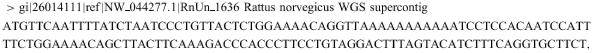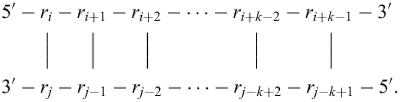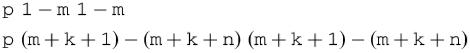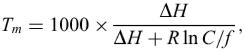Abstract
The abbreviated name, ‘mfold web server’, describes a number of closely related software applications available on the World Wide Web (WWW) for the prediction of the secondary structure of single stranded nucleic acids. The objective of this web server is to provide easy access to RNA and DNA folding and hybridization software to the scientific community at large. By making use of universally available web GUIs (Graphical User Interfaces), the server circumvents the problem of portability of this software. Detailed output, in the form of structure plots with or without reliability information, single strand frequency plots and ‘energy dot plots’, are available for the folding of single sequences. A variety of ‘bulk’ servers give less information, but in a shorter time and for up to hundreds of sequences at once. The portal for the mfold web server is http://www.bioinfo.rpi.edu/applications/mfold. This URL will be referred to as ‘MFOLDROOT’.
INTRODUCTION
The concept of RNA secondary structure began with the work of Doty and Fresco (1,2). The prediction of RNA secondary structure (folding) by energy minimization using nearest neighbor energy parameters began with Tinoco and colleagues (3–6) and also with Delisi and Crothers (7). Efficient algorithms for RNA secondary structure prediction using dynamic programming methods borrowed from sequence alignment were developed independently by a number of people (8–13).
My own early RNA folding programs (12,14) computed a single minimum energy folding of an RNA sequence. They were popular in the 1980s and a modified version was incorporated into the UWGCG (University of Wisconsin Genetics Computer Group) suite of programs (15); the acronym was shortened to GCG when this group left the University of Wisconsin to form a private company. Initially, they used free energy parameters that had been summarized by Salser (16). After 1986, free energies from the Turner group (Doug H. Turner, Department of Chemistry, University of Rochester, Rochester, NY) were used (17).
The ‘mfold’ software for RNA folding was developed in the late 1980s (18). The ‘m’ simply refers to ‘multiple’. The core algorithm predicts a minimum free energy, ΔG, as well as minimum free energies for foldings that must contain any particular base pair. Any base pair, ri−rj, between the ith nucleotide and the jth nucleotide that is contained in a folding no more than δδG from the minimum, is plotted in a triangular plot called the ‘energy dot plot’. The base pair ri−rj is plotted in row i and column j of this matrix. The free energy increment, δδG, is chosen a priori by the user, who selects a ‘percent suboptimality’, P. From this, δδG is computed to be P/100 |ΔG|. Base pairs within this free energy increment are chosen either automatically, or else by the user, and foldings that contain the chosen base pair are computed. They have minimum free energy conditional on containing the chosen base pair. The description and use of the mfold package has appeared in a number of articles (19–22). The closely related ‘RNAstructure’ program has also been described (23,24).
The Turner group has published numerous articles over the years that detail the development of the RNA folding parameters. A subset of these articles are what I would call ‘major works’ that summarize the current state of the art. Version 1 of the mfold package used free energies that were described by Freier et al. (17). Versions 2.1 to 2.3 used the parameters from Walter et al. (25), although the incorporation of coaxial stacking parameters into the minimization algorithm has not been accomplished. The current version 3 software uses free energy data from Mathews et al. (26).
DNA folding prediction with the mfold software began in 1996, when DNA specific parameters were added to the mfold package through a collaboration with the SantaLucia group (John SantaLuica Jr., Department of Chemistry, Wayne State University, Detroit, MI). These data have been described by SantaLucia (27). The DNA stacking (27), single mismatch (28–32) and dangling end (33) parameters have been measured in the SantaLucia laboratories. The remaining terminal stacking and loop parameters were estimated by SantaLucia and have been incorporated into the mfold package by personal communication from John SantaLucia. They remain unpublished. In 1999, corrections for [Na+], [Mg++] were incorporated into the mfold package (34).
The mfold web server was first created at Washington University School of Medicine during the fall of 1995. DNA folding parameters were added in the spring of 1996. From 1995 until the fall of 2000, the server ran on SGI workstations or multiprocessor servers (Silicon Graphics, Inc. 1600 Amphitheatre Pkwy., Mountain View, CA 94043), as well as on a dual processor ‘Intel/Solaris’ platform (Pentium 2 processors by Intel Corp., Solaris operating system by Sun Microsystems). The server was moved to Rensselaer Polytechnic Institute (RPI) in October 2000. It ran on a dual processor ‘Intel/Linux’ platform (Linux operating system as developed and marketed by Red Hat, Inc., 1801 Varsity Drive, Raleigh, NC 27606). Since July 2002, the mfold web server has been running on a cluster of 36 dual processor ‘Intel/Linux’ workstations that were donated to the joint RPI-Wadsworth Bioinformatics Center by IBM (IBM Research, P.O. Box 218, Yorktown Heights, NY 10598). This equipment was awarded as an SUR (Shared University Research) grant to RPI and the Wadsworth Center (PI: M. Zuker).
The use of the mfold web server has grown steadily since its inception. The ‘quikfold’ server was added while I was still at Washington University. The remaining servers that will be described have all been added within that past 2 years. The servers have been used extensively by researchers in universities, medical schools, non-profit organizations, US government and military laboratories and by companies all over the world. In addition, the servers are being used in the teaching of computational biology methods.
SERVER CONTENT AND ORGANIZATION
The mfold web server comprises a number of separate applications that predict nucleic acid folding, hybridization and melting temperatures (Tms).
