Abstract
Aeromonas caviae R-specific enoyl-coenzyme A (enoyl-CoA) hydratase (PhaJAc) is capable of providing (R)-3-hydroxyacyl-CoA with a chain length of four to six carbon atoms from the fatty acid β-oxidation pathway for polyhydroxyalkanoate (PHA) synthesis. In this study, amino acid substitutions were introduced into PhaJAc by site-directed mutagenesis to investigate the feasibility of altering the specificity for the acyl chain length of the substrate. A crystallographic structure analysis of PhaJAc revealed that Ser-62, Leu-65, and Val-130 define the width and depth of the acyl-chain-binding pocket. Accordingly, we targeted these three residues for amino acid substitution. Nine single-mutation enzymes and two double-mutation enzymes were generated, and their hydratase activities were assayed in vitro by using trans-2-octenoyl-CoA (C8) as a substrate. Three of these mutant enzymes, L65A, L65G, and V130G, exhibited significantly high activities toward octenoyl-CoA than the wild-type enzyme exhibited. PHA formation from dodecanoate (C12) was examined by using the mutated PhaJAc as a monomer supplier in recombinant Escherichia coli LS5218 harboring a PHA synthase gene from Pseudomonas sp. strain 61-3 (phaC1Ps). When L65A, L65G, or V130G was used individually, increased molar fractions of 3-hydroxyoctanoate (C8) and 3-hydroxydecanoate (C10) units were incorporated into PHA. These results revealed that Leu-65 and Val-130 affect the acyl chain length substrate specificity. Furthermore, comparative kinetic analyses of the wild-type enzyme and the L65A and V130G mutants were performed, and the mechanisms underlying changes in substrate specificity are discussed.
A wide variety of bacterial strains accumulate the biological polyesters polyhydroxyalkanoates (PHAs) as an intracellular carbon and energy storage material. PHA has attracted industrial attention because of its potential use as a practical biodegradable thermoplastic (28, 32). Aeromonas caviae is capable of accumulating PHA copolymer consisting of (R)-3-hydroxybutyrate (C4) and (R)-3-hydroxyhexanoate (C6), which is soft and flexible and has appropriate properties for preparing polymer films when fatty acids or plant oils are used as carbon sources. We have proposed that the PHA biosynthesis pathway in A. caviae consists of two enzymes, R-specific enoyl-coenzyme A (CoA) hydratase (PhaJAc) [referred to below as (R)-hydratase] for supplying monomer units and PHA synthase (PhaCAc) for polymerization of the monomer.
(R)-hydratases catalyze an R-specific hydrating reaction of the fatty acid β-oxidation intermediates trans-2-enoyl-CoAs to (R)-3-hydroxyacyl-CoAs. Many organisms have been found to possess (R)-hydratase as a monofunctional enzyme or as an (R)-hydratase domain (hydratase 2 domain) of a multifunctional enzyme. In yeast, peroxisomal multifunctional enzyme type 2 (MFE-2) has the (R)-hydratase domain, which is involved in β-oxidation of straight-chain fatty acids (10). In the case of A. caviae, (R)-hydratase (PhaJAc) plays a critical role in supplying C4 and C6 monomer units from β-oxidation to PHA synthesis (6, 7). Genes homologous to the gene encoding PhaJAc have been found in several other PHA-producing bacteria, such as Rhodospirillum rubrum (26), Aeromonas hydrophila (24), Pseudomonas aeruginosa (33), Pseudomonas putida (4), and Pseudomonas oleovorans (4). Some PhaJs favoring medium-chain-length (C6-14) enoyl-CoAs may contribute to the supply of monomers for PHA synthesis via β-oxidation in pseudomonads (34). Indeed, these PhaJs, including PhaJAc, actually function as monomer suppliers for PHA production in recombinant bacteria when fatty acids or plant oils are used as carbon sources (8, 18).
The levels of amino acid sequence similarity between eukaryotic and bacterial (R)-hydratases are relatively high. For example, the level of partial identity between the hydratase 2 domain of peroxisomal MFE-2 from Saccharomyces cerevisiae and PhaJAc is 38.4% over 73 amino acids (6). The eukaryotic (R)-hydratases commonly have a characteristic hydratase 2 motif, which has been defined by Qin et al. (25) as [YF]-X(1,2)-[LVIG]-[STGC]-G-D-X-N-P-[LIV]-H-X(5)-[AS]. A sequencesimilar to the hydratase 2 motif is also found in the bacterial (R)-hydratases (34). However, the major difference in enzymatic properties between eukaryotic and bacterial (R)-hydratases is acyl chain length substrate specificity. Eukaryotic (R)-hydratases, such as the S. cerevisiae MFE-2, have specificity for a substrate with an acyl chain length of 10 C atoms (10), whereas bacterial (R)-hydratases, such as PhaJAc, have specificity for substrates with acyl chain lengths of 4 to 6 C atoms (7). This difference in substrate specificity between eukaryotic and bacterial (R)-hydratases may be derived from the difference in the architecture of the substrate-binding pockets.
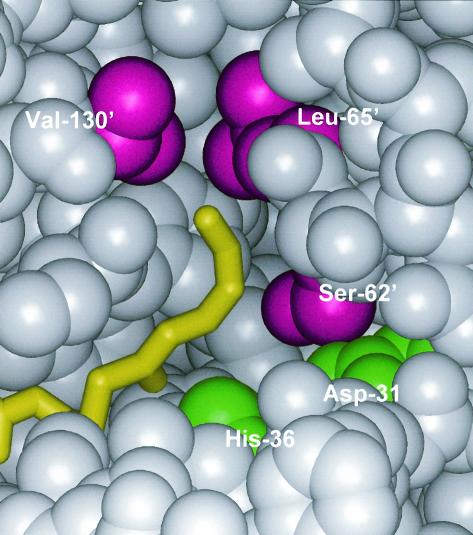
Recently, the crystal structure of PhaJAc at a resolution of 1.5 Å was determined (Protein Data Bank accession code 1IQ6) (11, 12). The monomer structure of the enzyme consists of a five-strand antiparallel β-sheet and five α-helices. Two of the monomers are associated with one another to form a functional homodimer with an extended 10-strand β-sheet. This structure is generally referred to as a hot dog fold, which was observed for the first time with β-hydroxydecanoyl thiol ester dehydratase (FabZEc) of Escherichia coli (21). In PhaJAc, Asp-31 and His-36 are located deep in the substrate-binding pocket (Fig. 1). These amino acids define the hydratase 2 motifs, are highly conserved among (R)-hydratases, and play critical roles in catalysis, as revealed by a mutational study (12). A structural docking model of this enzyme with a substrate provides information on its substrate recognition. The binding site for the acyl moiety of the substrate is surrounded by side chains consisting of Ser-62, Leu-65, Pro-70, Ser-74, Tyr-76, and Val-130. The side chains consisting of these residues define the depth and width of the acyl-chain-binding pocket, which specifically accepts C4 to C6 substrates but does not permit entrance of substrates longer than C6.
