Abstract
In 1997, 18 cases of influenza in Hong Kong (bird flu) caused by a novel H5N1 (chicken) virus resulted in the deaths of six individuals and once again raised the specter of a potentially devastating influenza pandemic. Slaughter of the poultry in the live bird markets removed the source of infection and no further human cases of H5N1 infection have occurred. In March 1999, however, a new pandemic threat appeared when influenza A H9N2 viruses infected two children in Hong Kong. These two virus isolates are similar to an H9N2 virus isolated from a quail in Hong Kong in late 1997. Although differing in their surface hemagglutinin and neuraminidase components, a notable feature of these H9N2 viruses is that the six genes encoding the internal components of the virus are similar to those of the 1997 H5N1 human and avian isolates. This common feature emphasizes the apparent propensity of avian viruses with this genetic complement to infect humans and highlights the potential for the emergence of a novel human pathogen.
Recurrent epidemics of influenza are fairly predictable annual events. In contrast, the future occurrence of a pandemic, caused by the emergence of a novel influenza A subtype against which the population has little or no immunity, is unpredictable as to timing or identity of the prospective agent. Influenza A viruses of aquatic birds comprise a diverse mix of antigenic subtypes [including 15 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes] and represent a large reservoir/source of novel antigens to which the human population is naïve (1, 2). The high species specificity of these viruses restricts transmission to mammalian species to a relatively rare event and only a few subtypes have circulated in mammalian species for extended periods. The consequences of interspecies transmission can be devastating. For example, an epizootic in harbor seals on the east coast of the U.S. in 1980 caused by an H7N7 virus resulted in mortality of some 600 seals (3). There have been several recent examples of the introduction of avian viruses into pig populations in different parts of the world, increasing the genetic diversity of swine viruses (4, 5).
Three pandemics occurred during the 20th century. The devastating pandemic of 1918–1919, which caused estimated deaths of some 50 million people worldwide, apparently followed the introduction of an avian H1N1 virus (6). The pandemics of 1957 and 1968 were the result of genetic reassortment whereby the circulating human virus acquired novel antigens, H2 and N2 in 1957 and H3 in 1968, from an avian source (7, 8). Subsequent pandemic threats have been sparked by deaths caused by unusual influenza infections. In early 1976, infection by a classical swine H1N1 virus caused the death of an army recruit at Fort Dix (9). In 1997, a highly pathogenic fowl plague (H5N1) virus claimed the lives of six of its eighteen victims in Hong Kong before the source of infection was removed by depopulation of the live poultry markets at the end of December (10–12). Surveillance of influenza viruses circulating in poultry during this outbreak in November and December identified several subtypes in addition to H5N1, of which H9N2 was the most frequent (13); there was no evidence of human infection by H9N2 viruses at that time despite intense surveillance. The isolation of H9N2 viruses from pigs in Hong Kong in August 1998 (D. Markwell and K. Shortridge, unpublished results) raised concern about their possible transmission to humans. Isolation in July and August of H9N2 viruses from five patients with influenza-like illness in the Guangdong province of China was reported early in 1999 (14).
In March 1999, two cases of mild influenza in Hong Kong were shown to be caused by infection with influenza A H9N2 viruses (15, 16). The two girls, aged 4 years and 13 months, lived in Kowloon and Hong Kong Island, respectively, and were admitted to different hospitals in early March; there is no known link between the two cases. Here, we describe the antigenic and genetic characteristics of the viruses and show that they are very closely related to a H9N2 virus isolated from a quail during the 1997 H5N1 outbreak in Hong Kong. Because their six internal genes are similar to those of the H5N1 viruses, the H9N2 viruses represent the reappearance of the H5N1 viruses in a different guise. Circulation of these viruses in domestic birds in southern China continues to pose a serious public health threat.
Materials and Methods
Viruses.