The basic ‘mfold’ server: input
The original applications on the mfold web server deal with folding a single RNA or DNA sequence per submission (job). The submission forms for RNA and DNA are separate for historical reasons only. RNA folding came first. These applications may be reached by following the ‘RNA Folding’ or ‘DNA Folding’ hyperlinks from the main portal or entrance page. The URL for the portal will likely remain stable, while those for separate applications will change. The default RNA folding form currently uses the latest version 3.0 free energies (26). These are recommended for most RNA folding. However, there is a link from this page to what is called the ‘RNA mfold version 2.3 server’. This server offers RNA folding using the older, version 2.3 energy parameters (25). Why use older and less accurate parameters? The reason is that we have enthalpies for these older parameters. As with the free energy parameters, the enthalpies were measured at 37°C. However, they are assumed to be constant within the range of temperatures that might occur in vivo or in the laboratory. This enables the server to extrapolate free energies to other temperatures and to fold at these temperatures.
Sequence name
A sequence name may be typed or pasted (entered) within the ‘Enter a name for your sequence:’ text field. Long names are truncated to 40 characters. Any ASCII characters may be used, including those with octal values greater than 200 Octal (O). The ‘dangerous’ characters, ”, <, > and ‘, are converted to ’, ≪(253 O), ≫(273 O) and ′ (264 O), respectively. The character, \, is eliminated. If no name is entered, then the sequence name becomes the ‘Job ID’, which is of the form yyMmmdd-hh-mm-ss, where yy is year, Mmm is month, dd is day, hh is hour, mm is minute and ss is second. For example, a job that comes into the server at 8:23:06 pm local time on 9 February 2003, will be assigned a Job ID of 03Feb09-20-23-06. If two jobs come in during the same second, the second one has the letter ‘a’ appended to it, and so on for more than two jobs during the same second.
Sequence
A sequence must be entered into the sequence text area box. All characters except for ‘A–Z’ and ‘a–z’ are removed. Lower case characters are converted to upper case. For RNA folding, ‘T’ or ‘t’ are converted to ‘U’, while ‘U’ or ‘u’ are converted to ‘T’ in DNA folding. If, for example, you enter
then the resulting sequence will be
rather than
The letter ‘N’ should be used for an unspecified base. It is not allowed to pair. The letters ‘B’, ‘D’, ‘H’ and ‘V’ denote ‘A’, ‘C’, ‘G’ and ‘U/T’ respectively. These nucleotides may pair only if their 3′ neighbor is unpaired. The purpose of this convention is to denote nuclease cleavage of the phosphodiesther bond linking the indicated nucleotide and its 3′ neighbor. It is used to constrain folding when nuclease digestion data specify susceptible bonds. In addition, the letters ‘W’, ‘X’, ‘Y’ and ‘Z’ also refer to ‘A’, ‘C’, ‘G’ and ‘U/T’, respectively. These nucleotides, if they pair, should do so only at the end of a helix. Thus, the mfold web server does not support the IUPAC (International Union of Pure and Applied Chemistry) ambiguous DNA character convention (35) shown in Table 1.
Table 1. IUPAC codes for incompletely specified bases in nucleic acid sequences.
| G/A | C/T | A/C | G/T | G/C | A/T | not A | not C | not G | not T | any base |
|---|---|---|---|---|---|---|---|---|---|---|
| R | Y | M | K | S | W | B | D | H | V | N |
Constraints
The text area in the constraints box allow for the optional incorporation of folding constraints. Each constraint consists of a single line in the box that must conform to a rigid format. When constraints are used, a hyperlink labeled ‘Explanation of sequence annotation’ appears on the primary results page.
The various types of constraints are itemized below. Multiple constraints of any form are allowed in any order.
-
Force a specific base pair or helix to form. The command
F i j k
-
will force the formation of the helix (single base pair if k=1)
The triple (i, j, k) refers to k consecutive base pairs, where ri·rj is the exterior closing base pair. If any of these base pairs cannot exist, then an error will be generated and the job will fail. The usual result is an output page that declares ‘Job aborted! No Structure!’. In the text area of the output page, successfully forced base pairs are underlined with (‘and’), while those that are forced to pair but cannot are underlined with ‘!’. Note that isolated base pairs are not allowed by the folding code. That is, even if ri·rj is a valid base pair, it will not be allowed to form unless either ri+1·rj−1 or ri−1·rj+1 is a valid base pair.
-
Prohibit a specific base pair or helix from forming. The command
P i j k
will prohibit every single base pair of the form ri+h·rj−h, 0≤h≤k, from occurring. Base pairs that are prohibited that could not form in any case will be underlined with ‘!’ in the text area of the primary output page, but the folding will proceed without error.
-
Force a string of consecutive bases to pair. The command
F i 0 k
(the second to last character is ‘zero’) will force nucleotides ri, ri+1, ri+2,…, ri+k−1 to pair. This is a single base when k=1. Forcing too many bases to pair or forcing a base labeled ‘N’ to pair will generate a fatal error.
-
Prohibit a string of consecutive bases from pairing. The command
P i 0 k
(the second to last character is ‘zero’) will prevent nucleotides ri, ri+1, ri+2,…, ri+k−1 from pairing. This is a single base when k=1. Forcing too many bases to be single stranded can generate a fatal error.
-
Prohibit a string of consecutive bases from pairing with another string. The command
P i j k l
will prevent the nucleotides ri, ri+1, ri+2,…, rj from pairing with nucleotides rk, rk+1, rk+2,…, rl (i≤j and k≤l). Note that if i=k and j=l, then the constraint is equivalent to forbidding all base pairs within the segment ri,…, rj.
Other folding parameters
RNA and DNA sequences may be linear or circular. The default is ‘linear’, but ‘circular’ may be chosen using an option button on the folding form. It is worth noting that the algorithm to fold circular nucleic acids is simpler than that for folding linear ones.
The folding temperature is fixed at 37°C for RNA folding using version 3.0 energy rules. For RNA folding with the version 2.3 parameters, or for DNA folding, any integral temperature between 0 and 100°C may be chosen.
-
Ionic conditions may be altered for DNA folding only. For RNA, the ionic conditions are fixed at [Na+]=1 M and [Mg++]=0 M. For folding, these are equivalent to physiological conditions. The following constraints apply:
[Na+]≥0.01 M,
[Mg++]≤0.1 M, and
[Na+]≤0.3 M if [Mg++]>0 M.