FIG. 1.
Schematic diagram of the acyl-chain-binding site of PhaJAc, represented by a space-filling model. The acyl tail of trans-2-hexenoyl-CoA is yellow, the side chains of the Ser-62, Leu-65, and Val-130 residues are magenta, and Asp-31 and His-36 are green. For clarity, residues 70 to 75, 132, and 133, which obscure the binding site, are not shown. Ser-62, Leu-65, and Val-130 were replaced by other amino acids with shorter side chains in this study. Asp-31 and His-36 were identified as a catalytic dyad in a previous study (12). A prime indicates that the amino acid residue is from another polypeptide chain. The atomic coordinates of this enzyme have been deposited in the Protein Data Bank (accession code 1IQ6).
The main objective of this work was to investigate the feasibility of altering the acyl chain length substrate specificity of PhaJAc through site-directed mutagenesis. Based on structural information, we selected three amino acid residues, Ser-62, Leu-65, and Val-130, for substitution. Nine single-mutation mutants and two double-mutation mutants of PhaJAc, in which there were amino acid substitutions at three positions, were generated and investigated. Here we describe the first successful case study of synthesis of PHA copolymers with desirable compositions based on rational alteration of the substrate specificity of PhaJAc.
MATERIALS AND METHODS
Site-directed mutagenesis.
Site-directed mutagenesis was performed by using a PCR method described by Imai et al. (13). The PCR primers used in this study are shown in Table 1. These primers were designed in inverted tail-to-tail directions to amplify pETNB3, an expression plasmid for phaJAc derived from pET-3a (7), together with the target sequence for amino acid substitution. Forward PCR primers contained the mutation sites (Table 1). Reverse PCR primers rS62/L65 and rV130 were used to replace Ser-62 or Leu-65 and Val-130, respectively. After PCR amplification with these primers, amplified linear DNA were phosphorylated and self-ligated by using a BKL kit (Takara Shuzo, Kyoto, Japan). Subsequently, the self-ligated PCR products were transformed into E. coli DH5α. The resulting plasmids were sequenced to confirm the DNA sequences of the open reading frames of mutated genes.
TABLE 1.
PCR primers used for construction of mutant PhaJAc molecules
| Primer direction | Primer | Amino acid substitution | Sequencea |
|---|---|---|---|
| Forward | fS62A | Ser-62 to Ala | 5′-CTCTTCGCCGGGCTGCTGGGCCAGCAGTTG-3′ |
| fL65I | Leu-65 to Ile | 5′-CTCTTCTCCGGGCTGATCGGCCAGCAGTTG-3′ | |
| fL65V | Leu-65 to Val | 5′-CTCTTCTCCGGGCTGGTGGGCCAGCAGTTG-3′ | |
| fL65A | Leu-65 to Ala | 5′-CTCTTCTCCGGGCTGGCGGGCCAGCAGTTG-3′ | |
| fL65G | Leu-65 to Gly | 5′-CTCTTCTCCGGGCTGGGGGGCCAGCAGTTG-3′ | |
| fV130A | Val-130 to Ala | 5′-GGGGAAGCCGTGGCCAAGCTGCCTTAA-3′ | |
| fV130G | Val-130 to Gly | 5′-GGGGAAGCCGTGGGCAAGCTGCCTTAA-3′ | |
| Reverse | rS62/L65 | Ser-62 to Ala, Leu-65 to Ile, Val, Ala, or Gly | 5′-GCTGGCGAGCAGCATGCCGTGGACTATGGG-3′ |
| rV130 | Val-130 to Ala or Gly | 5′-CGTCACGGCGAGGGCGCCGCCTTGGGT-3′ |
The underlined sequences are the mutation sites.
Introduction of second mutation.
A second mutation in the phaJAc gene was also introduced by the PCR method described above. To obtain an expression plasmid for a doubly mutated PhaJAc [pETNB3(L65A/V130G)], PCR was performed with the fL65A and rS62/L65 primers by using pETNB3(V130G) as a template. In the same manner, an expression plasmid for another doubly mutated PhaJAc [pETNB3(L65G/V130G)] was generated by using fL65G and rS62/L65 as the PCR primers.
Preparation of cell extracts.
Cell extracts were prepared from E. coli BL21(DE3) harboring the expression plasmid for PhaJAc or its mutant. The recombinant E. coli BL21(DE3) cells were inoculated into 100 ml of Luria-Bertani medium with ampicillin (100 μg/ml). The cells were cultivated in 500-ml flasks on a reciprocal shaker (130 strokes/min) at 30°C. After 3 h of cultivation, 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the T7 promoter. After an additional 2 h of cultivation, the cells were harvested and subjected to sonication. The disrupted cells were centrifuged (20,000 × g) for 20 min at 4°C, and the soluble fraction was used for a hydratase activity assay and enzyme purification.
Hydratase activity assay.
Enoyl-CoA hydratase activity was assayed by determining the hydration of trans-2-enoyl-CoA substrates. A 5-μl portion of an enzyme solution was added to 895 μl of 50 mM Tris-HCl (pH 8.0) containing 25 μM trans-2-enoyl-CoA in a quartz cuvette with 1.0-cm light path, and the decrease in absorbance at 263 nm was measured at 30°C. The ɛ263 of the enoyl-thioester bond is 6.7 × 103 M−1 cm−1. A C4 trans-2-enoyl-CoA substrate (crotonyl-CoA) was purchased from Sigma. C6, C8, C10, and C12 trans-2-enoyl-CoA substrates used for the hydratase assay were synthesized from a lithium salt of CoA and corresponding trans-2-alkenoic acids (Tokyo Kasei, Tokyo, Japan) based on a mixed-anhydride method (5). The C6 and C8 substrates were purified with a Sep-Pak C18 column (Waters, Milford, Mass.), as described by Valentin and Steinbüchel (35). The longer substrates (C10 and C12) were purified by precipitation at pH 1.0 and by washing the precipitate with pentane. In order to precipitate the C10 substrate, it was necessary to saturate the solution with NaCl at pH 1.0 (5).
