Abstract
In high throughput sequence analysis, it is often necessary to combine the results of contemporary bioinformatics tools, because no individual tool alone computes all the requested information. ESTAnnotator is a tool for the high throughput annotation of expressed sequence tags (ESTs) by automatically running a collection of bioinformatics applications. In the first step, a quality check is performed and repeats, vector parts and low quality sequences are masked. Then successive steps of database searching and EST clustering are performed. Already known transcripts present within mRNA and genomic DNA reference databases are identified. Subsequently, tools for the clustering of anonymous ESTs, and for further database searches at the protein level, are applied. Finally, the outputs of each individual tool are gathered and the relevant results presented in a descriptive summary. ESTAnnotator was already successfully applied for the systematic identification and characterisation of novel human genes involved in cartilage/bone formation, growth, differentiation and homeostasis. ESTAnnotator is available at http://genome.dkfz-heidelberg.de, contact: genome@dkfz.de.
INTRODUCTION
Expressed sequence tags (EST) (1) are single pass sequence reads from randomly selected cDNA clones that sample the diversity of genes expressed by an organism or tissue. They provide a highly cost-effective method of accessing and identifying expressed genes. In order to find new and unknown clones in a cDNA library there has to be a high throughput analysis of thousands of clones via EST sequencing and annotation. Since EST sequences represent parts of cDNA sequences even a simple BLAST search against suitable databases can result in the correct annotation. In other cases it might be neccessary to assemble the EST with homologous EST sequences out of a database to reconstruct the mRNA. Several protocols have been published (2,3) dealing with this issue.
Our tool combines these two approaches by switching automatically from the BLAST identification to EST assembly if necessary. The W3H task system (4) allows the straightforward implementation of a combination of bioinformatics tools. The system regulates the dataflow and produces output files in XML format for parsing and automatic processing and in HTML format for facilitating a final visual inspection of the results. Furthermore, the task system allows the immediate integration of ESTAnnotator into the W2H web interface (5). This interface allows easy access to sequence databases in addition to analysis programs. It provides a secure place for the storage of sequences and results together with secure access through HTTPS [SSL (http://www.nyx.net/%7Elmulcahy/ssl.html) and SSH (ftp://ftp.cert.dfn.de/pub/tools/net/ssh/)].
A scientific consortium within the framework of the German Human Genome Project (http://www.dhgp.de) faced the problem of annotating 5000 EST sequences derived from a human fetal cartilage cDNA library (6). ESTAnnotator was developed and used successfully for facilitating the semiautomatic analysis of this EST data.
MATERIALS AND METHODS
Preparation of input EST sequences
ESTAnnotator can process ABI or MegaBACE sequencer files by using Phred (7) for converting the trace files to high-quality sequences. The Repeatmasker program (Smit, A.F.A. and Green, P., http://repeatmasker.genome.washington.edu/) was used with either the database for repetitive elements included in the program, or the UniVec database for vector sequences (ftp://ftp.ncbi.nih.gov/pub/univec) in order to mask the input sequences.
BLAST similarity analysis
The EST sequences were run against four different databases using the gapped BLAST program (8). Beside the default search with the non-redundant nucleic acid database (ftp://ftp.ncbi.nih.gov/blast/db) additional BLAST searching strategies can be implemented using our list of >200 databases. In the cartilage EST annotation project, additional searches were performed against the human genomic sequence contig assembly database (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/H_sapiens), the human mRNA database (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/H_sapiens/RNA) and the RefSeq database (ftp://ftp.ncbi.nih.gov/refseq/cummulative).
Clustering of EST sequences
The clustering was started with a BLASTN (8) search against an organism-specific EST database, for example the human EST sequences from the EMBL database (ftp://ftp.ebi.ac.uk/pub/databases/embl). Only hits with an default expectation value less than e−20 were included in the assembly. The program CAP (9) was run for the assembly of the homologous EST sequences using a minimum overlap of 20 bp and a minimum sequence identity in overlaps or containments of 85%. The resulting consensus sequence was used for a repetition of the BLAST search in order to assemble further overlapping EST sequences. The clustering procedure was finished either if no additional sequences were found or three rounds of BLAST searches with assembly had been performed.
