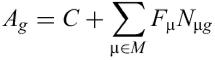Abstract
REDUCE is a motif-based regression method for microarray analysis. The only required inputs are (i) a single genome-wide set of absolute or relative mRNA abundances and (ii) the DNA sequence of the regulatory region associated with each gene that is probed. Currently supported organisms are yeast, worm and fly; it is an open question whether in its current incarnation our approach can be used for mouse or human. REDUCE uses unbiased statistics to identify oligonucleotide motifs whose occurrence in the regulatory region of a gene correlates with the level of mRNA expression. Regression analysis is used to infer the activity of the transcriptional module associated with each motif. REDUCE is available online at http://bussemaker.bio.columbia.edu/reduce/. This web site provides functionality for the upload and management of microarray data. REDUCE analysis results can be viewed and downloaded, and optionally be shared with other users or made publicly accessible.
BACKGROUND
In recent years, DNA microarrays have become a popular experimental tool for monitoring the mRNA transcript abundance for all genes in a cell simultaneously (1,2). The DNA sequence of the non-coding part of the genome contains a variety of regulatory signals that together are responsible for the differential regulation of gene expression. Through the combined analysis of transcriptome and genome, significant advances can therefore be made in understanding the molecular mechanisms underlying transcription control (3,4). A widely used strategy first clusters genes based on their expression profile across multiple conditions (5) and then searches for over-represented DNA motifs in the upstream regions of each gene cluster (6–10). One problem with the clustering approach to motif discovery, however, is that it requires distinct microarray experiments to be combined in an ad hoc fashion. Another problem is that it groups genes into disjoint subsets, while a promoter region usually has multiple transcription factor binding sites and receives input from a number of different signaling cascades (11).
Our laboratory has pioneered the motif-based regression analysis of a single transcriptome. Implemented as the algorithm REDUCE—an acronym that stands for regulatory element detection using correlation with expression—our method naturally takes into account the combinatorial nature of gene expression regulation and provides context-specific information about transcription factor activities (12). REDUCE works by fitting a multivariate predictive model to a single genome-wide expression pattern. The expression level of a gene is modeled as a sum of independent contributions from all transcription factors for which binding sites occur in the promoter region. A forward parameter selection strategy is used to select motifs from a large set of candidate motifs (e.g. all oligonucleotides up to a given length).
We have used REDUCE to analyze microarray data for mRNA expression in Saccharomyces cerevisiae (12,13) and Drosophila melanogaster (14). We have also successfully applied REDUCE to analyze genome-wide DNA–protein interaction data in Drosophila (14,15). Our motif-centered regression approach has been adopted and extended by other groups (16–18).
USERS AND DATA MANAGEMENT
Users must first register for an account via a fully automated procedure. The login system and tree-based data management structure allow for the safe and convenient storage of expression data and REDUCE analysis results. The uploaded data sets are private unless the user explicitly allows them to be shared within a group or made publicly accessible through our permissions management interface.
After registration and login, users are taken to the ‘Experiment View’ where they can upload and organize data (Fig. 1). The upload mechanism subjects incoming data to a series of filters and illegal expression values are discarded. Expression values specified as ratios or fold-changes are converted to log-ratios base two. The data can be uploaded one experiment at a time, as a two-column data file with each line containing an ORF and an expression value separated by a tab. It is also possible to upload multiple experiments as a single multi-column, tab-delimited file.
Figure 1.
‘Experiment View’ of the REDUCE analysis server. Uploaded expression data can be organized in folders and shared with other users or made publicly available.
MULTIVARIATE REGRESSION ANALYSIS
REDUCE is based on the following multivariate linear model for transcription initiation control:
The power of multiple-motif regression analysis is that the genome-wide transcriptional response measured in a microarray experiment can be decomposed in terms of distinct regulatory modules. This greatly facilitates the biological interpretation of transcriptome data. The regulatory modules that are significantly affected in a particular experiment will each be represented by one motif in the set M. The model tries to explain the log-ratio Ag for each gene g in terms of the number of occurrences Nμg of the motif μ in its promoter.