Construction of plasmids for PHA synthesis.
In order to express the phaJAc gene or a mutated gene under lac promoter control, the expression plasmids were constructed by using pUC19 (Takara Shuzo). The plasmid carrying the phaJAc gene (pETNB3) or the mutated gene was digested with XbaI and BamHI, and a 0.5-kb XbaI-BamHI fragment containing an open reading frame and its ribosome binding site was subcloned into the same sites of pUC19 to obtain the new pUCJ plasmids.
Synthesis and analysis of PHA.
E. coli LS5218 [fadR601 atoC2(Con)] was used for PHA synthesis experiments (14, 15, 27). The pUCJ plasmid carrying either the wild-type phaJAc gene or a mutated phaJAc gene was cotransformed into E. coli LS5218 together with pPPAC, which carried the PHA synthase 1 gene from Pseudomonas sp. strain 61-3 (phaC1Ps) in the broad-host-range vector pJRD215 (22, 29). The recombinant E. coli LS5218 cells were inoculated into 100 ml of M9 medium containing 0.25% sodium dodecanoate along with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). The cells were cultivated in 500-ml flasks on a reciprocal shaker (130 strokes/min) at 37°C for 96 h. The PHA content of dry cells and the composition were determined by gas chromatography after methanolysis of lyophilized cells in the presence of 15% sulfuric acid, as described previously (17). The polymers that accumulated in the cells were extracted with chloroform for 48 h at room temperature and were purified by reprecipitation with methanol. Molecular mass measurement by gel permeation chromatography was performed as described previously (1).
Overexpression and purification of enzyme.
For purification of the wild-type PhaJ enzyme or L65A, the soluble protein fraction was loaded directly onto a HiLoad Q-Sepharose HP16/10 column (Amersham Biosciences, Piscataway, N.J.) equilibrated with 20 mM HEPES buffer (pH 7.2) and was eluted with a linear gradient (250 ml) of 0 to 1.0 M NaCl (2.5 ml/min) (7). The enoyl-CoA hydratase assay was performed for each fraction (5 ml) by using crotonyl-CoA as the substrate, and active fractions were combined, concentrated, and desalted by ultrafiltration with a Vivaspine (Vivascience, Göttingen, Germany)
V130G was purified by a three-step procedure, as follows. First, the cell extract was brought to 40% (wt/vol) saturation with ammonium sulfate. Centrifugation was carried out at 10,000 × g for 10 min. The ammonium sulfate concentration of the resulting supernatant was then raised to 60% (wt/vol). Again, centrifugation was carried out. The resulting pellets that salted out with 40 to 60% ammonium sulfate were resuspended in HEPES buffer, and ammonium sulfate was added to the resulting supernatant to a final concentration of 1.0 M. Next, the solution was loaded onto a Phenyl Sepharose HR16/10 column (Amersham Biosciences) equilibrated with HEPES buffer containing 1.0 M ammonium sulfate. The enzyme was eluted with a 1.0 to 0 M ammonium sulfate gradient (250 ml) at a flow rate of 2.5 ml/min. The hydratase fractions were pooled, concentrated, and desalted. Finally, the hydratase was loaded onto a MonoQ HR5/5 column (Amersham Biosciences) that had been preequilibrated with the HEPES buffer. The enzyme was eluted with a linear gradient of 0 to 1.0 M NaCl (25 ml) at a flow rate of 1.0 ml/min. Fractions were concentrated and desalted and then frozen at −80°C for later analysis.
Determination of Km and kcat.
The kinetic parameters of the wild-type enzyme and mutants of this enzyme were obtained by monitoring the hydration reaction at 263 nm by using a trans-2-enoyl-CoA substrate with chain length of 4 to 12 carbon atoms in 900 μl (final volume) at 30°C. The trans-2-enoyl-CoA substrates used for the activity assay were synthesized as described above. Kinetic data were transformed to Lineweaver-Burk plots by using the GraFit, version 5, computer software (Erithacus Software, Surrey, United Kingdom). The Km values were calculated from the slopes of the curves, and the catalytic turnover values (kcat) were calculated by dividing the maximal reaction velocities by the total amount of enzyme in the reaction mixture.
CD spectroscopy.
Circular dichroism (CD) spectroscopy was performed with a Jasco J720 spectropolarimeter. The adsorption at 280 nm was determined for all samples and used for fine adjustment of the protein concentration of the samples used for CD spectroscopy. The far-UV spectra of the proteins were determined from 200 to 250 nm in 10 mM sodium phosphate buffer at 20°C and pH 7.0 with the following instrument settings: response, 1 s; sensitivity, 100 mdeg; and speed, 50 nm/min. The average for 25 scans was determined.
Other analytical procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed with 15% polyacrylamide gels. The molecular weight of the native enzyme was evaluated by gel filtration chromatography with a Superdex 75 HR 10/30 column (Amersham Biosciences) in 20 mM HEPES buffer containing 0.1 M NaCl. The molecular weight of the subunit was determined by SDS-PAGE. The thermal stability of the enzyme was evaluated by measuring the residual hydratase activity toward crotonyl-CoA (C4) after heating for 10 min at 50°C. The protein concentration was determined by the method of Bradford by using the Bio-Rad assay solution and bovine serum albumin as the standard.
RESULTS
Candidate amino acid residues responsible for formation of the substrate-binding pocket.