For the purposes of folding, Na+ may be considered equivalent to Li+, K+ and NH4+, while Mg++ is equivalent to Ca++.
The percent suboptimality, P, controls the free energy increment, δδG for displaying base pairs in the energy dot plot and for computing suboptimal foldings. Base pairs that can occur in foldings with free energies ≤ΔG+δδG will be plotted, and only foldings with free energies ≤ΔG+ δδG will be computed. Normally, δδG=P/100|ΔG|, but it is rounded up to 1 kcal/mol or down to 12 kcal/mol if outside this range.
The upper bound on the number of foldings is an absolute limit. The number of foldings computed may be less than this quantity. Normally, it should be less that this number because the number of computed foldings is more appropriately controlled through a proper choice of P and the window parameter.
The window parameter, W, controls the number of foldings that are computed. It may be thought of as a distance parameter. The distance between 2 bp, ri·rj and ri′·rj′ may be defined as max{|i−i′|, |j−j′|}. If k−1 foldings have already been predicted by mfold, the kth folding must have at least W base pairs that are at least a distance W from any of the base pairs in the first k−1 foldings. A new folding is not added to the output list unless this criterion is fulfilled As W increases, the number of predicted foldings decreases. If W is not specified, mfold selects a value by default based on sequence length.
If the maximum distance between paired bases parameter, M, is specified, then any base pair, ri·rj, in a folding of a linear molecule must satisfy j−i≤M. In a circular molecule, the condition becomes min{j−i, N +i−j}≤M, where N is the sequence length.
Immediate versus batch jobs
Folding sequences containing up to 800 bases may be done while the user waits. This is the default. At this time, folding results for immediate jobs are erased 24–26 h after they are submitted. For sequences from 801 to 6000 bases in length, the batch option must be selected from the appropriate option button. The user should enter a valid email address in this case, although email addresses are always welcome since they identify users. For batch jobs, pressing the ‘Fold RNA’ or ‘Fold DNA’ submission button takes the user to a notification page that indicates a URL for the results and the email address to which a notification will be sent when the folding is completed. If the link to the results pages is followed immediately, then the target page will be incomplete. It will contain only the sequence and some other input information. When the folding is complete, this page must be ‘refreshed’ or ‘reloaded’ for the results to be seen. The URL for the results remains valid for 48–51 h, after which the results are erased. Folding sequences of up to 10 400 bases is available at special request. Folding 6000 bases currently takes about 1.5 h. The hyperlink labeled ‘View Folding Results’ will enable a user to view any results on the server, provided that they have not been erased and that the user views from the same computer that generated the foldings. That is, the user's IP address must be the same.
Output parameters
The default, regular image resolution value, is 72×72 pixels per inch2 for png and jpg images. The low, medium and high values are 60×60, 110×110 and 200×200, respectively. This parameter has no effect on PostScript output files, as well as the pdf files that can be derived (‘distilled’) from them. These files can be scaled up as much as desired without loss in quality. Jpg and png images will always be displayed at 72×72 pixels per square inch. Changing the resolution will change the size of the entire image. Image sizes are given in Table 2.
Structure format plots are drawn showing individual bases or in an outline mode where bases do not appear. The outline option is suitable for images of very large folded molecules. The default choice of ‘Automatic’ will cause bases to be displayed in foldings of up to 800 nt and an outline to be drawn otherwise. Choosing the ‘Bases’ option for a large sequence might make sense if the user intends to magnify the structure to display a portion in which individual bases can be seen.
The grid lines in the energy dot plot may be turned off.
By default, bases will be numbered according to the length of the sequence. The default values are shown in Table 3. The user is free to alter this value, or to turn off base numbering altogether by choosing a frequency of zero.
The user may alter the automatically chosen orientation of the folded molecular by choosing a rotation angle. Positive values correspond to counter-clockwise.
Structure annotation has been described by Zuker and Jacobson (36). Bases in plotted structures may be annotated by ‘p-num’ values, which represent the number of ways that a base may pair in all foldings within δδG from the minimum energy. Low values indicate ‘well-defined’ bases. In particular, values of 0 or 1 indicate that a base is always single stranded or always paired to a unique partner, respectively. The number of times that a base is single stranded in the computed foldings is called its ‘ss-count’ number, and structure plots may also be annotated using these numbers. Color schemes are shown at MFOLDROOT/www-NAR03/doc/colors.html. Finally, for RNA folding only, the ‘high-light’ option allows the user to specify regions in the sequence to be highlighted. The selected bases are drawn in green, while the remaining bases are drawn in black. If the structure format is ‘Bases’, then the base characters will be drawn in color. If the structure format is ‘Outline’, then colored dots that represent bases will be superimposed on the ladder-like outline plot. If the default ‘Automatic’ option is used, then colored dots are used if the sequence size is >800. However, if the sequence size is ≤800, then both colored dots and bases are used. The bases appear in black or white inside an appropriately colored dot.
Table 2. Resolutions and images sizes of jpg and png images, the units are pixels per square inch and pixels, respectively.
| Resolution | 60×60 | 72×72 | 110×110 | 200×200 | 250×250 | 300×300 |
| Image size | 510×660 | 612×792 | 935×1210 | 1700×2200 | 2125×2750 | 2550×3300 |
Table 3. Default numbering increments for secondary structures.
| Sequence length | Numbering increment |
|---|---|
| 1–50 | 10 |
| 51–300 | 20 |
| >300 | 50 |
Folding results
Folding results may be viewed by following links from an initial results page. Some input information is reproduced at the top of this page in a text area. An example of the top portion of an initial results page is shown in Figure 1.
Figure 1.
This figure reproduces part of the initial results page for an RNA folding. Some input is reproduced in the text area. Various hyperlinks lead to the actual folding results.