A/Hong Kong/1073/99 (A/HK/1073/99) and A/Hong Kong/1074/99 (A/HK/1074/99) were isolated on March 5, 1999 from nasopharyngeal aspirates from a 4-year-old girl and a 13-month-old girl, respectively, by inoculation onto and subsequent passage in cultures of Madin–Darby canine kidney cells, in the presence of 2.5 μg/ml trypsin.
A/quail/Hong Kong/G1/97 (Qa/HK/G1/97), A/chicken/Hong Kong/G9/97 (Ck/HK/G9/97), and A/duck/Hong Kong/Y280/97 (Dk/HK/Y280/97) were isolated in Hong Kong in December 1997 and A/swine/Hong Kong/9/98 (Sw/HK/9/98) is one of two viruses isolated from pigs sampled in April 1998; they were passaged in the allantoic cavity of 10-day-old fertile hen eggs (13).
Antisera.
Both postinfection chicken sera and ferret sera were prepared as described (13, 17). Postinfection ferret antisera against A/HK/1073/99 and A/HK/1074/99 were obtained from J. Wood, National Institute for Biological Standards, South Mimms, U.K.
Antigenic Analyses.
Hemagglutination inhibition and neuraminidase inhibition tests were performed as described (17).
Rimantadine Sensitivity.
Susceptibility of virus replication to inhibition by rimantadine (0.01–1 μg/ml) was determined by ELISA, as described (18), by using appropriate reference ferret antiserum; drug-resistant control viruses showed no significant inhibition over this concentration range.
Gene Sequencing and Analyses.
Virus RNA was obtained from samples of infected cell culture fluid or allantoic fluid by phenol-chloroform extraction and ethanol precipitation. Reverse transcription–PCR used primers (sequences are available on request) specific for each of the eight RNA segments. PCR products were purified by agarose gel electrophoresis and a Geneclean II kit (Bio 101) and sequenced by using an ABI Prism dye terminator cycle sequencing kit and an ABI Model 377 DNA Sequencer (Perkin-Elmer, Applied Biosystems).
Sequence data were edited and analyzed by using the wisconsin sequence analysis package Ver. 8 (GCG). Phylogenetic analyses used paup (Phylogenetic Analysis Using Parsimony, Ver. 4.0, Swafford, Illinois Natural History Survey, Champaign, IL). The nucleotide sequences determined in this study are available from GenBank under accession numbers AJ404735, AJ404736, AJ289871–AJ289874, AJ278646–AJ278649, and AJ404626–AJ404637.
Results
Antigenic Characteristics.
The two viruses, A/HK/1073/99 and A/HK/1074/99, were identified initially by hemagglutination inhibition and neuraminidase inhibition tests by using hyperimmune rabbit antisera to HA subtypes 1–14 and NA subtypes 1–9 to be of the H9N2 subtype. Cells in the clinical specimens were shown by immunofluorescence with antinucleoprotein (anti-NP) antibody to be positive for influenza A NP. Furthermore, the demonstration that sera from the 4-year-old girl contained significant titers of anti-H9-neutralizing antibody provided further confirmation of the authenticity of infection by an H9 influenza A virus (W.L., unpublished results).
The antigenic relationships between the HAs of the two human isolates and H9N2 viruses recently isolated in Hong Kong from avian and porcine species were investigated by hemagglutination inhibition by using strain-specific postinfection chicken and ferret antisera. The HAs of the two human viruses were antigenically indistinguishable (by using ferret antisera to both viruses) and were closely related to the HA of the quail virus Qa/HK/G1/97 (Table 1). These three viruses were clearly distinguishable from the closely related chicken and swine H9N2 isolates, Ck/HK/G9/97 and Sw/HK/9/98, respectively; the two groups of viruses showed very limited crossreactivity in hemagglutination inhibition tests with ferret antisera.
Table 1.
