Abstract
Background
Although European Borrelia burgdorferi sensu lato isolates have been divided into five genospecies, specific tools for the serotype characterization of only three genospecies are available. Monoclonals antibodies (mAbs) H3TS, D6 and I17.3 identify B. burgdorferi sensu stricto (ss.), B. garinii and B. afzelii respectively, but no mAbs are available to identify B. valaisiana. In the same way, specific primers exist to amplify the OspA gene of B. burgdorferi ss., B. garinii and B. afzelii. The aim of the study was to develop species-specific mAb and PCR primers for the phenotypic and genetic identification of B. valaisiana.
Results
This study describes a mAb that targets OspA of B. valaisiana and primers targeting the OspA gene of this species. As the monoclonal antibody A116k did not react with strains NE231, M7, M53 and Frank and no amplification was observed with strains NE231, M7 and M53, the existence of two subgroups among European B. valaisiana species was confirmed.
Conclusions
The association of both monoclonal antibody A116k and primers Bval 1F and Bval 1R allows to specific identification of the B. valaisiana isolates belonging to subgroup 1.
Background
Lyme borreliosis (LB) caused by a spirochetal species Borrelia burgdorferi sensu lato (sl.) is a multisystemic illness distributed worldwide [1]. In North America and Eurasia, LB is the most common tick-borne disease [2]. On the basis of protein profiles, DNA-DNA relatedness and rRNA gene restriction pattern, B. burgdorferi sl. strains have been previously divided into three species: B. burgdorferi sensu stricto (ss.), B. garinii and B. afzelii [3,4]. B. burgdorferi ss. was found in ticks and patients infected in Europe as well as in North America, whereas B. garinii and B. afzelii were only found in ticks and patients infected in Eurasia [4,5]. B. andersonii and B. bissettii were described in North America [6,7], while B. japonica has been defined in Japan [8] and B. lusitaniae in Portugal [9].
First isolated in Valais-CH, from an I. ricinus tick, VS116 was characterized as B. valaisiana novel species [10]. As other species, it appeared that its geographical distribution was not uniform. In Switzerland, Scandinavia and the Netherlands, B. valaisiana was less frequently isolated than B. garinii and B. afzelii. In Ireland however, B. valaisiana was described as the most prevalent genospecies [11]. More recently, B. valaisiana has been isolated in Japan and East Asia from I. nipponensis, I. columnae and I. granulatus [12,13].
An association between clinical outcome and infection by strains of different genospecies has been suggested and suspicion of a pathogenic potential of B. valaisiana in humans was described [14,15]. Therefore, genospecies classification may increase the sensitivity of serologic and genetic analysis by appropriate choice of target antigens. Electrophoretic mobility of the major proteins allows us to distinguish most Borrelia genospecies, but monoclonal antibodies (mAb) are used to formally identify species on immunoblots. Up to now, only B. burgdorferi ss., B. garinii and B. afzelii among European isolates could be characterized by mAbs, H3TS was directed against OspA of B burgdorferi ss., D6 directed against a 12 kDa protein of B. garinii and I17.3 reacted with OspB of B. afzelii [16]; specific primers GI, GII and GIII were directed against ospA gene of Borrelia burgdorferi ss, B. garinii and B. afzelii, respectively [17]. In this study, the major outer surface protein OspA of the reference strain VS116 was selected as antigen for mAbs production, because of its particular protein profile in SDS-page and its potential discriminative utility. In the same way, we have designed primers specific to ospA gene of B. valaisiana. We described a mAb and primers specific to the major group of European B. valaisiana.
Methods
Bacterial strains
The Borrelia burgdorferi sl. strains used in this study are listed in Table 1 [see Additional file 1].