The energy dot plot
The energy dot plot is available in PostScript, png and jpg form. The ‘Text’ hyperlink leads to a plain text file that gives the basic dot plot information. The ‘istart’, ‘jstart’ and ‘length’ options correspond to the (i, j, k) numbers that define a helix, as defined previously. The ‘energy’ in an integer in units of tenth of a kcal/mol. Level 1 corresponds to helices in optimal foldings. Levels 2–4 correspond to the helices in increasingly suboptimal foldings. The levels from 1 to 4 correspond to the default coloring of the base pairs in the dot plot. The PostScript hyperlink leads to a static PostScript image of the energy dot plot. Letting δG be the minimum free energy of a folding containing a base pair, the default color (rgb Hex) of a base pair in the energy dot plot is given in Table 4.
Table 4. The default colors used in the energy dot plot.
| δG | Color |
|---|---|
| ΔG=δG | FFFFFF (black) |
| ΔG<δG≤ΔG+δδG/3 | D40721 (red) |
| ΔG+δδG/3<δG≤ΔG+2δδG/3 | 12CC24 (green) |
| ΔG+2δδG/3<δG≤ΔG+δδG | B6C614 (yellow) |
The png and jpg hyperlinks each lead to a new page containing a png or jpg image of the energy dot plot and buttons that allow the user to interact with or redraw the dot plot. It is called the primary dot plot page. A small ‘Details’ window, 550×40 (pixels) opens in the top left corner of the screen. With the ‘Click on a base pair to identify’ radio button selected (the default), clicking (left click) on the image will cause the (i, j) value of the selected base pair as well as its δG value to appear in the Details window. If a blank area of the plot is chosen, the value of δG will be declared ‘Undefined’.
Selecting the ‘Click on the image and redraw the dot plot with options below’ button activates the text area and five option buttons below. Clicking on a point in the dot plot will cause a new image to be created. If the magnification factor is chosen to be >1, then the new image will be magnified about the selected point; otherwise the entire dot plot will be redrawn. The value of the ‘Energy increment in kcal/mole’ option will be an upper bound on δG. For example, setting it to zero will cause only optimal base pairs to be plotted. The number of different colors used in the dot plot may be changed. If k is the value of the ‘Filter’ option, then only helices of size at least k will be plotted when the dot plot is redrawn. The ‘Image Width’ controls the resolution (size) of the redrawn image as explained above. If the PostScript ‘Output’ option is chosen, then a static PostScript image is created. If the png or jpg ‘Output’ option is chosen, then clicking on the plot will cause a new window to open containing the redrawn image and the same set of control buttons as before. This is called the secondary dot plot window. New png or jpg images created from this secondary window will be drawn within the secondary window. Similarly, new png or jpg images generated from the primary dot plot window will refresh the secondary window. A typical use would be to magnify about a region in the initial dot plot. Base pairs in the magnified dot plot can then be selected with precision, allowing the user to identify specific helices that may occur in optimal or suboptimal foldings. Closing the secondary window will not affect other windows. Closing the primary window will also close the Details window.
RNAML syntax
Computed secondary structures are also available in the RNAML (37) format. This format has been proposed as a syntax for exchanging RNA structure information. We are currently offering output that is compatible with the DTD (Document Type Definition) file maintained by the Major group at the Université de Montréal. This file may be found at: http://www-lbit.iro.umontreal.ca/rnaml/current/rnaml.dtd.
Some extra files
These are raw text files that contain information that might be useful to some users. Annotation by p-num value is available on the server, as described above. The raw values are also made available. The h-num link contains h-num values of each helix in the energy dot plot, sorted from lowest to highest value of h-num. The h-num value of a helix is the ‘well-definedness’ of a helix (38). Helices with lower h-num values tend to be more reliably predicted. The log file for a run is sometimes useful in finding the cause of a failed job.
Bulk downloading of results
All computed foldings may be downloaded at once in a single zipped file (.zip) or a compressed tar file (.tar.Z). The structures may be taken as PostScript, png or jpg plot files, or as raw text files in different formats.
The text files will be useful to those who wish to create structure plots on their own. Some of the text formats have been described before (21,22). A full description may be found at MFOLDROOT/www-NAR03/doc/structure-format.html. Redrawn structures or structures created using the new ‘sir_graph’ program can only be downloaded individually.
Single strand frequency
The ss-count file contains explicit statistics on single strandedness of each base in all of the computed foldings. The first line contains the number of foldings that were computed. The ith subsequent line contains the number of the ith base, the base itself and the number of times it is single stranded in all of the computed foldings. The ‘View plot’ button creates a plot of base number versus ss-count. When the ‘Averaging window’ is m>1, then the value that is plotted for the ith base is the average over the m bases centered at i. Fewer bases may be used near the ends of the plot. The magnification factor enables the user to zoom in on a region of interest.
Structure output
The hyperlink to ‘Structure i’ leads to a portion of a single html file that contains the ith structure in an easy to read text format. This format may be suitable for viewing small foldings. It may easily be pasted into text files. If the sequence is very large and the structure plot has low resolution, it might be more convenient to view a portion of the text file than to magnify a portion of the structure plot.
Selecting PostScript,.ct file, RnaViz ct, Mac ct, GCG connect or XRNA ss will lead to single structures in the designated format. Following the png or jpg link is similar to following the png or jpg links when viewing the energy dot plot. Both lead to a new page containing a png or jpg image of the structure plot and buttons that allow the user to interact with or redraw the folding. It is called the primary structure plot page. A small ‘Loop Free-Energy Decomposition’ window, 550×150 (pixels), opens in the top left corner of the screen. With the ‘Click on a base pair to view related structural details in the Loop Free-Energy Decomposition window’ radio button selected (the default), clicking (left click) on the image will cause the details of the selected base pair to appear in the small window. The window will show the type of loop, including a stack, that is closed by the selected base pair. The free energy of that loop is given, and the specific identity of the base pair is given. If the selected point does not unambiguously select a base pair, then the auxiliary window will print a message instructing the user to try again.
