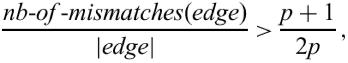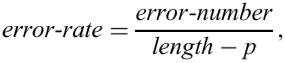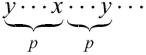Abstract
The presence of repeated sequences is a fundamental feature of genomes. Tandemly repeated DNA appears in both eukaryotic and prokaryotic genomes, it is associated with various regulatory mechanisms and plays an important role in genomic fingerprinting. In this paper, we describe mreps, a powerful software tool for a fast identification of tandemly repeated structures in DNA sequences. mreps is able to identify all types of tandem repeats within a single run on a whole genomic sequence. It has a resolution parameter that allows the program to identify ‘fuzzy’ repeats. We introduce main algorithmic solutions behind mreps, describe its usage, give some execution time benchmarks and present several case studies to illustrate its capabilities. The mreps web interface is accessible through http://www.loria.fr/mreps/.
INTRODUCTION
The presence of repeated sequences is a fundamental feature of genomes. From the genome explorer viewpoint, repeat is the simplest form of regularity and analyzing repeats gives first clues to discovering new biological phenomena in the same way as repeated words give a starting point to deciphering a script written in an unknown language.
Repeats in DNA are commonly classified in interspersed and tandem repeats (1). Beyond a simple syntactical distinction, these two types of repeats usually correspond to different evolutionary mechanisms: interspersed repeats typically result from replicative transposition, while tandem repeats usually result from replication slippage or from certain recombination events, such as unequal crossing-over or unequal sister chromatide exchange. This report focuses on tandem repeats and describes a computer tool that allows to identify them in a very efficient and exhaustive manner, and to represent them in a compact and biologically relevant way.
In eukaryotic genomes, tandem repeats are involved in various regulation mechanisms that are still being discovered. For example, tandem repeats participate in protein binding (2), affect the chromatin structure and are involved in heat-shock inducible expression mechanism (3).
Tandem repeats have been shown to be associated with recombination hot-spots in higher organisms. A relationship was established (4) between the recombination intensity and the density of GT repeats in human chromosomes. Tapper et al. (5) demonstrated an increase of the male recombination level in regions rich in tandem repeats with a pattern size between 10 and 100 bp, in contrast to female recombination that does not present such a correlation.
A well-documented noxious feature of tandem repeats is their involvement in human neurological disorders, such as Huntington's disease, fragile X syndrome, myotonic dystrophy and others (6,7). Those diseases are caused by an abnormal expansion of the number of repeated copies in trinucleotide tandem repeats located in either an intron or an exon of a gene [so-called trinucleotide repeat expansion diseases (TRED) (7)]. Instability of tandem repeats has also been shown to be associated with cancer development (8).
Tandem repeats are conserved in prokaryotes as well, both in plasmids and genomic DNA (9). A correlation has been observed between certain repeats and virulence factors of the bacteria (10). A major application of short tandem repeats is based on the inter-individual variability in copy number of certain repeats occurring in single loci. This feature makes tandem repeats a convenient tool for genetic profiling of individuals (11,12). The latter, in turn, is applied to pedigree analysis and establishing phylogenetic relationships between species, as well as to forensic medicine for instance (13).
In general, our knowledge of the origin and biological function of tandem repeats is still rudimentary. Attempts have been made to collect and systematically store in databases various information on tandem repeats. The mini-satellite database (http://minisatellites.u-psud.fr/) (14) collects and stores short tandem repeats of a certain number of species, computed with the Tandem Repeats Finder tool (15). A more specialized STDR database Short Tandem Repeat DNA Internet Database (http://www.cstl.nist.gov/biotech/strbase/) focuses on short tandem repeats involved in genetic mapping and identity testing. The Tandem Repeat Data Base (http://tandem.biomath.mssm.edu/cgi-bin/trdb/trdb.exe) (TRDB) is an integrated web-based environment that allows to compute, store and manipulate tandem repeats (comparison, forming clusters, displaying statistics), as well as some other types of repeats.
Related work
Software programs for finding tandem repeats in genomic sequences are available in the EMBOSS package (16):
equicktandem is a simple statistically-based program that identifies tandem repeat structures in DNA, for each pattern size up to a given bound. A possible consensus of the repeated pattern can then be computed by ETANDEM program.