All Borrelia isolates were grown in MKP medium [30] at 33°C. Extraction of DNA was performed as described previously [16]. B. burgdorferi sl. isolates were all characterized by RFLP of amplicons digested by Mse I (Boehringer, Mannheim, Germany) and Dra I (Life technology, CH) as described by Postic [18]. All the Borrelia isolates were submitted to reactivity of mAb H3TS, I 17.3, D6 and A116k [16]
B. turicatae, B. parkeri, B. coriaceae, B. anserina were kindly provided by T. Schwan (Rocky Mount. Lab, Hamilton, MT, USA). Other bacteria isolates listed on Table 2 were cultured and characterized by usual laboratory techniques in our department of bacteriology.
Table 2.
Bacteria, other than Borrelia burgdorferi sensu lato, evaluated in this study
| Bacterial species | Reactivity with mAb A116k | PCR with primers to OspA |
| Borrelia anserina | Neg | Neg |
| B. coriaceae | Neg | Neg |
| B. hermsii | Neg | Neg |
| B. parkeri | Neg | Neg |
| B. turicata | Neg | Neg |
| Aeromonas hydrophila | Neg | Neg |
| Candida albicans | Neg | Neg |
| Candida krusei | Neg | Neg |
| Escherichia coli | Neg | Neg |
| Klebsiella pneumoniae | Neg | Neg |
| Pseudomonas aeruginosa | Neg | Neg |
| Salmonella enteritidis | Neg | Neg |
| Staphyloccocus aureus | positive with 70 kDa and 75.3 kDa proteins | Neg |
| Staphyloccocus epidermidis | Neg | Neg |
ELISA for screening of hybridoma supernatants
Low-passages, whole-cell of VS215 isolate of B. burgdorferi ss., VS102 of B. garinii, VS461 of B. afzelii and VS116 of B. valaisiana were harvested in the log-phase growth, centrifuged and the resulting cell pellet was rinsed twice with PBS (Phosphate-Buffered Saline, pH 7.2, 0.1 M) with MgCl2 5 mM. The cells were suspended in distilled water. Concentrations were equally adjusted to 10 μg/ml and the four antigens were heated 5 min with 5% SDS at 37°C then 15 min at 95°C. The microwells of the ELISA plates were coated with the four strains in Na2CO3, 0.1 M, pH 9.6 at 4°C overnight. The wells were washed four times in PBS-T (PBS with 0.01% Tween) and blocked for 1 hour with BSA 1% in PBS and again washed four times. Supernatants of hybridomas were diluted 1/3 in PBS-T. Plates were incubated at 37°C for 1 hour and then washed four times with PBS-T. The conjugate (goat anti-mouse polyvalent conjugated to alkaline phosphatase (Sigma)) diluted 1/1000 in PBS-T was added and incubated at 37°C for 2 hours. After four washing steps, substrate (para-Nitrophenylphosphate, Virion, Rüschlikon, Switzerland) was added and plate incubated at room temperature for 15 min. ODs were read by a microplate reader at 405 nm (with reference OD at 620 nm).
Development of B.valaisiana specific monoclonal antibodies
The strain VS116 was used to produce mAbs specific to B. valaisiana. 200 μg of proteins were separated on a 12% Sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gel was cut into three parts. Both external parts were stained with Coomassie blue and used as references to cut the OspA band from the medium part of the gel. The OspA portion was thereafter used for immunization and the rest of the gel stained to confirm the extraction of the selected protein.
The immunization kit of ISL (Readysysteme, Zursach, AG-CH) was used. An estimated fraction of 20 μg proteins of a 12% gel containing the OspA band was incubated 48 h with monolayer of support cells provided with the kit. One mouse was sacrificed to extract splenocytes. The splenocytes were incubated four days in immunization medium with 20 μg of fresh antigen and pre-primed support cells.
Immunogen splenocyte suspension was then prepared and mixed with myeloma cells provided with the kit (ratio 1:10 with the splenocytes). The cells were fused with polyethylen glycol in HAT (Hypoxanthine, Aminopterin, Thymidine) medium and the resultant hybridomas were seeded into multi-well plates.