Selecting the ‘Click on the image and redraw the structure with options below’ button activates the text area and four option buttons below. Clicking on a point in the structure plot will cause a new image to be created. If the magnification factor is chosen to be >1, then the new image will be magnified about the selected point; otherwise the entire structure will be redrawn. The ‘annotation’ button allows the user to select p-num or ss-count annotation, even if these were not selected initially. The ‘annotation type’ button has no effect unless an annotation option is selected. In this case, an annotated structure will be drawn with colored base letters, with colored dots or with colored dots containing base letters, corresponding to the choices ‘Character’, ‘Dot’ or ‘Both’, respectively. The ‘Image Width’ controls the resolution (size) of the redrawn image, as explained above. If the PostScript ‘Output’ option is chosen, then a static PostScript image is created. If the png or jpg ‘Output’ option is chosen, then clicking on the plot will cause a new window to open containing the redrawn image. There are no buttons on this page. The ‘Color Table’ option leads to a table that explicitly shows the colors used in the structure plot versus the p-num or ss-count values. The p-num or ss-count file hyperlink leads to the corresponding raw text files. This is called the secondary structure window. New png or jpg images generated from the primary dot plot window will refresh the secondary window. A typical use might be to magnify about a region in the initial structure plot so that a portion of the structure might be printed at a suitable resolution. It could also help the user select base pairs with precision. Closing the secondary window will not affect other windows. Closing the primary window will also close the ‘Loop Free-Energy Decomposition’ window.
New form of structure output
The structure plots that are drawn by default and that can be downloaded at once are generated for the most part by the ‘naview’ program by Bruccoleri and Heinrich (39). The ‘naview’ program creates a device independent ASCII plot file (with suffix plt2). Software developed for the mfold package at Washington University creates PostScript, jpg and png images from these files. A new drawing program named ‘sir_graph’ was created at Washington University for creating displays of nucleic acid secondary structure. It is an interactive program that runs on a variety of Unix platforms. A non-interactive version, ‘sir_graph_ng’, creates displays for the mfold web server. The ‘Click Here for New Structure Viewing Options’ hyperlink on the primary results page leads to a ‘Structure Viewer’ page that is equivalent to the primary window page described above. This page is identical to the ones that may be reached by following the ‘new’ hyperlinks in the ‘View Individual Structures’ section.
The user has a wider selection of choices with this new software. The external loop, or exterior base pairs, may be drawn in the default way, arranged around a circle, or may be drawn ‘Flat’ with all the helices that meet it being parallel to one another. The ‘Flat_Alt’ option draws successive stems at 180° angles from the previous. The background may be the usual white or black. The base pair symbol may be a ‘dot’ which is usual for the mfold web server, or a ‘line’, which is common in most secondary structure displays. The ‘Algorithm’ may be the ‘Default’ or ‘Simple’ type, both of which avoid overlaps of stems. The ‘Default’ and ‘Simple’ algorithms draw the multi-branch loops differently. The ‘Natural’ option draws every loop as a perfect circle, with the stems coming out at appropriate angles. This may produce pleasing results for small foldings, but will usually produce a hopelessly tangled mess for large foldings. As with the display of structures using the older naview program, the ‘Loop Free-Energy Decomposition’ window opens and allows the user to identify precise loops or stacks, together with their free energies and closing base pairs. Also, the secondary structure window page is available for magnifications about chosen points.
Thermodynamic details
When temperature cannot be altered, the ‘Thermodynamic Details’ link leads to a portion of an html file that contains the entire decomposition of the particular folding into loops and stacks, together with their free energies and closing base pairs. Consecutive runs of base pairs are summarized as helices. Small portions of this file are displayed in the ‘Loop Free-Energy Decomposition’ window when the user is interacting with a png or jpg plot of a folding.
When temperature may be altered, then a more attractive page appears that gives the free energy, enthalpy, entropy and an estimated Tm. Tm is computed using a simple 2-state model. This assumes that the molecular is either folded as shown or else completely single stranded. Such an assumption is reasonable for short molecules. For larger molecules, it might be useful to refold near the predicted Tm. If the new Tm is ‘significantly’ larger, it indicates that the given folding can rearrange into another folding with a higher melting temperature. The server uses Tm=ΔH/ΔS, where ΔH and ΔS are the structure enthalpy and entropy, respectively. (Because of the units used for entropy, the actual formula is Tm=1000ΔH/ΔS−273.15 in °C.) The usual details table follows.
Structure dot plot
The ‘Dot plot folding comparisons’ option allows the user to view any subset of the computed foldings in a dot plot. This option is available when two or more foldings are computed. The user may select foldings to be displayed by selecting the corresponding radio buttons. The default is the display of the first two foldings. For convenience, an ‘All’ button has been supplied.
Base pairs that occur in all foldings are colored black. Those that occur in two or more, but not all foldings are colored grey. Otherwise, base pairs are assigned a unique color that depends on the structure. This system breaks down when more than 15 structures are computed.
Selecting the png or jpg option in the first button and pressing the ‘Do the Comparison’ option leads to a primary structure dot plot page that is very similar to the primary energy dot plot page. Clicking on a base pair with the default ‘Click on a base pair to identify’ radio button selected will show the identity of the base pair and list which of the computed structures contain that base pair. When the ‘Click on the image and redraw the dot plot with options below’ radio button is selected, a click on the dot plot will generate a new dot plot. The dot plot will be in a ‘secondary’ structure dot plot window if the png or jpg option is selected. This is very similar to the secondary dot plot window for the energy dot plot. The magnification, output format and image width parameters are the usual ones explained above for the energy dot plot. The ‘Multicolor Overlap’ option that may be selected will have the effect of drawing the otherwise grey dots in a multi-color mode that displays precisely what foldings contain that base pair. In the mfold web server, ‘dots’ are really plotted as (usually small) squares. A collection of trapezoids in different colors are drawn within the square region. These indicate which foldings contain that base pair. The dot plot must be drawn with a high enough magnification for this feature to be seen. It is not necessary when only two foldings have been computed. When three foldings are computed, the gray base pairs that are in structures 1&2, 1&3 and 2&3 are plotted in the lower left triangle in three distinct colors, so they may be identified without using the multicolor option.