The regression coefficients or slopes Fμ are uniquely determined by the fit to the log-ratios. They have a direct interpretation as the change in the concentration of the (active form of the) transcription factor in the nucleus. Note that it is not possible to determine whether a transcription factor is an activator or a repressor based on the sign of Fμ. For instance, both increased activation and decreased repression by a transcription factor will manifest itself as an increase in the expression level of its target genes. Only when an experiment is analyzed in which a wild-type strain is compared to a strain in which a transcription factor is over-expressed or has been deleted (19) is it possible to infer the character of the transcription factor from the sign of Fμ.
Motifs can be ranked in terms of how much they improve the fit of the model to the data (as described in detail in 12). This allows for an unbiased search of relevant motifs from a large set of candidates, e.g. all oligonucleotides up to length 7. From this set of 21 844 motifs, typically ∼100 motifs correlate strongly enough with expression to be significant. However, these motifs fall into ∼10 classes of closely related, partially overlapping motifs. The forward selection procedure as described (12) selects one representative from each such class. The set M will therefore typically contain ∼10 motifs.
Once a motif has been found to be of interest based on the analysis of a single microarray experiment, scoring it against a large set of other (published) experiments provides a useful way of ‘annotating’ it, since the conditions under which the corresponding (and often unknown) transcription factor changes activity can be identified. It the case of mutant versus wild-type data, factors that are upstream of the transcription factor in a signal transduction pathway will be identified this way. Our web site allows one to specify an IUPAC consensus motif, select any number of experiments and compute the value of Fμ for each of them. We also provide functionality to plot such inferred transcription factor activity profiles for a number of different motifs. The usefulness of this approach has already been demonstrated (12,13).
AN EXAMPLE
Figures 1–3 show the most important screens users will encounter when uploading and analyzing their own expression data. Using the ‘expression view’ shown in Figure 1, users first select the directory to which they want to save their experiment. We will use the 15 min time point of the heat shock experiment performed by Gasch et al. (20) as an example. Figure 2 shows how the two-column file containing the log-ratio for each gene is uploaded to the server. Once this step has been completed, the experiment can be submitted for analysis by checking the box in front of it and pressing the ‘REDUCE’ button. The user will be prompted for a choice of sequence data (in yeast, a typical choice is 600 bp upstream from the translation start site) and the maximum length of the candidate motifs. As soon as the analysis is complete—typically after 1–2 min—the user is notified that the results can be viewed. Figure 3 shows the resulting model. The motifs are colored red or green according to the sign of the regression coefficient Fμ. The number of genes with a match to the motif is listed as well, and by clicking on it, the user will see a list of all matched genes with a one-line description of their function, and sorted by their log-ratio.
Figure 3.
Typical result of a REDUCE analysis.
Figure 2.
The form for uploading a microarray experiment.
SYSTEM DESIGN
The computation server uses a modular architecture and employs only open source components. To plan for growing performance demands we have incorporated the CONDOR batch processing system so that a higher throughput can be achieved by just adding more servers (http://www.cs.wisc.edu/condor). Caching in the database makes CONDOR even more efficient, as we attempt to never perform the same operation twice. Algorithms that take more than a few seconds to run are disconnected from the application layer so that a user never waits.
Our application server is built on Apache and mod_perl (http://www.apache.org) using HTML::Mason (http://www.masonhq.com) for presentation logic. We use Postgres (http://www.postgresql.org/) as our database for session management caching and data storage. We use Apache::Session from a centralized database so that we can add more application servers without having to implement complicated load balancing.
The application itself is implemented as Perl (http://www.perl.com) objects. We store all the data necessary to instantiate these objects in the database. For example, a REDUCE batch job is entered into the queue as a list of motif, sequence and expression instance identifiers necessary to build the job. The computation servers know how to rebuild the job and then run the necessary functions.