Four days later, supernatants were screened for antibody production by ELISA as described above and specificity of positive supernatants was then confirmed by using an immunoblot assay with VS215 (B. burgdorferi ss.), VS102 (B. garinii), VS 461 (B. afzelii) and VS116 (B. valaisiana) as antigens [15]. The hybridomas which produced specific mAbs reactive to B. valaisiana were cloned by the limiting dilution technique.
Subtyping of mAbs was performed by immunoblot with peroxidase-conjugated rabbit antibodies specific for different isotypes of mouse immunoglobulins (Pierce, Rockford IL, USA)
PCR assay
Based on the comparison of ospA sequences, the following oligonucleotide primers specific to B. valaisiana were used in this study : Bval 1F 5'-TGCTGAAAATGCTACAAAAGCAGT-3' (forward primer) and Bval 1R 5'-CAAGACAAAACTTGTATTTACAAAAC-3' (reverse primer). Forward and reverse primers are located on the sequence of the ospA gene of B. valaisiana VS116 isolate at nucleotide position 453 to 476 and 696 to 721, respectively. PCR assays were performed with a Perkin Elmer 9600 thermocycler. Taq DNA polymerase, the four deoxynucleotide triphosphates and the reaction buffer were used as recommended by the supplier (Qiagen, Basel, Switzerland). Twenty pmol of each primer were used per 50 μl reaction mixture. About 100 ng of total genomic DNA was amplified for 40 cycles under the following conditions : 94°C, 3 min.; 40 cycles (60°C for 1 min, 72°C for 1 min, 94°C for 1 min) 60°C for 1 min, 72°C for 7 min, 4°C. PCR amplification products were resolved onto 1.0 % agarose electrophoresis gels and visualised under U.V. light using ethidium bromide. Physical containment measures warranted the absence of DNA contamination in the PCR procedure.
Computer analysis, nucleotide sequence and accession numbers
Sequence alignments was done with EMMA of the EMBOSS (European Molecular Biology Open Software Suite, http://www.hgmp.mrc.ac.uk/Software/EMBOSS/). EMMA calculates the multiple alignment of nucleic acid or protein sequences according to the method of Thompson et al [31]. The default parameter values were used.
The ospA sequences have been retrieved from Genbank with accession numbers Y10837, Y10838, AF095947, AF095943, Y10841, AF095942, Y10840, X68059, X85442, X14407, S88693, Y10897, Y10893, Y10892, X62387, Z29087, Y10896, X85441, Z29086, S48323, X80256, and X80254 for Borrelia strains PotiB1, PotiB2, M53, M7, UK, AR-2, VS116, 19857, 297, B31, 25015, DN127, HO14, IKA2, Pgau, VS461, VS100, WABSou, G25, Pbi, PBr, and T25, respectively. The ospA sequence of the Borrelia strain Ip90 has been published [32].
The nucleotide sequence of ospA gene from strain NE231 was determined by the dideoxy chain termination technique with the "Prism ready reaction dye deoxy-terminator cycle sequencing" kit (Applied Biosystem). The ospA sequence of NE231 has been entered in the EMBL/Genbank databases with accession number AJ249467.
Results and Discussion
The characterization of B. valaisiana was usually proceeded by rRNA gene restriction pattern after digestion by EcoRI and HindIII which contain three and four DNA fragments respectively [10], by RFLP of amplicons resulting from intergenic spacer of 23S-5S rRNA which contains four DNA fragments [18] or by hybridization on amplicons of 16s RNA gene [14]. Species specific primers designed on 16s RNA were described including Borrelia burgdorferi ss, B. garinii, B. afzelii and B. valaisiana [19]. The primers we designed in the OspA gene, as well as the monoclonal antibody evaluated here allowed us to specifically identified the major European group of B. valaisiana, as previously described as group I [20].