The other servers
The ‘quikfold’ server: folding many sequences at once
Many users are not interested in the sophisticated viewing options that are available on the mfold web server. Some may be content with the easy to read text format or with the ‘ct format’ files that may be downloaded and used locally to create secondary structure plots. Many wish to fold many sequences at once under the same conditions. For example, a user may wish to predict foldings and melting temperatures for hundreds of short molecular beacons.
For this reason, the ‘quikfold server’ was created. It uses the same ‘nafold2’ program that folds sequences in the regular server, but in a different, multiple molecule mode. Only immediate jobs are allowed, with each sequence containing no more than 600 nucleotides. The theoretical upper limit for the number of sequences that may be submitted at once is 25 000, but I would not advise submitting nearly that many! The upper limit depends on the Internet, browser settings (such as timeout) and other factors. I do know, for example, that 1000 sequences of length 100 each should pose no problem.
The input page contains buttons and text areas that have already been defined. The single new item is that one extra character is required for the sequence input box. Each of the input sequences must be separated by (at least) one semi-colon, ‘;’. All other characters, except for ‘A–Z’ and ‘a–z’, are removed in a preprocessing step.
The quikfold server gives text, ct, RNAML and thermodynamic details as output. In each case, the output is in a single, possibly large text file. The sequences are named by taking the given or default sequence name, and adding ‘_i’ to each name, where i ranges from 1 to the number of sequences. The folding results are erased within 40 min to several hours. An individual sequence that cannot fold will not abort the entire job. It will simply be skipped over in the output.
The ‘zipfold’ server: prediction of minimum free energy only
Some users want only the minimum folding energy of a sequence. For this reason, the zipfold server was created. The input page is similar to that of the quikfold server. Sequence lengths may be up to 800. In one instance, a user submitted many thousands of jobs; each containing 50 randomized sequences of length 500. The failure rate was less than 1%.
Because speed is important and so little information is being requested, the underlying code was simplified to run much faster. At the time of this writing, 378 tRNAs were processed in just over 11 s. The actual performance depends on the server load and difficult to control Internet related factors.
The underlying code has been stripped of its ability to handle constraints. Moreover, the ‘fill algorithm’ has been truncated so that only an optimal energy or a single optimal structure may be computed. Newer, experimental versions, streamline the sequence input as well.
All the output appears on a single, primary results page. The ΔG values are arranged in a list. Those sequences that cannot fold are given a large positive folding energy. The current value is 10 000.
The ‘Tm’ server for single stranded nucleic acids: prediction of two state melting temperature
Some users want a little more that just a minimum folding energy. They also desire an estimated Tm. In this case, the Tm server may be used. Only version 2.3 RNA folding parameters are available for this server, since enthalpies are required for estimating Tm. The input is the same as with the zipfold server.
The Tm server uses the same simplified folding program as the zipfold server, except that a minimum energy folding is computed in this case. The enthalpy, ΔH, of this folding is then computed using the appropriate nearest neighbor parameters. From this, it is easy to compute an entropy, ΔS and then Tm, using a 2-state model as described above.
In fact, this server was created to ‘service’ the ‘OligoArray 1’ program of Rouillard et al. (40). The OligoArray program ‘hits’ the server with single DNA sequences using a direct request created by a Java application. Subsequent versions of OligoArray will abandon this inefficient procedure.
The hybridization server: hybridization of two strands
The underlying mfold software has had numerous small changes added over the years to accommodate ‘special requests’ or applications. Many users have used the regular mfold software, either in ‘stand alone’ form or on the web, to simulate the hybridization of two strands of RNA or DNA. They have done so by taking two sequences, A=a1a2a3···am and B=b1b2b3···bm and creating a single sequence, S=a1a2a3···amn1n2···nkb1b2b3···bm, by concatenating A with B using some non-pairing characters, n1···nk as a linker.
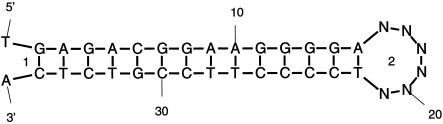
There are two problems with this approach. The first, less serious problem, is that the k linker residues end up in a loop of some sort. If A and B hybridize perfectly, then this loop will almost certainly be a hairpin loop. An example of such a folding is given in Figure 2. The problem is that an incorrect hairpin free energy is applied to loop ‘2’. Instead, it should be treated as an exterior loop, like ‘1’. In addition, an initiation free energy, ΔI, needs to be added. This is 4.1 kcal/mol for RNA at 37°C and 1.96 kcal/mol for DNA at 37°C.
Figure 2.
Two complementary DNA sequences have been concatenated by linking them with six Ns. The two loops are labeled ‘1’ and ‘2’. Loop ‘1’ is an exterior loop, which is correct, but loop ‘2’ is a hairpin loop instead of an exterior loop.
In order to accommodate such situations, the following feature was added to mfold. When three consecutive Ls occur in a sequence (L for Linker), they are recognized as a linker. If they occur in a hairpin loop, then this loop is treated instead as an exterior loop and ΔI is added as well. The output still contains the linker residues, but the value of ΔG is now correct.
The linker residues may be forced to be in a hairpin loop by forcing a simple hybridization of A with B. This may be accomplished by using the two constraints
These constraints forbid all intramolecular base pairs, so that the only allowed base pairs link A with B, forcing, among other things, the linker residues to be in a hairpin loop.
The more serious problem, however, is that any estimated Tm is nonsense, since total nucleic acid concentration, C, must be taken into account when two strands hybridize. The correct 2-state estimate for Tm is
where f=2 if A≠B and f=4 if A=B.
All of this is accomplished automatically by the hybridization server. The two sequences, A and B, are entered into the sequence box text area, separated by a semi-colon, ‘;’. The total nucleic acid concentration must be given. The result is a simple output page containing the job ID, ΔG, ΔH, ΔS and Tm in a single line. No units are given, although they are understood to be kcal/mol, kcal/mol, cal/(K · mol) and °C, respectively.