BLAST analysis at the protein level
Further database searching at the protein level was performed using either the output consensus sequence from the EST clustering step or the original input EST sequence (if the clustering procedure did not yield any overlapping sequences). To this end, BLASTX (8) was run against the SWISS-PROT database (ftp://ftp.ebi.ac.uk/pub/databases/swissprot/), and TBLASTX searched against all EMBL ESTs (ftp://ftp.ebi.ac.uk/pub/databases/embl).
Implementation under the W3H task system
ESTAnnotator was implemented under the W3H task system (4). This framework allows the integration of applications and methods to create tailor-made analysis task flows which can be used in a high throughput analysis without the usual necessity of customised programming. In such a task system it is necessary to describe the program flow and dependency of applications, the data flow and the merging of the individual outputs into a common output report. The system stores the results of the different programs together with graphical outputs. The final output of the task is an XML file which contains all relevant information generated by the task. The XML information is transformed by means of W2H's post-processing mechanism into an HTML page for the task report using the Extensible Style-sheet Language Transformations (XSLT; http://www.w3.org/TR/xslt).
WWW access by the web interface to HUSAR (W2H)
Using the W3H task system allowed the immediate integration of ESTAnnotator into the W2H web interface (5). HUSAR itself is a commercial package and authentication of the user will be required. Login and password can be obtained upon registration on our homepage (http://genome.dkfz-heidelberg.de). Test accounts for up to 4 weeks are free, regular accounts are available for a yearly fee. Members of the German Human Genome Project can use ESTAnnotator free of charge. Since we provide user accounts together with disk space, the sequences and the results can safely stay on our server, accessible only by the user. The security issue is important to users analysing novel EST data which should be kept private.
RESULTS
Program flow and application dependencies
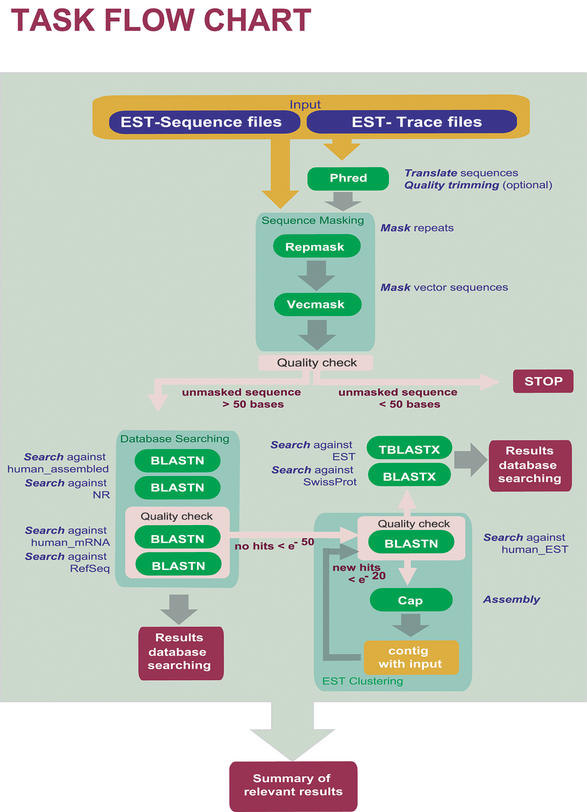
Figure 1 displays the flowchart of ESTAnnotator, showing program and data flow as well as the rules which are applied. ESTAnnotator can be divided into following subsections: preparation of quality-checked input EST sequences as well as the first quality check (shown at the upper part), the initial BLASTN analysis (shown at the left lower part) and the clustering of EST sequences as well as contig analysis (shown in the right lower part).
Figure 1.
Program flow in ESTAnnotator. Programs and rules used for high throughput annotation of ESTs.
There are three potential stopping points for the task: immediately after the preparation of input EST sequences, the next after BlastN analysis and the last (standard) exit after finishing all programs. At the first stop, sequences containing <50 readable bases after trimming and masking will be excluded from further processing, since no significant BLAST matches would be retrieved. The second stop, after the BLASTN analysis, indicates the successful annotation of an EST by finding corresponding matches in RefSeq or the human mRNA database using a default cutoff expectation value of <e−50 (adjustable by the user). The results for all BLAST hits will be summarised in the output. If no BLASTN hits above the cutoff value are found, the task automatically continues with the clustering of the EST sequences and with BLASTX analysis at the protein level. When finished, the summary will be presented. The different subsections of the ESTAnnotator tool will be described below, discussing the results from the human fetal cartilage EST project (6).