Phenotypic [16,21,22] and genetic [23] analyses of European isolates of B. burgdorferi sl. have demonstrated antigenic and structural heterogeneities of the OspA protein. B. burgdorferi ss. as exemplified by VS215 produced two major proteins at 31 kDa (OspA) and 34 kDa (OspB); B. garinii, isolate VS102, produced a single predominant OspA protein at 33 kDa and B. afzelii as the reference strain VS461 exhibited two major proteins OspA and OspB at 32 kDa and 35 kDa, respectively. Although the isolate VS116, B. valaisiana reference strain, had first been described as a new group, based on the OspA at 33.5 kDa [21], additional isolates of B. valaisiana showed more variability. The major outer surface membrane protein OspA of B. valaisiana were therefore in the range of 30 to 36 kDa [10,12]. Similarly, B. lusitaniae isolates were phenotypically heterogeneous within species. Such variability prevents a phenotypic characterization of this two species without specific markers such as mAbs. Although all B. valaisiana strains presented a dominant protein band of 32 to 34 kDa, none of them showed a reaction with OspA specific mAbs: H3TS, LA26 and LA31 as already described [10].
In this study five supernatants screened for antibody production by ELISA presented a preferential reactivity to B. valaisiana. The specificity was nevertheless confirmed on immunoblot by using total protein extracts from B. burgdorferi ss., B. garinii, B. afzelii and B. valaisiana. All the five mAbs produced were IgA specific to B. valaisiana and reacted with OspA of this species. Because of its strongest reaction with VS116 strain of B. valaisiana, A116k mAb was selected for further characterization.
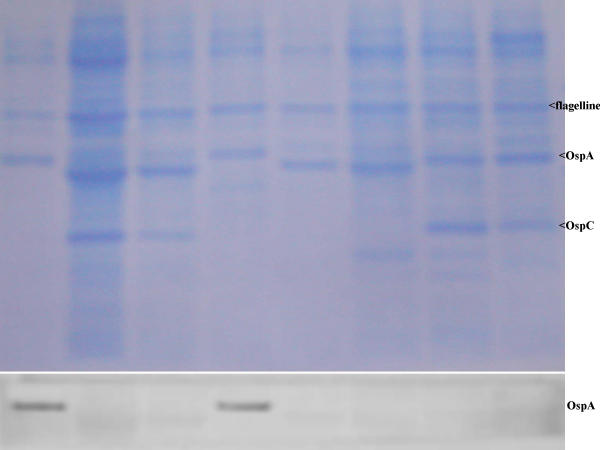
Sensitivity and specificity of A116k mAb was evaluated on immunoblot by testing following controls. A116k mAb did not bind to any protein of the 90 other B. burgdorferi sl. isolates belonging to B. burgdorferi ss. (n = 25), B. garinii (n = 25), B. afzelii (n = 25), B. lusitaniae (n = 5), B. japonica (n = 5), B. andersonii (n = 2) and B. bissettii (n = 3) showing 100% of specificity. Furthermore an immunoblot was performed with B. turicatae, B. parkeri, B. coriaceae, B. anserina and 12 unrelated bacterial species (Table 2). A116k mAb did not bind to any of them excepted with two proteins (70 kDa and 75.3 kDa) of Staphylococcus aureus. The reactions were nevertheless much weaker than with the OspA of B. valaisiana isolates accounting for highly probable non-specific cross-reaction. Twenty among 24 B. valaisiana isolates had been easily identified with A116k mAb. However, four isolates (NE231, M7, M53 and Frank) of B. valaisiana presented no reactivity with A116k mAb. Therefore, the sensitivity reached 83%. The OspA of these four strains presented a molecular weight lower than the reference strain VS116 (Figure 1) [22].
Figure 1.
(top) Protein profiles of B. burgdorferi sensu lato isolates after polyacrylamide gel electrophoresis and Western blot on polyvinylidene difluoride membrane stained with Coomassie blue. Lanes 1 to 8 B. valaisiana (VS116, NE231, Frank, UK, M7), B. burgdorferi sensu stricto (B31), B. garinii [20047], B. afzelii (VS461). (bottom) Area of the OspA Reactivity with monoclonal antibody A116k.