Antigenic relationships between the hemagglutinins of H9N2 viruses
| Viruses | Hemagglutination inhibition titer*
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hyperimmune rabbit sera
|
Postinfection chicken sera
|
Postinfection ferret sera
|
|||||||
| Ty/Wis 1/66 | Ck/HK G9/97 | Ty/Wis 1/66 | Qa/HK G1/97 | Ck/HK G9/97 | A/HK 1073/99 | Qa/HK G1/97 | Ck/HK G9/97 | Sw/HK 9/98 | |
| Ty/Wisconsin/1/66 | 1280 | < | 2560 | 80 | 80 | < | < | < | < |
| A/Hong Kong/1073/99 | 320 | < | 640 | 1280 | 640 | 1280 | 320 | < | < |
| A/Hong Kong/1074/99 | 320 | < | 640 | 2560 | 640 | 1280 | 320 | < | < |
| Qa/Hong Kong/G1/97 | 160 | < | 320 | 2560 | 320 | 640 | 640 | < | < |
| Ck/Hong Kong/G9/97 | 1280 | 640 | 80 | 640 | 5120 | 80 | 40 | 2560 | 640 |
| Sw/Hong Kong/9/98 | 640 | 320 | 160 | 640 | 5120 | 40 | < | 640 | 1280 |
Homologous titers in bold; < means < 40.
Neuraminidase inhibition assays with the strain-specific chicken and ferret antisera indicated greater crossreactivity between the N2s of the H9N2 viruses isolated from the different hosts (Table 2), although potential interference by antibody to the common H9 antigen complicates interpretation of the results. These N2s were distinguished from the N2s of representative H2N2 and H3N2 viruses circulating in the human population between 1957 and 1968 and since 1968, respectively (Table 2). Enzyme inhibition by the more broadly crossreactive hyperimmune rabbit antisera to these human viruses indicated a closer antigenic relationship of the N2s of the H9N2 human isolates with the N2 of an early H2N2 virus, A/Singapore/1/57, than with the N2 of a recent H3N2 virus, A/Beijing/32/92, which may reflect the closer relationship between the N2s of the 1957 virus and avian viruses (8).
Table 2.
Antigenic relationships between the neuraminidases of H9N2, H2N2, and H3N2 viruses
| Viruses | Subtype | Neuraminidase inhibition titer*
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Hyperimmune rabbit sera
|
Postinfection chicken sera
|
Postinfection ferret sera
|
|||||||
| Ty/Wis 1/66 | A/Sing 1/57 | A/Aichi 2/68 | A/Beij 32/92 | Qa/HK G1/97 | Ck/HK G9/97 | A/HK 1073/99 | Ck/HK G9/97 | ||
| Ty/Wisconsin/1/66 | H9N2 | 5120 | 640 | < | < | < | < | < | < |
| A/Singapore/1/57 | H2N2 | 640 | 2560 | < | < | < | 80 | < | < |
| A/Aichi/2/68 | H3N2 | 160 | 320 | 5120 | 640 | < | 80 | < | < |
| A/Beijing/32/92 | H3N2 | < | < | 160 | 1280 | ND | ND | ND | ND |
| A/Hong Kong/1073/99 | H9N2 | 640 | 320 | 160 | < | 320 | 320 | 320 | 160 |
| A/Hong Kong/1074/99 | H9N2 | 640 | 320 | 160 | < | 320 | 320 | 320 | 160 |
| Qa/Hong Kong/G1/97 | H9N2 | 320 | < | < | < | 640 | 160 | 80 | 160 |
| Ck/Hong Kong/G9/97 | H9N2 | 160 | 640 | 160 | < | 640 | 1280 | 80 | 640 |
Homologous titers in bold. Rabbit sera, < means < 160; chicken and ferret sera, < means < 80; ND, not done.
Genetic Relationships.