RepeatMasker (A.F.A. Smit and P. Green, RepeatMasker, http://ftp.genome.washington.edu/RM/RepeatMasker.html) is a well-known software for ‘masking’ repetitive and low complexity regions in DNA sequences, in order to suppress the ‘noise’ introduced by those regions in the search for similarity regions. As a part of this task, RepeatMasker identifies tandem repeats of a very limited type (certain micro-satellites). From the computer science perspective, designing efficient algorithms for finding tandem repeats in texts has been a subject of extensive research during the last decade. Algorithms were proposed (17) for finding all approximate tandem repeats in the case of substitution errors only (Hamming distance) and in the case when indels are allowed in addition (edit distance). The theoretic worst-case time complexity of proposed algorithms is O(nk log(k) log(n)+S) in the case of edit distance and O(nk log(n/k)+S) in the case of Hamming distance (k is the maximal distance between two tandemly repeated copies, and S is the number of repeats found). More recently, this work gave rise to another algorithm for finding tandem arrays of a certain type (18).
Other methods for finding tandem repeats have been proposed that, to our knowledge, have not been implemented in publicly available software. One method (19) finds tandemly repeated preselected patterns, with the aim of compressing the dna sequence in order to estimate its ‘information quantity’. An algorithm (20) proposes a heuristics for finding tandem repeats with an a priori specified size of repeated unit. Another proposed algorithm (21) uses a general combinatorial framework of ‘consensus repeat’ and makes use of some heuristic filtering steps to avoid exponential blow-up in time complexity.
A statistically founded heuristic algorithm, together with the associated Tandem Repeats Finder software has been presented (15). The general approach can be compared with the one used by the well-known Blast algorithm: it is based on first collecting the information about short (in practice, 5–7 bp) exact repeated fragments (seeds), and then extending those fragments, according to statistically-founded criteria, into approximate tandemly-repeated units.
To complete our survey, we mention two more recently published tools. One of them (23) is based on a seed-extension technique, similar to another (15), and tries to identify tandem repeats with an additional pattern structure. Another one, called TROLL (22), is a program for finding exact tandemly repeated copies of a priori specified patterns.
In this paper, we present a new algorithmic approach for finding approximate tandem repeats and a software program, named mreps, based on this approach. From the algorithmic viewpoint, this approach uses a new combined combinatorial/heuristic paradigm which differs from those used before (15,17,18).
In very general terms, it first finds, in an exhaustive manner, all approximate tandem arrays (under the Hamming distance model) which verify a certain combinatorial definition, similar to others (17,18).
This stage is done through a very efficient combinatorial algorithm (24) running in time O(nk log(k)+S), which improves on the algorithms (17) (for the Hamming distance case) and others (18). The repeats found are then further processed in order to eliminate redundancy, to get rid of certain artifacts of the mathematical definition used at the first stage, and to filter out statistically insignificant repeats. To summarize, our approach allows to compute tandem repeats in a flexible and biologically relevant way without renouncing the exhaustivity.
From a practical viewpoint, a distinguished feature of mreps is that it has no limitation whatsoever on the size of the repeated pattern. A single run of mreps allows to compute tandem repeats with all possible pattern sizes. Therefore, mreps is a universal tool for detecting all types of tandem repeats, from micro-satellites up to huge tandem duplications.
This makes a crucial difference with approaches that require specifying a possible pattern or its size a priori (19,20,22). This also improves on approaches (15,23) which usually have practical limitations on the pattern size, due to the memory size used by the algorithm. (Current version 3.2.1 of Tandem Repeats Finder limits the pattern size by 2000 bp, while later, we will show an example of tandem repeat with a much bigger pattern size identified by our program.)
Another important feature of mreps is that it allows to compute ‘loose’ repeats, that is repeats with big variability between repeated copies. This is currently done in a non-traditional way: instead of introducing a scoring function and specifying a threshold score value for found repeats, the user specifies a resolution parameter that determines the ‘fuzziness’ of found repeats. In metaphoric terms, this parameter plays the role of ‘magnifying glass’ allowing to ‘zoom in’ and ‘zoom out’ the genomic sequence and to find respectively more accurate or loose repeats. We will illustrate this feature later.