In order to improve the detection of strains belonging to B. valaisiana species, comparison of ospA gene sequences from isolates belonging to all known B. burgdorferi sensu lato species was performed and led us to the identification of oligonucleotide primers that were able to amplify a 269 bp DNA fragment specific to B. valaisiana. Forward and reverse primers were located on the sequence of the ospA gene of B. valaisiana VS116 isolate at nucleotide position 453 to 476 and 696 to 721, respectively.
The detection threshold of this PCR assay was determined by performing amplification reactions on a controlled number of cells. A stock sample of B. valaisiana strain VS116 was serially diluted in phosphate saline buffer. The number of Borrelia cells (± 2.0 × 107 cells/ml) in the stock sample was determined by dark-field microscopy, and aliquots of the dilution were tested for amplification using these primers. It was found that a template DNA input corresponding to about 5 spirochetes was sufficient to detect the expected amplified fragment (data not shown). No signal was observed when template DNA was omitted from the amplification reaction.
The specificity of the PCR assay was evaluated on the 90 B. burgdorferi sensu lato strains and 14 unrelated bacterial species (Table 1 [see Additional file 1] and Table 2), previously tested by using A116k mAb. Three B. garinii (SO1, BITS, and VS711) and one B. lusitaniae (BR41) strains were positive. The specificity was therefore 97 %. None of the 14 unrelated bacteria showed an amplified product with these primers. Twenty-one out of the 24 B. valaisiana strains had an amplified product at the expected size (Table 1 [see Additional file 1]). The strains NE231, M7 and M53 were not amplified with these primers.
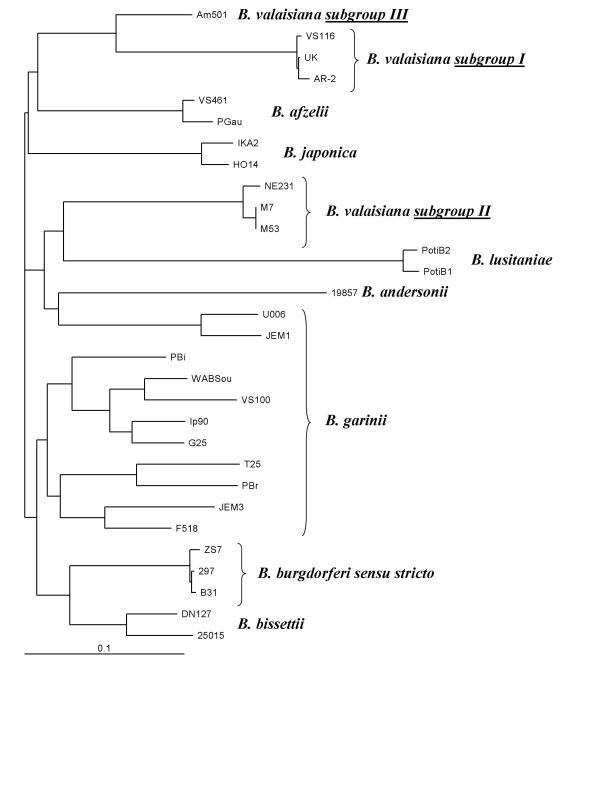
Comparison and alignment of the NE231 OspA deduced amino acid sequence with those of 28 B. burgdorferi sensu lato strains was performed. Based on this comparison, the 6 analysed B. valaisiana strains (UK, AR-2, VS116, M53, M7 and NE231) confirmed the existence of 2 subgroups (Figure 2). The subgroup I consists of three B. valaisiana isolates (UK, AR-2 and VS116). The amino acid sequences of OspA from these isolates were 98.9 % to 99.6 % homologous (Table 3 [see Additional file 2]). Subgroup II contains three B. valaisiana isolates (M53, M7 and NE231). OspA sequences of M7 and M53 were identical. These two sequences had a homology of 98.5 % (4 amino acid substitutions) with the OspA sequence of strain NE231. The average identity of the deduced amino acid sequences of the OspA proteins between subgroup I and II was about 69.9 %, and between B. valaisiana and representative isolates from other B. burgdorferi sensu lato species varied from 62.1 % to 75 %. These data were concordant with the two subgroups described by Wang [20], whose results were based on the OspA protein sequence of eight B. valaisiana isolates (subgroup I: VS116, M19, M49, M52, AR-2 and UK-c16; subgroup II: M7 and M53). In addition, a similar divergence has already been suggested by Masuzawa [12]. His hypothesis was based on molecular masses of the OspA and on sequences of the ospA gene of Asian strain Am501 compared to that of the reference strain VS116. However, a BLAST alignment of the Am501 OspA deduced amino acid sequence with that of M53 OspA revealed only 68% identities (data not shown). Taken together, these results could mean that some Asian B. valaisiana belong to a third group (Figure 2), distinct of group I (VS116) and group II (M53).