Comparison of the nucleotide sequences of all eight genes of the two H9N2 human isolates (99–100% homologous) with those of H9N2 viruses isolated from different types of birds in 1997 demonstrated a striking similarity, 99% or greater homology, between the genomes of the human and quail, Qa/HK/G1/97, isolates (Table 3). This contrasts with the differences (85–96% homology) between the genes of the human isolates and those of Dk/HK/Y280/97 and Ck/HK/G9/97 (with the exception of the polymerase protein PB1 and PB2 genes) isolates, which were shown to fall into a different lineage (13). Fig. 1 illustrates the phylogenetic relationships between the HA genes of H9N2 viruses recently isolated in East Asia and North America, and differentiates the three phylogenetic lineages of viruses, represented by Qa/HK/G1/97 (including A/HK/1073/99 and A/HK/1074/99), Ck/HK/G9/97 and Dk/HK/Y439/97, respectively, circulating in Hong Kong in 1997. The phylogenetic relationships between the N2s of these H9N2 viruses are similar (13). The 99% homology between the N2 genes of the human and quail H9N2 isolates contrasts with the differences between these genes and those of other avian H9N2 viruses (83–94% homology), chicken H5N2 viruses (80% homology), and human H2N2 and H3N2 viruses (84–81% homology), determined with sequences from GenBank.
Table 3.
Genetic relationship between H9N2 and H5N1 viruses
| Percent homology with A/HK/1073/99*
| ||||
|---|---|---|---|---|
| Gene | A/HK/97 (H5N1)† | Qa/HK/ G1/97 | Ck/HK/ G9/97 | Dk/HK/ Y280/97 |
| PB2 | 98 | 99 | 97 | 85 |
| PB1 | 99 | 99 | 98 | 90 |
| PA | 98 | 99 | 89 | 90 |
| H9 | – | 99 | 92 | 91 |
| NP | 99 | 99 | 90 | 90 |
| N2 | – | 99 | 93 | 94 |
| M | 98 | 99 | 96 | 96 |
| NS | 98 | 100 | 93 | 93 |
Similar relationships were obtained in comparisons with genes of A/HK/1074/99. Homology between the eight genes of A/HK/1073/99 and A/HK/1074/99 was 99% or greater.
Sequences used in these comparisons were PB2 and M of A/HK/485/97, PB1, PA, and NS of A/HK/156/97, and NP of A/HK/481/97.
Figure 1.
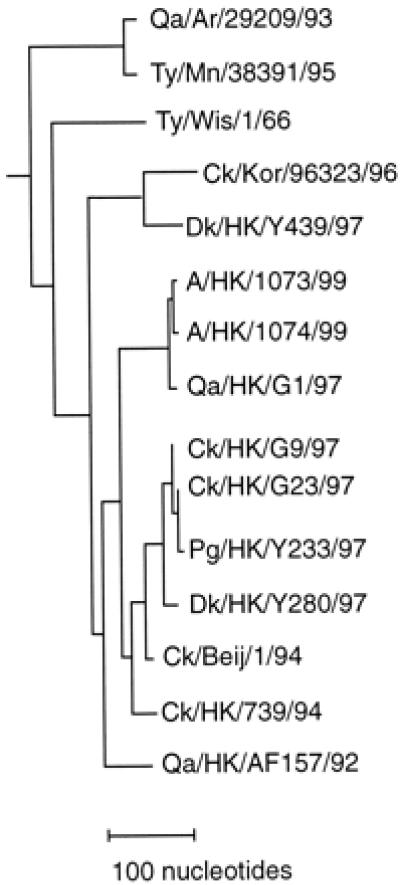
Phylogenetic relationships between the hemagglutinin genes of recent H9N2 viruses. Nucleotides 10–1,209 of the coding sequences were analyzed with PAUP by using a maximum parsimony algorithm; the tree is rooted to the HA sequence of A/duck/Alberta/60/76 (H12 N5). The lengths of the horizontal lines are proportional to the number of nucleotide differences (as indicated). With the exception of the sequences for A/HK/1073/99 and A/HK/1074/99, the sequences were obtained from GenBank, including those of A/quail/Arkansas/29209/93 (Qa/Ar/29203/93); A/turkey/Minnesota/38391/95 (Ty/Mn/38391/95); A/turkey/Wisconsin/1/66 (Ty/Wis/1/66); A/chicken/Korea/96323/96 (Ck/Kor/96323/96); A/duck/Hong Kong/Y439/97 (Dk/HK/Y439/97); A/chicken/Hong Kong/G23/97 (Ck/HK/G23/97); A/pigeon/Hong Kong/Y233/97 (Pg/HK/Y233/97); A/chicken/Beijing/1/94 (Ck/Beij/1/94); A/chicken/Hong Kong/739/94 (Ck/HK/739/94); and A/quail/Hong Kong/AF157/92 (Qa/HK/AF157/92).