METHODS
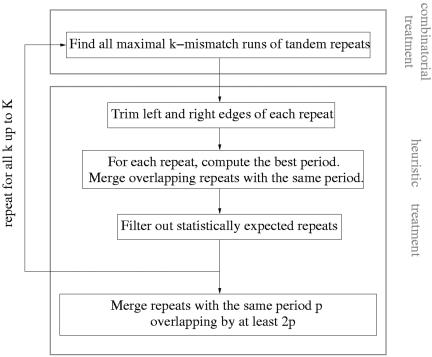
In this section, we describe the main steps of the mreps algorithm. The structure of the algorithm is shown in Figure 1. It consists of two main parts: the first one (upper frame) collects certain repeated sequences through an efficient combinatorial algorithm. Those sequences serve as ‘raw material’ for the second part (lower frame), which applies to them an heuristic treatment in order to obtain biologically relevant repeats.
Figure 1.
Flowchart of the algorithm.
Combinatorial treatment
The core of the program is an efficient combinatorial algorithm for finding all repetitive structures of a certain kind in a given sequence. Mathematical foundations of this algorithm have been described in publications (24,25), and we only outline its main principles here.
Let us first introduce some basic terminology. An exact repeat is a string that can be represented as a smaller string repeated contiguously twice or more. For example, ACACAC is a repeat, as it can be represented as string ac repeated three times. The length of the repeated pattern is called the period (2 for the case of ACACAC), and the number of pattern copies is called the exponent (3 for ACACAC). If the exponent is 2, the repeat is usually called a tandem repeat. Importantly, however, the exponent of a repeat is not necessarily an integer number. For example, ACACAC is considered as a repeat with exponent 2.5, as it can be thought of as ‘AC repeated 2.5 times’. Note that in the case of non-integer exponent, a pattern associated with the repeat is not defined uniquely: acaca can be equally written as (ac)2a, or as a(ca)2. Considering non-integer exponents is useful for several reasons: this provides a more consistent model of tandem arrays and even allows to speed up search algorithms (25); on the other hand, considering repeats with non-integer exponents is biologically relevant, as they do occur in genomic sequences and the ‘repeated unit’ is often artificially defined. Note that terms period and exponent (sometimes called order) are borrowed from the area of word combinatorics, where tandemly repeated structures (called periodicities) have been studied for a long time (see Chapter 8 in 26).
Given a sequence, our goal is to identify all repeats occurring in it. It is natural to always extend each repeat to the right and to the left as much as possible, as far as the periodicity is respected. We call such repeats maximal. For example, aatcatcatatagc contains the repeat atc atc at (period 3, exponent 8/3). This is a maximal repeat, as it cannot be further extended to the left/right preserving the period 3.
Maximality is an important idea that will be followed throughout our development. It provides a natural definition of repeats occurring in a sequence (it is natural to consider atata as a single repeat, rather than to consider two distinct repeats atat and tata). Another important consideration is that the set of all maximal repeats in a sequence comprises all tandem repeats occurring in this sequence. Note that the maximality of a repeat is not a property of the repeat itself but of the context it occurs in, as the same repeat can be maximal in one context and non-maximal in another.
Since we always have to tolerate errors between repeated copies, the next step is to extend the notion of maximal repeat to the approximate case. The basic notion we use is called the maximal run of k-mismatch tandem repeats. Given an error threshold k, a run of k-mismatch tandem repeats of period p is a string such that any substring of size 2p is a tandem repeat with at most k substitution errors. For example, gcac acac ag is a run of 1-mismatch tandem repeats of period 4 (the corresponding tandem repeats are gcac acac, caca caca, and acac acag). As in the case of exact repeats, the maximality condition means that each considered run of k-mismatch tandem repeats is extended to the left/right as far as it still verifies the definition. As an example, consider the sequence gcgatgaagtgggc. The substring cga tga ag is a maximal run of 1-mismatch tandem repeats of period 3, as there is at most one mismatch in each of the tandem repeats cga tga, gat gaa, and atg aag. On the other hand, this run is maximal since extending it to the left/right by one letter introduces the second mismatch in the corresponding tandem repeat.
From the algorithmic point of view, a remarkable feature of exact and approximate repeats, as defined above, is that those structures can be found extremely fast in a given sequence. Formally, algorithms designed in (24,25) allow to identify all exact maximal repeats in a sequence of length n in time O(n), and all maximal runs of k-mismatch tandem repeats in time O(nk log(k)+S) (S the number of repeats found). Those algorithms are based on advanced string processing techniques and describing them is beyond the scope of this paper. The existence of so efficient (linear in the sequence length) algorithms capable of computing so rich information about tandemly repeated patterns in the sequence is at the origin of the mreps program.