Figure 2.
Dendrogram of B. burgdorferi sensu lato isolates (n = 30). The B. valaisiana isolates clustered into 3 separated branches confirming the results previously published by Wang [20] and Masuzawa [12].
The heterogeneity of B. valaisiana species has been previously evidenced by several authors and on diverse targets such as rrf-rrl spacer, rrs, fla and OspC genes but was not revealed by using standard genotyping methods such as ribotyping [10], RAPD [24] and RFLP [25]. The heterogeneity of the ospA gene sequences was previously reported within the B. garinii species [17,23,25]. Our data, as well as those from previous publications [12,20] indicated that such major ospA gene differences also occur within another Borrelia genospecies. The ospA gene encodes an outer membrane lipoprotein that is differentially expressed by B. burgdorferi sensu lato in ticks and in mammals [26]. Although the role of the OspA protein is not fully understood, OspA may be important in the bacterium-host interaction. Studies have indicated that OspA protein possesses B-cell mitogenic and cytokine-stimulatory properties [27,28]. It seems also that OspA may play a role in the adherence of B. burgdorferi sensu lato to different eucaryotic cells [29]. Taking into account all these features, the heterogeneity observed within the OspA protein sequences could be a key factor in tissue specificity or in the infection of specific hosts.
Conclusion
The existence of two subgroups among European B. valaisiana species was confirmed, as well as the existence of a third group of B. valaisiana in Asia, as it has already been described. The heterogeneity was first observed on protein profile as the OspA band of NE231, M7, M53 and Frank presented a molecular weight lower than the reference strain VS116 [22]. In addition, Wang [20] described the presence of two subgroups based on the OspA protein sequence of eight strains. One monoclonal antibody and PCR primers presented in this study are therefore useful serologic and genetic tools to classify strains of B. valaisiana group I. The lack of reactivity of strain Frank with monoclonal antibody A116k was not concordant with the result of the PCR-amplification of ospA gene. Therefore, further analyses have to be done. The sequence analysis of the complete OspA gene should explain this contradiction.
List of abreviations
Borrelia burgdorferi ss:. Borrelia burgdorferi sensu stricto
Borrelia burgdorferi sl.: Borrelia burgdorferi sensu lato
OspA: outer surface protein A
MAb: monoclonal antibody
PBS: phosphate buffer saline
RFLP: restriction fragment length polymorphism
RAPD: randomly amplified polymorphic DNA
kDa: kilo-Dalton
SDS: Sodium-dodecyl-sulfate
HAT: Hypoxanthine, Aminopterin, Thymidine
OL: our laboratory
Competing interests
None declared.
Authors'contributions
Karine Ryffel has performed all the experimental parts of the study and wrote the first draft of the manuscript.
Eric Dayer has helped in preparing and advising for the monoclonal antibody production
Anne-Gabrielle Bretz has cultured and prepared all the Borrelia isolates used in the study
Edmond Godfroid has designed the primers used for PCR, performed with his team the sequencing of the OspA genes
Olivier Péter, I has designed the whole study on Borrelia valaisiana, advising for the serological analysis (ELISA and Immunoblots), have writen the intermediate and final drafts of the manuscript.