It is of particular interest that the six “internal” genes of the two human isolates, like those of Qa/HK/G1/97 (13), were similar to the internal genes of the H5N1 viruses which cocirculated in poultry late in 1997 and which caused fatal human infections (Table 3). Thus, the six genes encoding the internal virus components represent a common feature of the H9N2 and H5N1 avian viruses which infected humans.
More detailed analyses of the genetic variation between A/HK/1073/99, A/HK/1074/99, and Qa/HK/G1/97 showed that the differences between the corresponding genes of the two human isolates were comparable to the differences between the genes of the quail and the human isolates accumulated over some 15 months. Although we do not know the extent of mutation after human infection, the data are consistent with different sources of infection and may reflect the genetic variation among similar viruses circulating in avian (and animal) species at that time. The degree of heterogeneity among the genes of the three H9N2 viruses, in both nucleotide and amino acid sequences, was comparable with that reported by Zhou et al. (19) for the corresponding genes of H5N1 viruses isolated from chickens in Hong Kong in 1997 and H5N2 viruses isolated from chickens in Mexico in 1994–1995. Although the coding sequences of only three H9N2 viruses were available for comparison, it was of interest to note the marked variation in the proportion of nucleotide differences which resulted in differences in amino acid sequences; the low proportion for M1 (0 of 28) and NP (2 of 26) contrasted with the high proportion for NS1 (4 of 6), NS2 (2 of 2), and M2 (6 of 6) proteins, indicating significant differences in selective pressure. The proportion of coding changes in the HA genes (32%) was greater than that observed among the avian H5N1 viruses in 1997 and was closer to that of the human H5N1 and chicken H5N2 viruses (19).
Amino Acid Sequence Comparisons.
Comparisons of the amino acid sequences of the proteins of the two H9N2 and various H5N1 human isolates indicated no consistent differences between them for PB1, PB2, NP, M1, or NS1; the greater similarity between the NP and NS1 proteins of A/HK/481/97 (H5N1) and the two H9 viruses was particularly striking. Comparison with published data for other H9N2 viruses (13) indicated that a number of amino acids were characteristic of the protein sequences of the quail and human H9N2 and human H5N1 viruses and differentiated them from the proteins of other H9N2 viruses. Of particular note were differences in translation termination of PB1 and NS1 proteins. The PB1 protein possessed an extra amino acid at its C terminus, a penultimate Gly-757, and the NS1 protein (230 amino acids) was 12 amino acids longer than the proteins of certain other H9N2 viruses.
Typically, the M2 proton channel showed greater variability than the M1 protein (20), reflecting a greater proportion of coding changes. Notably for the human and quail isolates, all nucleotide differences in the M2 coding sequence produced changes in amino acids, whereas none of the nucleotide differences in the M1 coding sequence altered the amino acid sequence. Two amino acids, Leu-10 and Asn-82, present in the M2s of the quail and human H9N2 and human H5N1 viruses were absent from the sequences of other H9N2 viruses. On the other hand, two amino acids, Glu-16 and Lys-18, of the two human H9N2 viruses differed from those of the quail H9N2 and the H5N1 proteins and reflected more closely the amino acid sequences of the proteins of some other H9N2 viruses. Because the sequences in this region of the quail H9N2 and human H5N1 proteins, containing Gly-16 and Arg-18, correspond to those of the M2 proteins of H1N1 and H3N2 human viruses, changes in these amino acids are unlikely to be associated with adaptation of H9N2 to humans. Replication of the human H9N2 isolates was shown to be sensitive to inhibition by rimantadine; in this respect, none of the amino acids 26, 27, 30, 31, or 34 of the M2 proteins of the H9N2 viruses correspond to residues which confer resistance to the anti-influenza drugs, amantadine and rimantadine (21).