Computing maximal runs of k-mismatch tandem repeats is, on its own, a good way to detect tandemly repeated sequences in genomes. However, several limitations of this approach have still to be lifted in order to obtain a fully adequate algorithm for detecting relevant tandem repeats in genomic DNA. One such limitation is that the parameter k, bounding the maximal number of mismatch errors between two tandemly repeated copies, should be specified beforehand. As k is an absolute number that cannot be expressed as a fraction of the period, this implies that the user has to have an a priori knowledge of the period of repetitions she/he is looking for in order to be able to specify a proper number of allowed errors. This restricts the power of the approach.
Among other limitations, there are certain artifacts of the definition of maximal runs of k-mismatch tandem repeats, that produce some unnatural ‘side effects’ in computed repeats (we will explain this below in more details). Also, the definition sometimes turns out to be too rigid: for example, two long stretches of a's separated by just one c have to be viewed as two distinct repeats and not as one repeat with a substitution error. Another illustration of the rigidity is that indels (insertion and deletion errors) are not directly accounted for. Finally, another ‘limitation’ is that all repetitions are output, including those, like simply aa, which have obviously no significance in the genome.
A further processing is then needed to deal with all those limitations. This justifies the heuristic treatment (lower frame in Fig. 1) that we will describe now.
Heuristic treatment
As shown in Figure 1, the heuristic part is composed of several steps that we describe.
Trimming edges
The required maximality of runs of k-mismatch tandem repeats implies that errors are sometimes ‘artificially’ added to the ends of a repeat, in order to always reach k mismatches on the extremities. For example, for k=1, we could find the repeat gacacat with period 2, in which bases g t are obviously redundant. Therefore, we process repeats in order to get rid of this artifact of the mathematical definition.
This processing is subtler than just cutting off those bases, which form a mismatch. We cut off, from each side, the longest edge that satisfies the inequality
where nb-of-mismatches(edge) is the number of mismatches introduced by edge and p is the period. This formula allows to cut off an edge containing ‘too many’ mismatch errors in it, and on the other hand, insures that the remaining part of the repeat is sufficiently big to still make sense. For example, repeat gaaggac aacggac agcggac aatg found for k=2 and p=7 will be reduced to ggacaacggacagcggaca, although among the three bases deleted from each end there are only two which produce a mismatch.
Computing the best period and merging
The primary goal of this step is to cope with another artifact of the definition of runs of k-mismatch tandem repeats: the same region can be represented as several distinct runs of tandem repeats, computed for different periods. For example, for k=1, the sequence atatatatataaa is computed as a repeat with period 2 (at at at at at aa a), with period 4 (atat atat ataa a), and with period 6 (atatat atataa a). We then have to determine the ‘best’ period of a repeat and for this, we need to measure the quality of a repeat. We measure it by the error-rate:
where length is the length of the repeat, and error-number is the number of mismatches in the repeat, except that two mismatches formed by a nucleotide are counted for one, if the mismatched nucleotides are the same. In other words, the following situation is accounted for one mismatch:
For example, the above repeat has the error rate 1/11=0.09 for p=2, 1/9=0.11 for p=4, and 1/7=0.14 for p=6.
For each repeat with a period p, we compute the period from the interval [1,…, p] realizing the minimal error rate. This period is considered as the true period of the repetition. For example, each of the three above repeats has the true period 2, which makes of them a single actual repeat.
After computing the true period of each repeat, repeats having the same period and overlapping by at least two periods are merged into a single repeat.
An important consequence of this step is that it changes the meaning of the k parameter: it does not specify anymore the maximal number of mismatches between two adjacent copies of length p. For example, consider the repeat atatgg atatag atat with period 6 identified for k=1. Its true period will be computed as 2 (at at at gg at at ag at at at at), although there are two mismatches between adjacent at and gg. From now on, we will call it the resolution parameter, as its meaning corresponds to the ‘degree of fuzziness’ of computed repetitions. Increasing the resolution allows to identify large fuzzy repeats, that cannot be ‘seen’ with small resolution values.