The crystal structure analysis of PhaJAc suggested that side chains consisting of eight amino acids, Asp-31, His-36, Ser-62, Leu-65, Pro-70, Ser-74, Tyr-76, and Val-130, surround the acyl moiety of the substrate in the form of the enzyme-substrate complex. Figure 1 shows the positions of Asp-31, His-36, Ser-62, Leu-65, and Val-130. We previously demonstrated through a mutational study the involvement of Asp-31 and His-36 in catalysis of the hydration reaction (12). Analysis of the PhaJAc structure revealed that Ser-62 is located within hydrogen-bonding distance of Asp-31 and may define the width of the substrate-binding site. Leu-65 and Val-130, which are located at the bottom of the substrate-binding site, may define the depth of the acyl-chain-binding pocket. We therefore focused on these three residues to verify how they determine the specificity for the acyl chain length of substrates.
Alteration of the substrate specificity of PhaJAc.
Seven single-amino-acid-substitution mutants were generated, in which Ser-62, Leu-65, or Val-130 was replaced by other amino acids. The genes were expressed individually in E. coli BL21(DE3), and production of each recombinant protein was checked by SDS-PAGE analysis by using whole-cell extracts. Similar amounts of recombinant proteins were produced in all cases (data not shown). Next, the hydratase activities of the cell extracts were assayed by using crotonyl-CoA (C4) and octenoyl-CoA (C8) as substrates.
The results are summarized in Table 2. The strain harboring the wild-type enzyme gene showed hydratase activities toward crotonyl-CoA and octenoyl-CoA of 1,594 and 0.86 U/mg, respectively. The ratio of the hydratase activity with the C8 substrate to the hydratase activity with the C4 substrate (C8/C4 activity ratio) was calculated to be 5.4 × 10−4. When S62A was expressed, reduced activity with the C4 substrate was observed, whereas the activity with the C8 substrate was almost the same as that of the wild-type enzyme. This suggests that the bulkiness of an amino acid at this position might be important for stabilization of the C4 substrate within the substrate-binding pocket.
TABLE 2.
Hydratase activities with trans-2-crotonyl-CoA (C4) or trans-2-octenoyl-CoA (C8)
| Expressed PhaJAc | Formation of inclusion bodya | Sp act (U/mg) with:
|
C8/C4 activity ratio (10−3) | |
|---|---|---|---|---|
| Crotonyl- CoA (C4) | Octenoyl- CoA (C8) | |||
| None | − | ND | NDb | ndc |
| Wild type | − | 1,594 | 0.86 | 0.54 |
| S62A | − | 67.0 | 0.55 | 8.2 |
| L65I | − | 1,880 | 2.25 | 1.2 |
| L65V | − | 1,538 | 7.92 | 5.1 |
| L65A | − | 1,256 | 69.8 | 56 |
| L65G | + | 15.8 | 1.98 | 130 |
| V130A | − | 1,288 | 6.59 | 5.1 |
| V130G | + | 68.5 | 21.2 | 310 |
| L65A/V130G | + | 0.08 | ND | nd |
| L65G/V130G | + | 0.05 | ND | nd |
To estimate the amount of inclusion body formation, insoluble fractions of the cell extract were subjected to SDS-PAGE analysis. −, not detectable or trace formation of inclusion body (less than 5% of the total PhaJAc enzyme produced); +, mostly formed as inclusion body (almost 100%).
ND, not detected.
nd, not determined.
For the mutated PhaJAc molecules with a Leu-65 substitution, the C8/C4 activity ratio increased as the size of the side chain of the amino acid residue decreased. In particular, the L65A mutation led to increased activity (69.8 U/mg) with the C8 substrate, which was up to approximately 60-fold higher than that of the wild-type enzyme. For the L65G mutant enzyme, most PhaJ proteins were produced as inclusion bodies. A soluble and active fraction of L65G also exhibited a high C8/C4 activity ratio (1.3 × 10−2).
Replacement of Val-130 by Gly led to a significant change in the enzyme activity with the C8 substrate. The C8/C4 activity ratio for V130G reached the highest value, 3.1 × 10−1. However, most V130G mutant proteins were also observed as inclusion bodies.
Furthermore, we generated two doubly mutated genes with mutations at the positions encoding Leu-65 and Val-130 (L65A/V130A and L65A/V130G), which were subsequently expressed in E. coli. We also checked the level of recombinant protein production from each expression plasmid by analysis of whole-cell extracts by SDS-PAGE. Similar levels of recombinant protein production were observed in all cases (data not shown). However, very small amounts of doubly mutated PhaJs appeared in the cell extracts, suggesting that most of the mutant PhaJAc molecules were produced as inclusion bodies. This was probably due to incorrect folding of the protein as a result of the double mutation. Consequently, very weak or no hydratase activity was detected in these samples.
PHA biosynthesis with mutant PhaJAcmolecules.
To investigate how the changed substrate specificities of PhaJAc affect in vivo PHA synthesis, expression plasmids harboring phaJAc or phaJAc mutant genes were cotransformed with pPPAC. The pPPAC plasmid harbors the phaC1Ps gene encoding the Pseudomonas sp. strain 61-3 PHA synthase 1 protein, which has a broad substrate specificity, ranging from C4 to C12 (22). The recombinant E. coli LS5218 cells were grown in the presence of dodecanoate, and their ability to accumulate PHA was assayed. Table 3 summarizes the results obtained. The positive control strain harboring both the phaC1Ps gene and the wild-type phaJAc gene accumulated 27% (wt/wt) PHA, while the negative control strain harboring the phaC1Ps gene alone accumulated 4% (wt/wt) PHA. When the various single-mutation enzymes were expressed individually, the strains accumulated PHA at levels ranging from 12 to 22% (wt/wt), suggesting that they still had the ability to supply monomers for PHA production. In particular, V130G and L65I exhibited higher monomer-supplying activities than the other mutant enzymes examined (22 and 21% [wt/wt] PHA, respectively). The double-mutation enzymes (L65A/V130G and L65G/V130G) exhibited no significant monomer-supplying activities.
TABLE 3.