Acknowledgments
This research was supported by the Swiss National Science Foundation (No 32-52739.97), the Valais Foundation for applied research in Valais hospital medicine. This work is part of the PhD dissertation of Karine Ryffel.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table 1 : NAME : A mabPCRTab1.doc (list of Borrelia used in the study)
Table 3 : NAME : A mabPCRTab3.xls (amino acid identity matrix…)
Contributor Information
Karine Ryffel, Email: Karine_Ryffel@hotmail.com.
Olivier Péter, Email: olivier.peter@consilia-sa.ch.
Eric Dayer, Email: eric.dayer@consilia-sa.ch.
Anne-Gabrielle Bretz, Email: olivier.peter@consilias-sa.ch.
Edmond Godfroid, Email: Edmond.Godfroid@ulb.ac.be.
References
- Steere AC. Lyme disease [see comments] N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- Bennett CE. Ticks and Lyme disease. Adv Parasitol. 1995;36:343–405. doi: 10.1016/s0065-308x(08)60494-7. [DOI] [PubMed] [Google Scholar]
- Canica MM, Nato F, du Merle L, Mazie JC, Baranton G, Postic D. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti JC, Assous M, et al. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- van Dam AP, Kuiper H, Vos K, Widjojokusumo A, de Jongh BM, Spanjaard L, et al. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis. 1993;17:708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- Marconi RT, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic D, Ras NM, Lane RS, Hendson M, Baranton G. Expanded diversity among Californian borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127) J Clin Microbiol. 1998;36:3497–3504. doi: 10.1128/jcm.36.12.3497-3504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- Le Flèche A, Postic D, Girardet K, Péter O, Baranton G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol. 1997;47:921–925. doi: 10.1099/00207713-47-4-921. [DOI] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Le Flèche A, Postic D, Péter O, Baranton G, et al. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19) Int J Syst Bacteriol. 1997;47:926–932. doi: 10.1099/00207713-47-4-926. [DOI] [PubMed] [Google Scholar]
- Kirstein F, Rijpkema S, Molkenboer M, Gray JS. The distribution and prevalence of B. burgdorferi genomospecies in Ixodes ricinus ticks in Ireland. Eur J Epidemiol. 1997;13:67–72. doi: 10.1023/A:1007360422975. [DOI] [PubMed] [Google Scholar]
- Masuzawa T, Fukui T, Miyake M, Oh HB, Cho MK, Chang WH, et al. Determination of members of a Borrelia afzelii-related group isolated from Ixodes nipponensis in Korea as Borrelia valaisiana. Int J Syst Bacteriol. 1999;49:1409–1415. doi: 10.1099/00207713-49-4-1409. [DOI] [PubMed] [Google Scholar]
- Masuzawa T, Takada N, Imai Y, Yanagihara Y. Molecular epidemiology of Borrelia burgdorferi sensu lato in the East Asia. In VIII International Conference on Lyme borreliosis and other tick-borne diseases. 1999;Abstr O5:5. June 20–24 1999 Munich. [Google Scholar]
- Rijpkema SG, Tazelaar DJ, Molkenboer M, Noordhoek GT, Plantinga G, Schouls LM, et al. Detection of Borrelia afzelii, Borrelia burgdorferi sensu stricto, Borrelia garinii. Clinical Microbiology and Infection. 1997;3:109–116. doi: 10.1111/j.1469-0691.1997.tb00259.x. [DOI] [PubMed] [Google Scholar]
- Ryffel K, Péter O, Rutti B, Suard A, Dayer E. Scored antibody reactivity determined by immunoblotting shows an association between clinical manifestations and presence of Borrelia burgdorferi sensu stricto, B. garinii, B. afzelii, and B. valaisiana in humans. J Clin Microbiol. 1999;37:4086–4092. doi: 10.1128/jcm.37.12.4086-4092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz AG, Ryffel K, Hutter P, Dayer E, Péter O. Specificities and Sensitivities of Four Monoclonal Antibodies for Typing of Borrelia burgdorferi Sensu Lato Isolates. Clin Diagn Lab Immunol. 2001;8:376–384. doi: 10.1128/CDLI.8.2.376-384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfroid E, Ben Messaoud A, Poliszczak A, Lobet Y, Bollen A. Assignment of Borrelia burgdorferi strains G25 and VS461 to the Borrelia garinii and Borrelia afzelii genospecies, respectively: a comparison of OspA protein sequences. DNA Seq. 1995;5:251–254. doi: 10.3109/10425179509030975. [DOI] [PubMed] [Google Scholar]