Filtering out statistically expected repeats
The next important step consists in filtering out repeats which are statistically expected, keeping only those that are statistically significant. This step is guided by the following principle, general in bioinformatics: only those observations that have a small probability to appear as a random event can be biologically significant. A formal implementation of this principle requires introducing a formal probabilistic model of corresponding random events. In our case, we would need a probabilistic model of DNA sequence (usually, a Bernoulli or Markov model) and a method for estimating the probability of observing a given repeat in a random sequence generated by the model.
Unfortunately, a corresponding theory is not readily available, and elaborating such a method (especially for the case of repeats with errors) is a non-trivial problem, still largely open. Therefore, we used a computer simulation, and experimentally characterized repeats typically occurring in a ‘random genomic sequence’. Here is the way we did it.
Intuitively, there are two reasons why a repeat can be statistically insignificant: one is its small length, and another is its high error rate. In the first case, a repeat might be perfect but too short (like aaa for example), and thus having big chances to appear in a random sequence. In the second case, the repeat might be too ‘noisy’, and so, indistinguishable from the ‘background noise’.
For these reasons, we introduced two distinct filters: a length filter and a quality filter. The length filter we came up with is very simple: it eliminates any repeat with the length smaller than p+9 ( p the period). Our experiments showed that this simple filter discriminates short repeats occurring randomly very well.
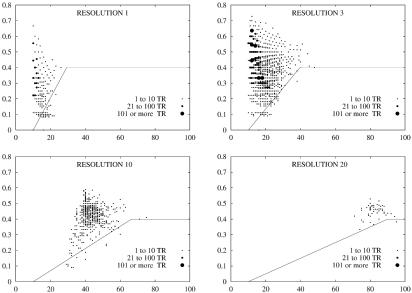
After applying the length filter, we need to discriminate random repeats on the basis of their error rate, and depending on their length and on the resolution for which the corresponding repeat has been computed. For this purpose, we first obtained several pseudo-random DNA sequences by shuffling real genomic sequences, i.e. by mixing up its letters in a pseudo-random way. Then, we run mreps on those sequences, with different resolution values, and registered the parameters (length and error rate) of the repeats remaining after applying the length filter. On the basis of these data, we established an empirical threshold function in order to discriminate, in a most accurate way, the repeats typically found in a random sequence. The results of this procedure are illustrated on Figure 2.
Figure 2.
Threshold error rate for filtering our statistically expected repeats (quality filter). Typical simulation results, together with the threshold error rate, are shown for values 1, 3, 10, 20 of the resolution parameter. Each picture shows the threshold error rate function and the error rate of repeats found in a pseudo-random DNA sequence. The three thickness levels of points distinguish the number of found repeats with the same length and error rate (see the legend).
Gathering the results
The goal of the final step is to put together repeats found for different resolution values. This is done by iterating the whole algorithm for all resolution values, up to a certain value K that defines the final resolution level (Fig. 1).
Groups of collected repeats with the same period are then processed again so that repeats overlapping by at least two periods are merged into a single one.
RESULTS
Software
The mreps software is written in ANSI C and is distributed under GPL licence. It is currently run under Linux, SunOS, Digital Unix and Windows systems. The sequence to be processed can be stored in a file or be input directly in the command line. The sequence file can be in plain or fasta format, in the latter case it can contain one or several sequences. The following parameters can be specified: start and end positions of the region to be processed; a length interval, a period interval, and a minimal exponent of the repetitions to report; a resolution level. Besides, there exists a sliding window option allowing to process the sequence by overlapping sliding windows of a given size. Currently, this option is used for genomic sequences bigger than 30 Mb (e.g. whole chromosomal sequences), as smaller sequences can be processed as a whole in a single run of mreps on a regular computer. Another option allows to suppress the filtering of small repeats by using a much weaker filter for small length values. This can be useful in some situations, when small repeats are of interest too.
mreps execution times for different sequence sizes and resolution values are given in Table 1.
Table 1. Execution time (in seconds) depending on the sequence size and the resolution value.
| Size resolution | 10 Kb | 10 Kb | 2 Mb | 4.65 Mb |
|---|---|---|---|---|
| 0 | 0.34 | 0.38 | 2.38 | 5.14 |
| 1 | 0.36 | 0.61 | 683 | 15.63 |
| 3 | 0.37 | 0.86 | 13.10 | 31.81 |
| 10 | 0.47 | 2.07 | 41.79 | 99.92 |
| 20 | 0.68 | 4.49 | 100.34 | 244.79 |
The output is a list of all repeats, each of which being characterized by the following parameters: start and end positions of the repeat in the sequence, overall size of the repeat, period, exponent, error level, the repeat sequence itself.