Accumulation of PHA in recombinant E. coli LS5218 harboring the phaC1Ps gene with a mutated phaJAc gene in the presence of dodecanoatea
| Expressed PhaJAc | PHA content (%, wt/wt) | PHA compositionb (mol%)
|
||||
|---|---|---|---|---|---|---|
| 3HB (C4) | 3HHx (C6) | 3HO (C8) | 3HD (C10) | 3HDD (C12) | ||
| None | 4 | 2 | 16 | 47 | 21 | 14 |
| Wild type | 27 | 11 | 76 | 7 | 4 | 2 |
| S62A | 12 | 20 | 50 | 16 | 10 | 4 |
| L65I | 21 | 17 | 66 | 10 | 4 | 3 |
| L65V | 12 | 19 | 50 | 20 | 7 | 4 |
| L65A | 16 | 12 | 49 | 24 | 12 | 3 |
| L65G | 15 | 14 | 44 | 26 | 11 | 5 |
| V130A | 22 | 14 | 69 | 10 | 5 | 2 |
| V130G | 16 | 19 | 29 | 32 | 15 | 5 |
| L65A/V130G | 4 | 7 | 18 | 44 | 19 | 12 |
| L65G/V130G | 4 | 7 | 15 | 49 | 18 | 11 |
Cells were cultivated for 96 h at 37°C in M9 medium containing sodium dodecanoate (0.25%, wt/vol). All values are averages from triplicate tests.
Abbreviations: 3HB, 3-hydroxybutyrate; 3HHx, 3-hydroxyhexanoate; 3HO, 3-hydroxyoctanoate; 3HD, 3-hydroxydecanoate; 3HDD, 3-hydroxydodecanoate.
The substrate specificities of the monomer-supplying enzymes employed affected the monomer compositions of PHA. When L65A, L65G, or V130G was used as a monomer supplier, a significant change in the composition of PHA occurred. The molar percentages of the 3-hydroxyoctanoate unit (C8) and the 3-hydroxydecanoate unit (C10) in the PHA were considerably higher than those in other PHAs. These results indicate that the L65A, L65G, or V130G mutation changed the substrate specificity toward the C8 and C10 monomers compared to the substrate specificity of the wild-type enzyme, resulting in the formation of PHA with higher C8 and C10 contents. To measure the molecular weights and polydispersities of polymers, the PHAs were extracted from the cells harboring the phaC1Ps and phaJAc genes (wild-type enzyme, L65A, or V130G) or the phaC1Ps gene alone. As a result, all extracted samples were found to have high number average molecular masses, ranging from 1.5 × 105 to 4.4 × 105 Da. The polydispersities were similar in all cases, with values of approximately 1.8.
Purification of mutant enzymes.
To compare the enzymatic properties of the wild-type enzyme and two mutants, L65A and V130G, we purified them from cell extracts of the corresponding recombinant E. coli BL21(DE3) strains. L65A was purified by a one-step procedure by using exchange chromatography in the same manner as the wild-type enzyme (7). V130G was purified by a three-step procedure by using ammonium sulfate cuts and hydrophobic and exchange chromatography, as shown in Table 4. The V130G mutant enzyme was purified with 14-fold-higher specific activity and a level of recovery of 28%. The purified wild-type enzyme, L65A, and V130G were subjected to SDS-PAGE analysis. These enzymes were homogeneous, and each enzyme had a molecular mass of approximately 15 kDa (data not shown). In addition, the molecular masses of both mutant enzymes in the native state were estimated to be 30 kDa by gel filtration chromatographic analysis. This indicates that these proteins form homodimers, as demonstrated previously for the wild-type enzyme (7).
TABLE 4.
Purification of mutated PhaJAc (V130G) from recombinant E. coli BL21(DE3)
| Step | Total activitya (U) | Total protein (mg) | Sp acta (U/mg) | Purifi- cation (fold) | Recovery (%) |
|---|---|---|---|---|---|
| None | 3.9 × 103 | 62 | 62 | 1 | 100 |
| (NH4)2SO4 precipita- tion (40-60% cut) | 2.8 × 103 | 36 | 78 | 1.3 | 73 |
| Phenyl Sepharose | 1.5 × 103 | 4.0 | 373 | 6.0 | 38 |
| MonoQ | 1.1 × 103 | 1.2 | 883 | 14 | 28 |
Enoyl-CoA hydratase activity toward crotonyl-CoA.
Kinetic analysis of the wild-type enzyme and mutants.
To gain insight into the mechanisms underlying changes in substrate specificity, we performed a comparative kinetic analysis of the wild-type enzyme and the mutants (L65A and V130G) using purified samples. The kinetic constants with the various lengths of trans-2-enoyl-CoA are summarized in Table 5. The data show that the wild-type enzyme had almost the same Km values for the C6 to C12 substrates; for example, the Km values were 40 μM for the C6 substrate and 43 μM for the C12 substrate. However, the Km for the C4 substrate (24 μM) was considerably lower than the values for the other substrates. In contrast, the kcat of the wild-type enzyme decreased as the substrate chain length decreased in the range from C4 to C12, except for C10. These kinetic parameters imply that the wild-type enzyme is specific for the substrates with shorter acyl chains (C4 and C6).
TABLE 5.
Kinetic constants of wild-type PhaJAc and mutants L65A and V130G at 30°C
| Substrate | Wild-type enzyme
|
L65A
|
V130G
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | |
| Crotonyl-CoA (C4) | 24 ± 2 | 1,922 ± 94 | 8.0 × 107 | 33 ± 3 | 899 ± 51 | 2.7 × 107 | 154 ± 11 | 3,141 ± 127 | 2.0 × 107 |
| Hexenoyl-CoA (C6) | 40 ± 5 | 294 ± 23 | 7.4 × 106 | 18 ± 2 | 89 ± 3 | 4.9 × 106 | 102 ± 12 | 402 ± 25 | 3.9 × 106 |
| Octenoyl-CoA (C8) | 42 ± 5 | 0.58 ± 0.04 | 1.4 × 104 | 21 ± 2 | 27 ± 1 | 1.3 × 106 | 76 ± 6 | 405 ± 14 | 5.3 × 106 |
| Decenoyl-CoA (C10) | 42 ± 4 | 0.65 ± 0.04 | 1.5 × 104 | 27 ± 4 | 18 ± 1 | 6.7 × 105 | 13 ± 2 | 108 ± 5 | 8.3 × 106 |
| Dodecenoyl-CoA (C12) | 43 ± 5 | 0.15 ± 0.01 | 3.5 × 103 | 34 ± 3 | 1.7 ± 0.1 | 5.0 × 104 | 5 ± 1 | 16 ± 1 | 3.2 × 106 |
L65A showed a kinetic profile that was very different from the wild-type enzyme profile. The Km of L65A for the C4 substrate (33 μM) was slightly higher than that of the wild-type enzyme (24 μM), whereas the values for the C6 to C12 substrates (range, 18 to 34 μM) were lower than those of the wild-type enzyme (40 to 43 μM). The kcat values of L65A for C4 and C6 (899 and 89 s−1, respectively) were less than one-half the wild-type kcat values (1,922 and 294 s−1, respectively). However, the kcat values of L65A for substrates longer than C6 (1.7 to 27 s−1) were considerably higher than the corresponding values of the wild-type enzyme (0.65 to 0.15 s−1). From the comparison of kinetic parameters of L65A and the wild-type enzyme, we concluded that the increases in the activities of L65A with the substrates longer than C6 resulted mainly from the increases in kcat values.
