-242. [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchesi, J. R., and A. J. Weightman. 2000. Modified primers facilitate rapid screening of 16S rRNA gene libraries. BioTechniques 29:48-50. [DOI] [PubMed] [Google Scholar]
- 33.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 34.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1996. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature? Appl. Environ. Microbiol. 62:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nardi-Dei, V., T. Kurihara, C. Park, N. Esaki, and K. Soda. 1997. Bacterial dl-2-haloacid dehalogenase from Pseudomonas sp. strain 113: gene cloning and structural comparison with d- and l-2-haloacid dehalogenases. J. Bacteriol. 179:4232-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palleroni, N. J. 1997. Prokaryotic diversity and the importance of culturing. Antonie Leeuwenhoek 72:3-19. [DOI] [PubMed] [Google Scholar]
- 38.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 39.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poelarends, G. J., J. E. Hylckama Vlieg, J. R. Marchesi, L. M. Freitas Dos Santos, and D. B. Janssen. 1999. Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J. Bacteriol. 181:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rainey, F. A., and J. Wiegel. 1996. 16S ribosomal DNA sequence analysis confirms the close relationship between the genera Xanthobacter, Azorhizobium, and Aquabacter and reveals a lack of phylogenetic coherence among Xanthobacter species. Intl. J. Syst. Bacteriol. 46:607-610. [Google Scholar]
- 42.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reij, M. W., J. Kieboom, J. A. de Bont, and S. Hartmans. 1995. Continuous degradation of trichloroethylene by Xanthobacter sp. strain PY2 during growth on propene. Appl. Environ. Microbiol. 61:2936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Schwarze, R., A. Brokamp, and F. R. J. Schmidt. 1997. Isolation and characterization of dehalogenases from 2,2-dichloropropionate-degrading soil bacteria. Curr. Microbiol. 34:103-109. [DOI] [PubMed] [Google Scholar]
- 47.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and O. J. Van der. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slater, J. H., A. J. Weightman, and B. G. Hall. 1985. Dehalogenase genes of Pseudomonas putida PP3 on chromosomally located transposable elements. Mol. Biol. Evol. 2:557-567. [DOI] [PubMed] [Google Scholar]
- 49.Smith, M. R., W. J. J. van den Tweel, and J. A. M. de Bont. 1991. Degradation of 3-chloro-2-methylpropionic acid by Xanthobacter sp. CIMW 99. Appl. Microbiol. Biotechnol. 36:246-251. [DOI] [PubMed] [Google Scholar]
- 50.Spiess, E., C. Sommer, and H. Gorisch. 1995. Degradation of 1,4-dichlorobenzene by Xanthobacter flavus 14P1. Appl. Environ. Microbiol. 61:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanier, R. Y., N. J. Palleroni, and M. Douderoff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 46:159-271. [DOI] [PubMed] [Google Scholar]
- 52.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tay, S. T. L., H. F. Hemond, M. F. Polz, C. M. Cavanaugh, and L. R. Krumholz. 1999. Importance of Xanthobacter autotrophicus in toluene biodegradation within a contaminated stream. Syst. Appl. Microbiol. 22:113-118. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 56.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Peer, Y., and R. Dewachter. 1993. TREECON—a software package for the construction and drawing of evolutionary trees. Comput. Applic. Biosci. 9:177-182. [DOI] [PubMed] [Google Scholar]
- 58.van der Ploeg, J. R., G. van Hall, and D. B. Janssen. 1991. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J. Bacteriol. 173:7925-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, K., and P. W. Baker. 2000. Environmentally relevant microorganisms. J. Biosci. Bioeng. 89:1-11. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weightman, A. J., J. H. Slater, and A. T. Bull. 1979. The partial purification of two dehalogenases from Pseudomonas putida PP3. FEMS Microbiol. Lett. 6:231-234. [Google Scholar]
- 62.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]