Access
The mfold web server is open to all users. No restrictions are applied to commercial users. However, users should be aware that the server is not secure and that data flowing both in and out may be detected by others. Moreover, query information is stored indefinitely in log files on the server. These log files are treated as confidential information, although gross statistics on usage are collected and disseminated. Furthermore, some of the submissions are selected as examples for teaching, but only if database searches reveal that the sequence is already in a public database.
The regular mfold server may not be hit by ‘GET’ requests. Only the ‘POST’ operation is permitted. The other applications that have been described may also be hit with ‘GET’ requests. Hitting these other applications directly using, for example, Java applications is permitted and will continue to be permitted until such time as the server is overloaded. This has not happened yet. At Washington University, perhaps 100–200 folding jobs were submitted each work day, with far fewer on weekends. For the month of December, 2002 (for example), I counted 21 688 submissions to the regular mfold web server, 3128 to quikfold, 35 904 to zipfold, 193 428 to Tm and 542 to the hybridization server.
EQUIPMENT AND ORGANIZATION
The current web server is running on equipment donated to RPI by IBM Research in the fall of 2001. The detailed hardware specifications are given in Table 5. The total value of the donated equipment comes to about $214 000. We contributed another $5000–$10 000 for a switch and a custom designed power source. All equipment was originally assembled and housed at the Academy of Electronic Media (http://www.academy.rpi.edu/). The server is expected to be moved to the Voorhees Computing Center during the spring or summer of 2003.
Table 5. The mfold web server is a major application on the RPI-Wadsworth Bioinformatics Center web site. It is housed on a high-performance web server consisting of a dual 1 GHz Pentium processor with 4 Gb memory and a 73 Gb disk, connected to a cluster of 35 other dual 1 GHz Pentiums, each with 1 Gb of memory and a 36 Gb disk.
| Description | Quantity |
|---|---|
| X series 330 1U single processor | 36 |
| #33l315 1 Gb ecc Rdimm | 38 |
| Additional 1 GHz Pentium proc | 36 |
| Flat panel monitor 9511AG1 | 1 |
| Flat panel monitor rack kit | 1 |
| Keyboard 28l3644 | 1 |
| Rack 9306910 | 1 |
| HD 36 Gb drive 37L7206 | 36 |
| DLT tape drive 20/40 Gb | 1 |
| Linux | 1 |
| 100baseT ethernet switch | 3 |
| APC Smart UPS 1400RMB (for web server only) | 1 |
| Miscellaneous cables and mounting hardware | 1 |
FUTURE DIRECTIONS
It is our intention to keep the RNAML output up-to-date and in accord with the DTD file: http://www-lbit.iro.umontreal.ca/rnaml/current/rnaml.dtd. Within the next month or two, the RNAML output will contain the ‘x, y’ co-ordinates that correspond to the structure plots produced by the ‘sir_graph’ program, when they are available. The next year or two should see the incorporation of important, novel applications that are currently being developed.
Hybridization with partition functions
New algorithms for the folding and hybridization of two separate strands of RNA or DNA have been created (41). Initial versions of the software are now being carefully reprogrammed and updated. We are investigating theoretical and experimental ways to improve the accuracy of predications. We are also beginning to consider dealing with the thermodynamics of hybridization of probes to nucleic acids immobilized on chips (42).
Database searching
We are developing a new ‘BLAST-like’ algorithm for searching nucleic acid databases that is based on the computer science technique of hashing, for speed, but searches for regions of best complementarity between two sections of DNA or RNA. Scoring is based on nearest neighbor free energy parameters for RNA or DNA. An early version is now being used to search for putative binding sites for micro-RNAs (miRNAs) in databases of mRNAs, and to search for (undesirable) alternative binding sites for gene specific DNA probes.
Development of user interface
The current user interface for the mfold web server is now several years old and would benefit from either partial or total rewriting. The submission of data through forms is still reasonable. A ‘browse’ option for uploading sequence files could easily be added. It would be useful to make it easier to enter constraint information. Even more valuable would be a preprocessor that would check the constraint information for consistency and reasonableness. Up to now, many constrained foldings have failed because of minor errors in constraints.
The output pages could benefit substantially by being redesigned. As a first step, the relatively new ‘sir_graph’ program should replace the older naview program for creating secondary structure plots. It would be desirable to create an interactive structure drawing feature. Such a feature already exists in the stand alone version of the ‘sir_graph’ program. We are fortunate here at RPI to have the cutting edge Academy of Electronic Media; a group that might collaborate with us in creating a really first rate user interface for the web server.
CITING THE MFOLD WEB SERVER
Authors who make use of the mfold web server should cite this article as a general reference and should also include the URL to the entrance page, http://www.bioinfo.rpi.edu/applications/mfold. The web server pages will list additional articles for citation that relate to the free energy parameters that are used and the underlying software.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online and can also be found at MFOLDROOT/www-NAR03/supp.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported, in part, by grant #GM54250 from the National Institutes of Health. I thank Gerald Johns for hardware and systems support in the early days of the server at Washington University. All of the graphics applications (except for naview) were written by Darrin Stewart, who also wrote the ‘cgi’ scripts for interactive viewing of plots. I thank Art Sanderson (Vice President of Research at RPI), for connecting me with the Academy of Electronic Media and for supporting this project; Bill Shumway, for initiating and facilitating interactions with IBM Research; and Alex Yu, who has done so much work in assembling the hardware, organizing the web server layout, porting applications and in keeping the server running day in and day out. Finally, I thank IBM Research for the SUR grant that gave us a thirty-fold increase in computer power for housing this valuable resource.