FUTURE DIRECTIONS
One of our priorities is to bring our system into compliance with the MIAME and MAGE-ML standards (21–24). This will enable us to integrate with other databases and data curation tools. We will also expand the list of organisms and sequence types for which REDUCE analysis can be performed and make it possible for users to upload their own sequence data in FASTA format. Currently, users can select upstream and downstream sequences for S.cerevisiae, Caenorhabditis elegans and D.melanogaster. The analysis of transcription regulation in human and mouse is inherently more difficult since enhancer regions are spread over distances of 10 kb or more from the coding region. Ongoing research in our laboratory is trying to answer the questions whether in its current incarnation REDUCE can be used for there organisms; if so, we will add the corresponding upstream sequences to our server. Finally, the use of weight matrices was already shown to have the potential of greatly increased performance of REDUCE (12) and we are working on putting this approach on a more solid basis. In general, the infrastructure provided by our upload server and the expandable way in which it was designed will allow us to quickly add new functionality as new tools are developed.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andre Boorsma, Barrett Foat, Feng Gao and Marcel van Batenburg for stimulating discussions and feedback, and Laura Rogers for her help with the filtering and conversion modules. H.J.B. was partially supported by NIH grant LM007276.
REFERENCES
- 1.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee N. and Zhang,M.Q. (2002) Functional genomics as applied to mapping transcription regulatory networks. Curr. Opin. Microbiol., 5, 313–317. [DOI] [PubMed] [Google Scholar]
- 4.Fickett J.W. and Wasserman,W.W. (2000) Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol., 11, 19–24. [DOI] [PubMed] [Google Scholar]
- 5.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 7.Neuwald A.F., Liu,J.S. and Lawrence,C.E. (1995) Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci., 4, 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Helden J., Andre,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 9.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 10.Hertz G.Z., Hartzell,G.W.,III and Stormo,G.D. (1990) Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci., 6, 81–92. [DOI] [PubMed] [Google Scholar]
- 11.Yuh C.H., Bolouri,H. and Davidson,E.H. (1998) Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science, 279, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 12.Bussemaker H.J., Li,H. and Siggia,E.D. (2001) Regulatory element detection using correlation with expression. Nature Genet., 27, 167–171. [DOI] [PubMed] [Google Scholar]
- 13.Koerkamp M.G., Rep,M., Bussemaker,H.J., Hardy,G.P., Mul,A., Piekarska,K., Szigyarto,C.A., De Mattos,J.M. and Tabak,H.F. (2002) Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell., 13, 2783–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orian A., Van Steensel,B., Delrow,J., Bussemaker,H.J., Li,L., Sawado,T., Williams,E., Loo,L.W., Cowley,S.M., Yost,C. et al. (2003) Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev., 17, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Steensel B.D., and J.Bussemaker,H.J. (2003) Genome-wide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl Acad. Sci. USA, 100, 2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keles S., van der Laan,M. and Eisen,M.B. (2002) Identification of regulatory elements using a feature selection method. Bioinformatics, 18, 1167–1175. [DOI] [PubMed] [Google Scholar]
- 17.Wang W., Cherry,J.M., Botstein,D. and Li,H. (2002) A systematic approach to reconstructing transcription networks in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 99, 16893–16898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang D.Y., Brown,P.O. and Eisen,M.B. (2001) Visualizing associations between genome sequences and gene expression data using genome-mean expression profiles. Bioinformatics, 17 (Suppl. 1), S49–S55. [DOI] [PubMed] [Google Scholar]
- 19.Hughes T.R., Marton,M.J., Jones,A.R., Roberts,C.J., Stoughton,R., Armour,C.D., Bennett,H.A., Coffey,E., Dai,H., He,Y.D. et al. (2000) Functional discovery via a compendium of expression profiles. Cell, 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 20.Gasch A.P., Spellman,P.T., Kao,C.M., Carmel-Harel,O., Eisen,M.B., Storz,G., Botstein,D. and Brown,P.O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell., 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C. et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. (2001) Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 22.Ball C.A., Sherlock,G., Parkinson,H., Rocca-Sera,P., Brooksbank,C., Causton,H.C., Cavalieri,D., Gaasterland,T., Hingamp,P., Holstege,F. et al. (2002) Standards for microarray data. Science, 298, 539. [DOI] [PubMed] [Google Scholar]
- 23.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brazma A., Parkinson,H., Sarkans,U., Shojatalab,M., Vilo,J., Abeygunawardena,N., Holloway,E., Kapushesky,M., Kemmeren,P., Lara,G.G. et al. (2003) ArrayExpress-a public repository for microarray gene expression data at the EBI. Nucleic Acids Res., 31, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]