V130G exhibited kinetic constants that were completely different than those of the wild-type enzyme. For a C4 substrate, both the Km and kcat values (154 μM and 3,141 s−1, respectively) were markedly higher than the values of the wild-type enzyme (24 μM and 1,922 s−1, respectively). As the acyl chain length of the substrate increased, the Km and kcat values of V130G tended to gradually decrease. For the C10 and C12 substrates, the Km values of V130G (13 and 5 μM, respectively) were less than one-half the Km values of the wild-type enzyme, while the kcat values of V130G (108 and 16 s−1, respectively) were more than 100-fold higher than the kcat values of the wild-type enzyme. These kinetic data obviously show that the change in the substrate specificity of V130G was due to changes in both the Km and kcat values.
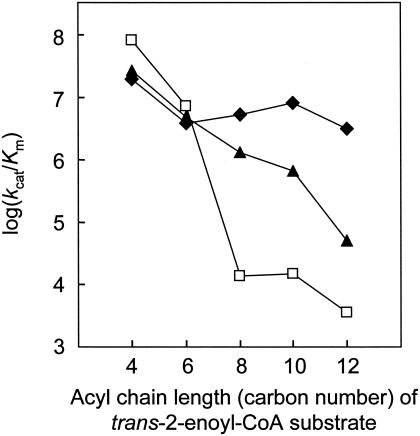
The specific constants (kcat/Km) of the wild-type enzyme and mutants (L65A and V130G) are also given in Table 5. In addition, to easily explain the dependence of the specific constants on the acyl chain length of the substrate, the logarithmic values of the specific constants are shown in Fig. 2. For the C8 substrate, L65A and V130G exhibited specific constants that were approximately 3 and 4 orders of magnitude higher than those of the wild-type enzyme. Even for the C12 substrate, V130G exhibited a specific constant that was nearly 3 orders of magnitude higher than that of the wild-type enzyme. The specific constants of L65A tended to decrease logarithmically with an increase in the acyl chain length of the substrate. In contrast, V130G exhibited high specific constants almost independent of the acyl chain length of the substrate.
FIG. 2.
Relationship between log(kcat/Km) and acyl chain length of the trans-2-enoyl-CoA substrate for the wild-type enzyme (□), L65A (▴), and V130G (⧫). The values for kcat, Km, and kcat/Km are shown in Table 5.
Thermal stability and CD analysis of mutant enzymes.
Protein stability generally correlates with the thermal stability of a protein. Thus, in order to evaluate protein stability, the residual activities of purified enzymes (wild-type enzyme, L65A, and V130G) after heat treatment at 50°C for 10 min were assayed. As a result, the wild-type enzyme, L65A, and V130G showed residual activities of 29, 25, and 13%, respectively (the hydratase activity in each sample before heat treatment was defined as 100%). L65A exhibited a slightly lower activity than the wild-type enzyme, whereas the activity of V130G was less than one-half the wild-type activity. This suggests that the structural stability of V130G is perceptibly lower than that of the wild-type enzyme or L65A.
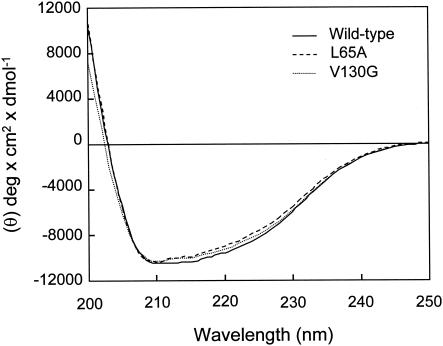
Figure 3 shows CD spectra for the wild-type enzyme, L65A, and V130G in the far-UV region (200 to 250 nm). This analysis revealed that the spectra for the wild-type and mutant enzymes were similar, suggesting that the secondary structures of the mutants were not significantly changed.
FIG. 3.
CD spectra of purified wild-type enzyme, L65A, and V130G: far-UV spectra of the wild-type enzyme, L65A, and V130G.
DISCUSSION
We succeeded in identifying two important residues, Leu-65 and Val-130, in PhaJAc responsible for determination of the acyl chain length substrate specificity. These two amino acid residues are located at the bottom of the proposed substrate-binding pocket, as predicted by X-ray crystal structure analysis (12). Interestingly, although Val-130 is located at the C-terminal end of PhaJAc, it was found to be very important in both acyl chain length substrate specificity and structural stability. Among bacterial PhaJs, the A. caviae enzyme has the smallest molecular size (133 amino acids), suggesting that PhaJAc might be the minimum unit for expressing hydratase activity.
In the kinetic analysis, the specificity constant (kcat/Km) of the wild-type enzyme for the C8 substrate was 3 orders of magnitude lower than the specificity constant for the C6 substrate (Fig. 2 and Table 5). In contrast, the specificity constant of L65A for the C8 substrate was only 1 order of magnitude lower than the specificity constant for the C6 substrate. This implies that the side chain of Leu-65 prohibits substrates longer than C6 from entering into the substrate-binding pocket. Therefore, replacement of Leu-65 by Ala (a shorter side chain amino acid residue) allows C8 and longer substrates to enter the substrate-binding pocket. Based on a computer-aided structural inspection of the enzymes with Leu-65 replaced by other amino acids, only L65G facilitated the acceptance of substrates with chain lengths longer than C6 if the main chain structure of the enzyme was not changed. For L65A, the Cβ atom of alanine is predicted to prohibit longer substrates from entering the substrate-binding pocket. However, if the mutation caused some structural changes in L65A, the substrate-binding pocket was calculated to be 5.6 Å deeper at the maximum depth than the substrate-binding pocket in the wild-type enzyme. This depth is sufficient to accept the long C8 substrate because the acyl moiety in octenoyl-CoA (C8 substrate) is only 2.4 Å longer than that in hexenoyl-CoA (C6 substrate). Although the comparison of the CD spectra of the wild-type enzyme and L65A showed that the molecules have the same secondary structure (Fig. 3), the substrate-binding pocket of L65A is somewhat deeper than that of the wild-type enzyme judging from the kinetic analysis.