REFERENCES
- 1.Doty P., Boedtker,H., Fresco,J.R., Haselkorn,R. and Litt,M. (1959) Secondary structure in ribonucleic acids. Proc. Natl Acad. Sci. USA, 45, 482–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fresco J.R., Alberts,B.M. and Doty,P. (1960) Some molecular details of the secondary structure of ribonucleic acid. Nature, 188, 98–101. [DOI] [PubMed] [Google Scholar]
- 3.Borer P.N., Dengler,B., Tinoco,I.,Jr and Uhlenbeck,O.C. (1974) Stability of ribonucleic acid double-stranded helices. J. Mol. Biol., 86, 843–853. [DOI] [PubMed] [Google Scholar]
- 4.Tinoco I., Jr and Uhlenbeck,O.C. (1971) Estimation of secondary structure in ribonucleic acids. Nature, 230, 362–367. [DOI] [PubMed] [Google Scholar]
- 5.Tinoco I. Jr, Borer,P.N., Dengler,B., Levine,M.D., Uhlenbeck,O.C., Crothers,D.M. and Gralla,J. (1973) Improved estimation of secondary structure in ribonucleic acids. Nature New Biol., 246, 40–41. [DOI] [PubMed] [Google Scholar]
- 6.Uhlenbeck O.C., Borer,P.N., Dengler,B. and Tinoco,I.,Jr (1973) Stability of RNA hairpin loops: A6-Cm-U6. J. Mol. Biol., 73, 483–496. [DOI] [PubMed] [Google Scholar]
- 7.Delisi C. and Crothers,D.M. (1971) Prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA, 68, 2682–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterman M.S. and Smith,T.M. (1978) RNA secondary structure: a complete mathematical analysis. Math. Biosci., 42, 257–266. [Google Scholar]
- 9.Waterman M.S. (1978) Secondary structure of single-stranded nucleic acids. In Rota,G.-C. (ed.), Studies in Foundations and Combinatorics number 1 in Advances in Mathematics, Supplementary Studies. Academic Press, NY, pp. 167–212. [Google Scholar]
- 10.Nussinov R., Pieczenik,G., Griggs,J.R. and Kleitman,D.J. (1978) Algorithm for loop matchings. SIAM J. Appl. Math., 35, 68–82. [Google Scholar]
- 11.Nussinov R. and Jacobson,A.B. (1980) Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl Acad. Sci. USA, 77, 6309–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuker M. and Stiegler,P. (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankoff D., Kruskal,J.B., Mainville,S. and Cedergren,R.J. (1983) Fast algorithms to determine RNA secondary structures containing multiple loops. chapter 3. In Sankoff,D. and Kruskal,J.B. (eds), Time Warps, String Edits, and Macromolecules: The Theory and Practice of Sequence Comparison, Addison-Wesley Reading, MA, pp. 93–120. [Google Scholar]
- 14.Zuker M. (1989) Computer prediction of RNA structure. Methods Enzymol., 180, 262–288. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J., Haeberli,P. and Smithies,O. (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res., 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salser W. (1977) Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harbor Symp. Quant. Biol., 42, 985–1002. [DOI] [PubMed] [Google Scholar]
- 17.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.M., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger J.A., Turner,D.H. and Zuker,M. (1989) Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA, 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger J.A., Turner,D.H. and Zuker,M. (1990) Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol., 183, 281–306. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. (1994) Prediction of RNA secondary structure by energy minimization. chapter 23. In Griffin,A.M. and Griffin,H.G. (eds), Computer Analysis of Sequence Data, 25, Part II, Humana Press, Inc., Totowa, NJ, pp. 267–294. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In Barciszewski,J. and Clark,B.F.C. (ed.), RNA Biochemistry and Biotechnology, number 70 in NATO Science Partnership Sub-Series: 3: High Technology. chapter 2, Kluwer Academic Publishers Dordrecht, The Netherlands, pp. 11–43. [Google Scholar]
- 23.Mathews D.H., Andre,T.C., Kim,J., Turner,D.H. and Zuker,M. (1998) An updated recursive algorithm for RNA secondary structure prediction with improved free energy parameters. chapter 15. In Leontis,N.B. and SantaLucia,J.,Jr (eds), American Chemical Society Symposium Series 682, American Chemical Society; Washington, DC, pp. 246–257. [Google Scholar]
- 24.Mathews D.H., Turner,D.H. and Zuker,M. (2000) RNA secondary structure prediction. chapter 11.2. In Beaucage,S., Bergstrom,D.E., Glick,G.D. and Jones,R.A. (eds), Current Protocols in Nucleic Acid Chemistry, John Wiley & Sons; New York, NY, pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter A.E., Turner,D.H., Kim,J., Lyttle,M.H., Müller,P., Mathews,D.H. and Zuker,M. (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA, 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 27.SantaLucia J., Jr (1998) A unified view of polymer, dumbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 29.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 30.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A·C mismatches in DNA: Sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 32.Peyret N., Senevirtne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G, and T·T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 33.Bommarito S., Peyret,N. and SantaLucia,J.,Jr (2000) Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res., 28, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyret N. (2000) Prediction of Nucleic Acid Hybridization: Parameters and Algorithms. PhD Theris, Wayne State University Department of Chemistry, Detroit, MI.
- 35.Cornish-Bowden A. (1985) Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res., 13, 3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. and Jacobson,A. (1998) Using reliability information to annotate RNA secondary structures. RNA, 4, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waugh A., Gendron,P., Altman,R., Brown,J.W., Case,D., Gautheret,D., Harvey,S.C., Leontis,N., Westbrook,J., Westhof,E. et al. (2002) RNAML: A standard syntax for exchanging RNA information. RNA, 8, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker M. and Jacobson,A.G. (1995) ‘Well-determined’ regions in RNA secondary structure prediction. application to small and large subunit rRNA. Nucleic Acids Res., 23, 2791–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruccoleri R. and Heinrich,G. (1988) An improved algorithm for nucleic acid secondary structure display. Comput. Appl. Biosci., 4, 167–173. [DOI] [PubMed] [Google Scholar]
- 40.Rouillard J.-M., Herbert,C.J. and Zuker,M. (2002) OligoArray: Genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrov R.A. and Zuker,M. (2003) Prediction of hybridization and melting for double stranded nucleic acids. Biophys. J., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.