Preparation of input EST sequences
ESTAnnotator was designed for processing raw trace or sequence text files. Since EST sequences are single pass reads which may contain a substantial number of sequencing errors, the Phred base calling algorithm (7) can be used to trim the EST sequences according to the quality in the trace file. Since EST sequences are generated by complex cDNA cloning procedures, they are often contaminated with vector and linker sequences. For generating high-quality input EST sequences vector sequences and naturally occurring repetitive elements are masked by default. If too much sequence information is lost due to quality trimming or masking, no further analysis of the input sequence occurs and the task automatically stops. This was the case for ∼2.6% of the EST reads in the cartilage EST project. The average usable read length obtained exceeded 500 bp. The EST sequences were not pre-filtered for redundancy in order to quantify the transcripts and trap alternatively spliced genes.
BLASTN similarity analysis
Even with short EST sequences, it is possible in many cases to identify a corresponding known cDNA already present in the databases. Among the cartilage EST sequences 69.6% showed significant similarity to known genes in the human RefSeq collection, and another 4.8% were found to human model RNAs. Approximately 23% of the cartilage EST sequences could not be identified as known transcripts or proteins, but showed significant similarity to genomic regions and/or anonymous ESTs (6). A subset of these potentially novel gene sequences is currently under detailed experimental scrutiny for expression in cartilage tissue using RT–PCR, northern blotting and mRNA in situ hybridization.
Using the NCBI human assembly database a corresponding genomic location could be identified for >90% of the EST sequences. This information will be valuable in selecting possible candidate genes from regions of the human genome, to which diseases related to malformations of the skeleton have been mapped genetically.
Though the initial focus of ESTAnnotator was the identification of human EST sequences for the cartilage project, the variable selection of databases allows the user to encompass many different organisms.
Clustering and BLAST analysis at the protein level
The BLAST algorithm was used to find homologous, potentially overlapping EST sequences for clustering. A cutoff value of e−20 was chosen to remove spurious matches while still identifying the regions of similarity between the EST sequences, which represents a ∼50 bp region of similarity using human EST sequences. The consensus sequence of a cluster assembled by CAP containing the input EST sequence is saved as a consensus contig sequence. As the clustering may turn out to be quite time consuming, it can be switched off by the user and it was rarely used in the cartilage project. Comparing the resulting consensus sequence or the input sequence against SWISS-PROT using BLASTX to check for an annotated protein or all ESTs using TBLASTX to detect homologous EST sequences of other organisms were the final steps for retrieving annotation.
The ESTAnnotator report
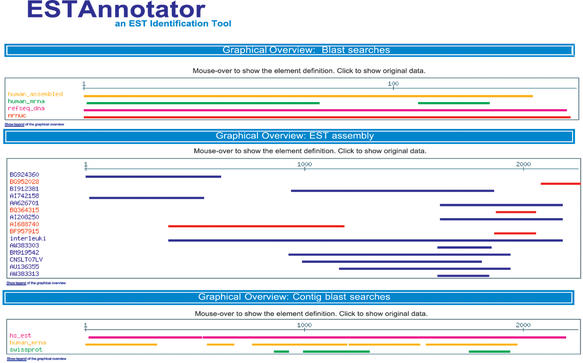
The final ESTAnnotator report is an HTML page that displays the database id and the description line of the top three hits of the BLAST search results if their expectation value is <0.01. Additionally a link to the original BLAST output is provided. To illustrate the position of the BLAST hits and the clustered EST sequences, corresponding graphical outputs are displayed in the lower part (Fig. 2). The clustered sequences are color-coded in blue and red depending if the plus or minus strand was used in the assembly. All hits above an expectation value of 0.01 are summed up in the BLAST graphical outputs. The alignment information can be accessed by clicking on the hits within the graphical output. By downloading the XML report file from the server, the BLAST results (expectation value and description line) for each EST sequence can easily be parsed into a database file.
Figure 2.
Graphical output of ESTAnnotator. The results of the BLAST similarity searches, the clustering and the BLAST results using the consensus contig sequence are displayed.
Performance
In the fetal cartilage project the sequences or trace files were processed in batches of 50–100 and each batch took about 1–2 days to be completed when run on our SUN Enterprise with six processors depending on the overall machine load. The XML report files were downloaded and the data transferred to a local database.