The distribution of mreps is available at its web site (http://www.loria.fr/mreps/). A web-based interface of mreps is available at the same site too. In addition to the previous information, the web-based interface allows the user to display an alignment of each repeat showing the mismatch errors, as well as the nucleotide composition of the repeat.
Case studies
We now present several case studies we carried out using mreps in order to illustrate its capabilities.
Cluster of tandem repeats in Neisseria meningitidis MC58
N.meningitidis (Meningococcus) is a virulent bacterium that causes septicemia and meningitidis diseases. The genome of N.meningitidis strain MC58 (serogroup B), of size 2 272 351 bp (27), contains many repetitive elements of a very broad size range (from several bases to kilobases), some of which are involved in antigenic variation and genome fluidity. A variation of the exponent (copy number) of micro-satellite tandem repeats modifies the transcriptional status of genes associated with surface cellular proteins, suggesting an effect in pathogenicity (10). Short palindromic repetitions are involved in chromosomic rearrangement, as observed in N.meningitidis genome. This phenomenon concerns in particular the deletion of porA gene that has a cluster of surrounding palindromic repeats responsible for the gene deletion (28).
As a result of running mreps on the whole genome of N.meningitidis MC58, we identified a tandem repeat which is itself repeated over 30 times throughout the whole genome. The tandem repeat is of period 7 and size 31, and its occurrences have a strongly conserved consensus ttttagg tttctga ttttggt tttctgt ttt. It was identified by running mreps with resolution 3. A closer analysis revealed that the repeat is a part of a larger conserved repeated region of size about 100 bp. The copies of this region are conserved with about 75% of similarity, and the aforementioned tandem repeat represents a highly conserved ‘core’ of the region.
All copies of the tandem repeat occur in intergenic regions. Some of them are close to genes that are directly involved in pathogenicity, such as a gene of competence protein ComA (gene NMB0702), or genes of the PilS cassette (genes NMB0019-26). This suggests that the found tandem repeat may act as a binding site for a protein involved in transcription regulation.
A further analysis could be to demonstrate whether there exists a co-regulation of the genes containing the tandem repeat in their promoter region. Another hypothesis is that the occurrences of that tandem repeat are related to recombination sites, as it was already conjectured for other repeated sequences in Neisseria species (29).
Large tandem duplication in N.meningitidis MC58
In the same N.meningitidis MC58 genome, mreps reveals a huge tandem duplication: a 32 036 bp sequence repeated more than twice without errors ( positions 1 135 353 to 1 199 546). There are 36 protein-coding genes located in the repeated region. Elucidating a role in the genome and the nature of this huge repeat is an open issue that requires a further study.
Note that identifying this repeat took mreps only about 2 s on a regular 1 GHz Pentium IIITM computer.
Polymorphic STR in human genome
Short Tandem Repeats are widely used as genetic markers in forensic applications. This usage hinges on the fact that the size of micro- and mini-satellites is highly polymorphic among individuals, which makes them a perfect fingerprinting tool. Besides, STRs are particularly easy to amplify by PCR and remain stable even when the DNA is decomposed, as in post-mortal tissues.
Locus D21S11 is a complex STR located on the human chromosome 21. The repeat has a complex irregular multi-period repeat structure, e.g. one of its alleles allele 28, see http://www.cstl.nist.gov/biotech/strbase/ is (tcta)4(tctg)6(tcta)3 ta(tcta)3 tca(tcta)2 tcca ta(tcta)10.
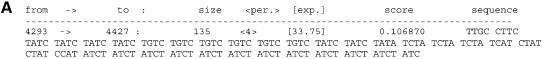
We were able to detect this locus de novo by running mreps on the human sequence AP000433 (57 399 bp) containing the locus. In particular, running mreps with resolution 5 identifies the repeat which matches the above allele as shown in Figure 3A.
Figure 3.
Excerpts of mreps output. (A) Resolution 5 and (B) resolution 18.
This example illustrates, in particular, the capacity of mreps to detect repeats with irregular and ‘fuzzy’ repetitive structure.
Repeats in flocculation genes in Saccharomyces cerevisiae
Repeats in flocculation genes in the S.cerevisiae genome is an interesting example of tandem arrays occurring inside coding sequences, and it has been used as a test example for tandem repeat finding software.