CD spectral analysis of soluble V130G revealed that it has the same secondary structure as the wild-type enzyme (Fig. 3). However, the thermal stability of soluble V130G was lower than those of the wild-type enzyme and L65A. Therefore, replacement of Val-130 by Gly led to a decrease in the structural stability of the enzyme, probably due to disruption of the hydrophobic core which is formed by about 10 amino acids, including Leu-65 and Val-130, at the bottom of the substrate-binding pocket. This hydrophobic core is important for correct folding of this enzyme. On the other hand, the specificity for the acyl chain length of the substrate was altered significantly by introducing the amino acid substitution at Val-130. The alteration of the substrate specificity of V130G is apparent from the changes in both the Km and kcat values (Table 5). Based on the computer-aided structural inspection, the substrate-binding pocket of V130G was calculated to be 4.5 Å deeper at the maximum depth than that of the wild-type enzyme, assuming that the overall structure of V130G is nearly identical to that of the wild-type enzyme. This indicates that the extended acyl-chain-binding site of V130G is not deeper than that of L65A, which is inconsistent with the observation that the acyl chain length substrate specificity of V130G is significantly broader than that of L65A. Interestingly, the binding site of V130G is much wider than that of L65A, which may explain this apparent contradiction. The kinetic analysis of V130G revealed that the Km decreases with an increase in the chain length of the substrate (Table 5). This also accounts for the fact that a longer substrate fits in the extended acyl-chain-binding site of V130G.
As demonstrated in a previous study (12), PhaJAc has a characteristic hot dog fold, which was also observed in E. coli FabZEc (21). The manner of substrate entry into the substrate-binding pocket of FabZEc is similar to that of PhaJAc; that is, the acyl moiety of the substrate enters the substrate-binding pocket, whereas the CoA or acyl carrier protein moiety of the substrate is outside the enzyme. Hence, the substrate specificity of FabZEc could be altered by introducing an amino acid substitution at the region adjacent to the bottom of the substrate-binding pocket. As for other enzymes, such as lipase (9, 16, 20) and desaturase (3, 36), there have been several reports concerning the successful alteration of acyl chain length substrate specificities by burrowing of the cavity of the substrate-binding pocket. Among PHA biosynthesis enzymes, PhaA and PhaJ are the only enzymes with crystal structures that have been determined so far (12, 23). Therefore, like PhaJAc, it may be possible to change the acyl chain length substrate specificity of PhaA to a desirable substrate specificity based on the structural information.
Recently, workers in our laboratory and other workers have succeeded in producing many PHA synthases which have higher activities or altered substrate specificities by means of in vitro or in vivo evolution (2, 19, 30, 31). In this study, we also produced monomer-supplying enzymes with substrate specificities different from those of the wild-type monomer-supplying enzyme, PhaJAc. We now believe that a systematic combination of the PHA synthases produced and the monomer-supplying enzymes developed in our laboratory should allow us to construct an artificial PHA biosynthesis pathway capable of producing high-performance PHA which cannot be produced by wild-type enzymes. This new approach for polymer synthesis has the potential to improve industrial production of PHA by recombinant microorganisms and transgenic plants.
Acknowledgments
We thank C. Nomura for critical reading of the manuscript.
This work was supported by SORST (Solution Oriented Research for Science and Technology) of Japan Science and Technology Corporation (JST), by grant-in-aid for scientific research 15710053 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.T.), and by the Special Postdoctoral Researchers Program of RIKEN Institute (T.T.).
REFERENCES
- 1.Abe, H., Y. Doi, T. Fukushima, and H. Eya. 1994. Biosynthesis from gluconate of a random copolyester consisting of 3-hydroxybutyrate and medium-chain-length 3-hydroxyalkanoates by Pseudomonas sp. 61-3. Int. J. Biol. Macromol. 16:115-119. [DOI] [PubMed] [Google Scholar]
- 2.Amara, A. A., A. Steinbüchel, and B. H. Rehm. 2002. In vivo evolution of the Aeromonas punctata polyhydroxyalkanoate (PHA) synthase: isolation and characterization of modified PHA synthases with enhanced activity. Appl. Microbiol. Biotechnol. 59:477-482. [DOI] [PubMed] [Google Scholar]
- 3.Cahoon, E. B., and J. Shanklin. 2000. Substrate-dependent mutant complementation to select fatty acid desaturase variants for metabolic engineering of plant seed oils. Proc. Natl. Acad. Sci. USA 97:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiedler, S., A. Steinbüchel, and B. H. Rehm. 2002. The role of the fatty acid β-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch. Microbiol. 178:149-160. [DOI] [PubMed] [Google Scholar]
- 5.Fong, J. C., and H. Schulz. 1981. Short-chain and long-chain enoyl-CoA hydratase from pig heart muscle. Methods Enzymol. 70:390-398. [DOI] [PubMed] [Google Scholar]
- 6.Fukui, T., and Y. Doi. 1997. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J. Bacteriol. 179:4821-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukui, T., N. Shiomi, and Y. Doi. 1998. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J. Bacteriol. 180:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukui, T., S. Yokomizo, G. Kobayashi, and Y. Doi. 1999. Co-expression of polyhydroxyalkanoate synthase and (R)-enoyl-CoA hydratase genes of Aeromonas caviae establishes copolyester biosynthesis pathway in Escherichia coli. FEMS Microbiol. Lett. 170:69-75. [DOI] [PubMed] [Google Scholar]