DISCUSSION
ESTAnnotator first performs a quality check on EST sequence reads and then uses BLAST searches against different nucleotide databases in order to annotate EST sequences. If this initial search step does not identify a known gene or mRNA, an EST clustering step with subsequent translated BLAST searches tries to find matches to related annotated proteins or nucleic acid sequences. The combination of both steps according to certain rules speeds up the medium to large-scale EST annotation by avoiding time consuming clustering after a successful primary annotation by BLAST or manual intervention and re-inputting of sequences to a separate EST clustering tool. To our knowledge, ESTAnnotator is the first publicly available tool for such an automated EST analysis. It was shown that ESTAnnotator successfully led to an immediate bioinformatical identification of ∼75% of 5000 EST sequences originating from a human fetal cartilage cDNA library (6).
The default databases in ESTAnnotator were chosen to best suit the characterisation of human ESTs. Currently, three databases in the BLAST analysis part as well as the EST database for clustering can be chosen freely from our set of >200 up-to-date local databases. In the near future, we will implement an organism parameter, which automatically switches the databases used by ESTAnnotator to fit the special requirements of model organisms.
Efforts to assemble and annotate raw DNA sequence data have been developed employing clustering of ESTs as in STACKdb (10) or in Unigene (11). Getting high similarity matches with Unigene or STACKdb sequences could also be used to gain the full mRNA sequence from an EST, in many cases possibly replacing the time consuming clustering step.
The implementation in the W2H environment enables the users to securely transfer sequences to our server using HTTPS or SSH, start ESTAnnotator and then log out. The process can be checked or the results can be retrieved by logging in again later. Sequences and results are always in a space belonging to he user and cannot be viewed publicly. The users data is kept strictly private, but the user still has the advantage of using the tool through the web without having to administer software and update databases locally.
Acknowledgments
ACKNOWLEDGEMENTS
T.H. and E.R.S. gratefully acknowledge financial support from BMBF/German Human Genome Project grant 01KW9984 and wish to thank Dr B. Lee (Baylor College of Medicine, Houston, TX) for construction of the human fetal cartilage cDNA library and the laboratories of Professor Dr B. Zabel (Mainz) and Professor Dr A. Winterpacht (Erlangen) for fruitful cooperation. Preparation of the manuscript was aided by Christopher Previti.
REFERENCES
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1661. [DOI] [PubMed] [Google Scholar]
- 2.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res., 28, 3657–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayoubi P., Jin,X., Leite,S., Liu,X., Martajaja,J., Abduraham,A., Wan,Q., Yan,W., Misawa,E. and Prade,R.A. (2002) PipeOnline 2.0: automated EST processing and functional data sorting. Nucleic Acids Res., 30, 4761–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst P., Glatting,K.H. and Suhai,S. (2003) A task framework for the web interface W2H. Bioinformatics, 19, 278–282. [DOI] [PubMed] [Google Scholar]
- 5.Senger M., Flores,T., Glatting,K., Ernst,P., Hotz-Wagenblatt,A. and Suhai,S. (1998) W2H: WWW interface to the GCG sequence analysis package. Bioinformatics, 14, 452–457. [DOI] [PubMed] [Google Scholar]
- 6.Zabel B., Schlaubitz,S., Stelzer,C., Luft,F., Schmidt,E.R., Hankeln,T., Hermanns,P., Lee,B., Jakob,F., Noeth,U., Mohrmann,G., Tagariello,A. and Winterpacht,A. (2002) Molecular identification of genes and pathways involved in skeletogenesis by EST sequence analysis and microarray expression profiling of human mesenchymal stem cell differentiation. Abstract 13th Ann. Meeting German Soc. Hum. Genetic., Leipzig. Medizinische Genetik, 14, 245. [Google Scholar]
- 7.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X. (1992) A contig assembly program based on sensitive detection of fragment overlaps. Genomics, 14, 18–25. [DOI] [PubMed] [Google Scholar]
- 10.Christoffels A., van Gelder,A., Greyling,G., Miller,R., Hide,T. and Hide,W. (2001) STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res., 29, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou D., Zhao,W.D., Wright,F.A., Yang,H.Y., Wang,J.P., Sears,R., Baer,T., Kwon,D.H., Gordon,D., Gibbs,S. et al. (2001) Assembly, annotation and integration of UNIGENE clusters into the human genome draft. Genome Res., 11, 904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]