Chromosome 1 of S.cerevisiae (230 203 bp) contains intron- free FLO9 and FLO1 genes located on complementary strands and on opposite ends of the chromosome (positions 24 001–27 969 and 203 389–208 002, respectively). The two genes have a very high similarity, and both contain a 135 bp element tandemly repeated over 10 times in FLO9 and almost 18 times in FLO1. Figure 3B shows an excerpt of mreps output (run with resolution 18) revealing those repeats:
Thus, mreps identifies in both genes a 135 bp pattern tandemly repeated over 10 times, with exactly the same overall size of 1423 bp. Aligning those regions reveals their almost perfect similarity. In addition, mreps finds three other overlapping repeats of a 135 bp pattern. Together with the ‘main’ repeat, they can actually be seen as parts of a single tandem repeat structure, with a DNA pattern that changes several times along the repeat. On the protein level, the repeat is translated to a domain of 45 amino acids tandemly repeated almost 18 times, with several ‘jumps’ in the similarity of repeated copies.
As reported (15), a 135 bp tandem repeat of the same kind occurs in the FLO5 gene located on chromosome 8 (562 639 bp, positions 525 388–528 615). There are also other tandem repeats which are conserved across those genes (15) and can be identified by mreps.
In chromosome 8, mreps was able to discover a sequence of 1998 bp tandemly repeated twice without errors (positions 212 254–216 251). In particular, this sequence contains a metallothionein gene that occurs then in two proximate copies (YHR053C and YHR055C).
DISCUSSION
In this paper, we have presented a new software tool for a de novo identification of tandemly repeated structures in a genomic sequence. A remarkable feature of the program is its efficiency, as it is able to identify all types of tandem repeats through in a single and fast run on a whole genomic sequence.
Another important characteristic of the program is its ability to identify loose repeats through a special resolution parameter. These features make of mreps a flexible and powerful tool that could be used for locating a particular type of tandem repeats, or to make a fast genome-wide analysis of tandemly repeated patterns. The software is open-source; it can be freely downloaded or queried through a web-based interface.
Acknowledgments
ACKNOWLEDGEMENTS
We are greatly indebted to Mathieu Giraud who was at the origin of mreps implementation. We thank Ralph Rabbat and Mariana-Liliana Ibanescu for their contributions at certain stages of mreps design. We thank Gary Benson for helpful discussions, Laurent Noé for his help in analyzing repeated elements in Neisseria and Marie-Dominique Devignes for commenting on the manuscript. At different stages, this work was supported by the REMAG working group of INRIA and the French Action Spécifique ‘Algorithmes et Séquences’ of CNRS. Large part of this work has been done during stays of Roman Kolpakov at LORIA in 2000–2002, supported by INRIA.
REFERENCES
- 1.Brown T.A. (1999) Genomes. BIOS Scientific Publishers, Oxford, UK. [Google Scholar]
- 2.Richards R.I., Holman,K., Yu,S. and Sutherland,G.R. (1993) Fragile X syndrome unstable element, p(CCG)n, and other simple tandem repeat sequences are binding sites for specific nuclear proteins. Hum. Mol. Genet., 2, 1429–1435. [DOI] [PubMed] [Google Scholar]
- 3.Leibovitch B.A., Lu,Q., Benjamin,L.R., Liu,Y., Gilmour,D.S. and Elgin,S.C. (2002) GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol., 22, 6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majewski J. and Ott,J. (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res., 10, 1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tapper W.J., Morton,N.E., Dunham,I., Ke,X. and Collins,A. (2001) A sequence-based integrated map of chromosome 22. Genome Res., 11, 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caskey C.T., Pizzuti,A., Fu,Y.H.,Jr, Fenwick,R.G. and Nelson,D.L. (1992) Triplet repeat mutations in human disease. Science, 256, 784–789. [DOI] [PubMed] [Google Scholar]
- 7.Mitas M. (1997) Trinucleotide repeats associated with human disease. Nucleic Acid Res., 25, 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibodeau S.N., Bren,G. and Schaid,D. (1993) Microsatellite instability in cancer of the proximal colon. Science, 260, 816–819. [DOI] [PubMed] [Google Scholar]