- 9.Gaskin, D. J., A. Romojaro, N. A. Turner, J. Jenkins, and E. N. Vulfson. 2001. Alteration of lipase chain length specificity in the hydrolysis of esters by random mutagenesis. Biotechnol. Bioeng. 73:433-441. [DOI] [PubMed] [Google Scholar]
- 10.Hiltunen, J. K., B. Wenzel, A. Beyer, R. Erdmann, A. Fossa, and W. H. Kunau. 1992. Peroxisomal multifunctional β-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J. Biol. Chem. 267:6646-6653. [PubMed] [Google Scholar]
- 11.Hisano, T., T. Fukui, T. Iwata, and Y. Doi. 2001. Crystallization and preliminary X-ray analysis of (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate biosynthesis. Acta Crystallogr. D Biol. Crystallogr. 57:145-147. [DOI] [PubMed] [Google Scholar]
- 12.Hisano, T., T. Tsuge, T. Fukui, T. Iwata, K. Miki, and Y. Doi. 2002. Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeomonas caviae involved in polyhydroxyalkanoate biosynthesis. J. Biol. Chem. 278:617-624. [DOI] [PubMed] [Google Scholar]
- 13.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins, L. S., and W. D. Nunn. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins, L. S., and W. D. Nunn. 1987. Regulation of the ato operon by the atoC gene in Escherichia coli. J. Bacteriol. 169:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joerger, R. D., and M. J. Haas. 1994. Alteration of chain length selectivity of a Rhizopus delemar lipase through site-directed mutagenesis. Lipids 29:377-384. [DOI] [PubMed] [Google Scholar]
- 17.Kato, M., H. J. Bao, C. K. Kang, T. Fukui, and Y. Doi. 1996. Production of a novel copolymer of 3-hydroxybutyric acids and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl. Microbiol. Biotechnol. 45:363-370. [Google Scholar]
- 18.Kichise, T., T. Fukui, Y. Yoshida, and Y. Doi. 1999. Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int. J. Biol. Macromol. 25:69-77. [DOI] [PubMed] [Google Scholar]
- 19.Kichise, T., S. Taguchi, and Y. Doi. 2002. Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl. Environ. Microbiol. 68:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, R. R., G. King, R. A. Moreau, and M. J. Haas. 1997. Altered acyl chain length specificity of Rhizopus delemar lipase through mutagenesis and molecular modeling. Lipids 32:123-130. [DOI] [PubMed] [Google Scholar]
- 21.Leesong, M., B. S. Henderson, J. R. Gillig, J. M. Schwab, and J. L. Smith. 1996. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure 4:253-264. [DOI] [PubMed] [Google Scholar]
- 22.Matsusaki, H., S. Manji, K. Taguchi, M. Kato, T. Fukui, and Y. Doi. 1998. Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. strain 61-3. J. Bacteriol. 180:6459-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modis, Y., and R. K. Wierenga. 2000. Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase. J. Mol. Biol. 297:1171-1182. [DOI] [PubMed] [Google Scholar]
- 24.Park, S. J., W. S. Ahn, P. R. Green, and S. Y. Lee. 2001. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by metabolically engineered Escherichia coli strains. Biomacromolecules 2:248-254. [DOI] [PubMed] [Google Scholar]
- 25.Qin, Y. M., A. M. Haapalainen, S. H. Kilpelainen, M. S. Marttila, M. K. Koski, T. Glumoff, D. K. Novikov, and J. K. Hiltunen. 2000. Human peroxisomal multifunctional enzyme type 2. Site-directed mutagenesis studies show the importance of two protic residues for 2-enoyl-CoA hydratase 2 activity. J. Biol. Chem. 275:4965-4972. [DOI] [PubMed] [Google Scholar]
- 26.Reiser, S. E., T. A. Mitsky, and K. J. Gruys. 2000. Characterization and cloning of an (R)-specific trans-2,3-enoylacyl-CoA hydratase from Rhodospirillum rubrum and use of this enzyme for PHA production in Escherichia coli. Appl. Microbiol. Biotechnol. 53:209-218. [DOI] [PubMed] [Google Scholar]
- 27.Rhie, H. G., and D. Dennis. 1995. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl. Environ. Microbiol. 61:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudesh, K., H. Abe, and Y. Doi. 2000. Synthesis, structure and properties of polyhydroxyalkanoate: biological polyesters. Prog. Polym. Sci. 25:1503-1555. [Google Scholar]
- 29.Taguchi, K., Y. Aoyagi, H. Matsusaki, T. Fukui, and Y. Doi. 1999. Co-expression of 3-ketoacyl-ACP reductase and polyhydroxyalkanoate synthase genes induces PHA production in Escherichia coli HB101 strain. FEMS Microbiol. Lett. 176:183-190. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi, S., H. Nakamura, T. Hiraishi, I. Yamato, and Y. Doi. 2002. In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis. J. Biochem. (Tokyo) 131:801-806. [DOI] [PubMed] [Google Scholar]
- 31.Takase, K., S. Taguchi, and Y. Doi. 2003. Enhanced synthesis of poly(3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis and in vitro recombination of type II polyhydroxyalkanoate synthase gene. J. Biochem. (Tokyo) 133:139-145. [DOI] [PubMed] [Google Scholar]
- 32.Tsuge, T. 2002. Metabolic improvements and use of inexpensive carbon sources in microbial production of polyhydroxyalkanoate. J. Biosci. Bioeng. 94:579-584. [DOI] [PubMed] [Google Scholar]
- 33.Tsuge, T., T. Fukui, H. Matsusaki, S. Taguchi, G. Kobayashi, A. Ishizaki, and Y. Doi. 2000. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol. Lett. 184:193-198. [DOI] [PubMed] [Google Scholar]
- 34.Tsuge, T., K. Taguchi, S. Taguchi, and Y. Doi. 2003. Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: metabolic tools for polyhydroxyalkanoate synthesis via fatty acid β-oxidation. Int. J. Biol. Macromol. 31:195-205. [DOI] [PubMed] [Google Scholar]
- 35.Valentin, H. E., and A. Steinbüchel. 1994. Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl. Microbiol. Biotechnol. 40:699-709. [Google Scholar]
- 36.Whittle, E., and J. Shanklin. 2001. Engineering Δ9 16:0-acyl carrier protein (ACP) desaturase specificity based on combinatorial saturation mutagenesis and logical redesign of the castor Δ9 18:0-ACP desaturase. J. Biol. Chem. 276:21500-21505. [DOI] [PubMed] [Google Scholar]