- Postic D, Assous MV, Grimont PA, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Sohns B, Bautsch W. Detection and typing of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks attached to human skin by PCR. J Clin Microbiol. 1998;36:3355–3358. doi: 10.1128/jcm.36.11.3355-3358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Dankert J. Two distinct ospA genes among Borrelia valaisiana strains. Res Microbiol. 2000;151:325–331. doi: 10.1016/S0923-2508(00)00155-8. [DOI] [PubMed] [Google Scholar]
- Péter O, Bretz AG. Polymorphism of outer surface proteins of Borrelia burgdorferi as a tool for classification. Zentralbl Bakteriol. 1992;277:28–33. doi: 10.1016/s0934-8840(11)80867-4. [DOI] [PubMed] [Google Scholar]
- Ryffel K, Péter O. From the discovery to clinical implications: Borrelia valaisiana . J Spiroch Tick-borne Dis. 2001;7:64–72. [Google Scholar]
- Wilske B, Preac-Mursic V, Gobel UB, Graf B, Jauris S, Soutschek E, et al. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J Clin Microbiol. 1993;31:340–350. doi: 10.1128/jcm.31.2.340-350.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Spanjaard L, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J Clin Microbiol. 1998;36:768–776. doi: 10.1128/jcm.36.3.768-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema SG, Herbes RG, Verbeek-De Kruif N, Schellekens JF. Detection of four species of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected from roe deer (Capreolus capreolus) in The Netherlands. Epidemiol Infect. 1996;117:563–566. doi: 10.1017/s0950268800059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva AM, Fikrig E. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J Clin Invest. 1997;99:377–379. doi: 10.1172/JCI119169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten RM, Modur VR, McIntyre TM, Weis JJ. Borrelia burgdorferi outer membrane protein A induces nuclear translocation of nuclear factor-kappa B and inflammatory activation in human endothelial cells. J Immunol. 1996;157:4584–4590. [PubMed] [Google Scholar]
- Potter MR, Rittling SR, Denhardt DT, Roper RJ, Weis JH, Teuscher C, et al. Role of osteopontin in murine Lyme arthritis and host defense against Borrelia burgdorferi. Infect Immun. 2002;70:1372–1381. doi: 10.1128/IAI.70.3.1372-1381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, de Silva AM, Montgomery RR, Fish D, Anguita J, Anderson JF, et al. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J Clin Invest. 2000;106:561–569. doi: 10.1172/JCI9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preac-Mursic V, Wilske B, Schierz G. European Borrelia burgdorferi isolated from humans and ticks culture conditions and antibiotic susceptibility. Zentralbl Bakteriol Mikrobiol Hyg [A] 1986;263:112–118. doi: 10.1016/s0176-6724(86)80110-9. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. W Clustal: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson M, Noppa L, Barbour AG, Bergstrom S. Heterogeneity of outer membrane proteins in Borrelia burgdorferi : comparison of osp operons of three isolates of different geographic origins. Infect Immun. 1992;60:1845–1853. doi: 10.1128/iai.60.5.1845-1853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 : NAME : A mabPCRTab1.doc (list of Borrelia used in the study)
Table 3 : NAME : A mabPCRTab3.xls (amino acid identity matrix…)