- 9.Van Belkum A., Scherer,S., Van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in procaryotic genomes. Microbiol. Molec. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders N.J., Jeffries,A.C., Peden,J.F., Hood,D.W., Tettelin,H., Rappuoli,R. and Moxon,E.R. (2000) Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol., 37, 207–215. [DOI] [PubMed] [Google Scholar]
- 11.Jeffreys A.J., Wilson,V. and Thein,S.L. (1985) Individual-specific ‘fingerprints’ of human DNA. Nature, 316, 76–79. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y., Leppert,M., O'Connell,P., Wolff,R., Holm,T., Culver,M., Martin,C., Fujimoto,E., Hoff,M., Kumlin,E. et al. (1987) Variable number of tandem repeat (VNTR) markers for human gene mapping. Science, 235, 1616–1622. [DOI] [PubMed] [Google Scholar]
- 13.Butler J.M. (2001) Forensic DNA Typing: Biology and Technology Behind STR Markers. Academic Press, London, UK. [Google Scholar]
- 14.Le Fleche P., Hauck,Y., Onteniente,L., Prieur,A., Denoeud,F., Ramisse,V., Sylvestre,P., Benson,G., Ramisse,F. and Vergnaud,G. (2001) A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol., 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson G. (1999) Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: The european molecular biology open software suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 17.Landau G. and Schmidt,J. (1993) An algorithm for approximate tandem repeats. In Apostolico,A., Crochemore,M., Galil,Z., and Manber,U. (eds), Proceedings of the 4th Annual Symposium on Combinatorial Pattern Matching, Number 684 in Lecture Notes in Computer Science, Springer-Verlag, Berlin. Padova, Italy, pp. 120–133. [Google Scholar]
- 18.Landau G., Schmidt,J. and Sokol,D. (2001) An algorithm for approximate tandem repeats. J. Comp. Biol., 8, 1–18. [DOI] [PubMed] [Google Scholar]
- 19.Rivals E., Delgrange,O., Delahaye,J.-P., Dauchet,M., Delorme,M.-O., Hénaut,A. and Olivier,E. (1997) Detection of significant patterns by compression algorithms: the case of approximate tandem repeats. Comput. Appl. Biosci., 13, 131–136. [DOI] [PubMed] [Google Scholar]
- 20.Benson G. and Waterman,M.A. (1994) method for fast database search for all k-nucleotide repeats. Nucleic Acids Res., 22, 4828–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagot M.-F. and Myers,E.W. (1998) Identifying satellites in nucleic acid sequences. In Istrail,S., Pevzner,P. and Waterman,M., (eds), Proceedings of the 2nd Annual International Conference on Computational Molecular Biology (RECOMB 98), ACM Press, pp. 234–242. [Google Scholar]
- 22.Castelo A.T., Martins,W. and Gao,G.R. (2002) TROLL—tandem repeat occurrence locator. Bioinformatics, 18, 634–636. [DOI] [PubMed] [Google Scholar]
- 23.Hauth A.M. and Joseph,D.A. (2002) Beyond tandem repeats: complex pattern structures and distant regions of similarity. Bioinformatics, 18, S31–S37. [DOI] [PubMed] [Google Scholar]
- 24.Kolpakov R. and Kucherov,G. (2001) Finding approximate repetitions under Hamming distance. In Meyer auf der Heide,F. (ed.), 9th European Symposium on Algorithms (ESA 2001), Aarhus, Denmark, Volume 2161 of Lecture Notes in Computer Science, pp. 170–181. [Google Scholar]
- 25.Kolpakov R. and Kucherov,G. (1999) Finding maximal repetitions in a word in linear time. In Proceedings of the 1999 Symposium on Foundations of Computer Science, New York (USA), IEEE Computer Society, pp. 596–604. [Google Scholar]
- 26.Lothaire M. (2002) Algebraic Combinatorics on Words. Cambridge University Press. [Google Scholar]
- 27.Tettelin H., Saunders,N.J., Heidelberg,J., Jeffries,A.C., Nelson,K.E., Eisen,J.A., Ketchum,K.A., Hood,D.W., Peden,J.F., Dodson,R.J. et al. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science, 287, 1809–1815. [DOI] [PubMed] [Google Scholar]
- 28.Van der Ende A., Hopman,C.T. and Dankert,J. (1999) Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect Immun., 67, 2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Correia F.F., Inouye,S. and Inouye,M. (1986) A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J. Bacteriol., 167, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]