Hemagglutinin.
Features of the hemagglutinin of particular interest with regard to the host range and pathogenicity of the virus include receptor-binding specificity (22, 23) and susceptibility to proteolytic activation (24, 25). A feature of the hemagglutinins of H5N1 viruses known to be responsible for pathogenicity in chicken and mice, and which may have contributed to its virulence in humans, is the sequence (in italics) of basic amino acids, PQRERRRKKR↓G, at the site of cleavage to HA1 and HA2, which renders it susceptible to furin-like enzymes in cells of a wide variety of tissues (25). The sequence, PARSSR↓G (residues 324–330, H3 numbering), of the H9 hemagglutinin corresponds to those of other nonpathogenic avian viruses, consistent with the low pathogenicity of these H9N2 viruses in experimental infections of chickens (26). Furthermore, there was no loss of glycosylation of Asn-11 which has been associated with enhanced intracellular cleavability and pathogenicity (27). Two additional potential glycosylation sites, Asn-94 and Asn-198 (numbered according to H3), close to the receptor-binding site at the top of the HAs of the quail and human viruses may contribute to the difference in antigenicity from the chicken and pig isolates and may influence receptor-binding characteristics.
The host range of influenza A viruses is associated with differences in the specificity of HA for attachment to sialic acid-containing receptors on susceptible cells (22, 23). For example, HAs of avian and equine viruses exhibit a preference for sialic acid (SA) linked to the penultimate galactose by an α2,3 glycosidic linkage (SAα2,3Gal), whereas the HAs of human viruses have a preference for SAα2,6Gal, reflecting the preponderance of these terminal sugar moieties in the different hosts. Substitution of Leu-226 by Gln in the HA of early human H3N2 viruses was associated with a change in preference for binding to SAα2,3 Gal rather than to SAα2,6Gal moieties (22). With regards to their ability to infect humans, the HAs of the human and quail (as well as the chicken and pig) H9N2 viruses possessed Leu-226 in contrast to the HAs of several other contemporary H9N2 avian viruses, which have Gln at this position (Table 4). A number of amino acids within the receptor-binding site which are involved directly in binding sialic acid are highly conserved among the different HA subtypes (28). These include Tyr-98, Ser-136, Trp-153, Leu-194, and Tyr-195, which are conserved in the HAs of the various H9N2 viruses. Two other conserved residues, His-183 and Glu-190, are not maintained among all H9N2 viruses isolated from ducks, chickens, and pigeons in Hong Kong (Table 4). In fact, of the H9N2 viruses studied, only the Qa/HK/G1/97 and the two human isolates possessed the combination His-183, Glu-190, and Leu-226 (numbered according to H3) which was typical of early human H3N2 viruses (although not of H1N1 viruses).
Table 4.
Variation of “conserved” residues in the receptor-binding sites of hemagglutinins of H9N2 viruses
| Virus | Amino acid residue*
|
||
|---|---|---|---|
| 183 | 190 | 226 | |
| A/HK/1073/99 | H | E | L |
| Qa/HK/G1/97† | H | E | L |
| Ck/HK/G9/97† | N | A | L |
| Dk/HK/Y280/97† | N | T | L |
| Ck/Beij/1/94† | N | V | Q |
| Dk/HK/Y439/97† | H | E | Q |
| Ty/Wis/1/66 | H | E | Q |
| Human H3N2 | H | E/D | L |
| Human H1N1 | H | D (N/V) | Q |
Numbering according to H3.
Different phylogenetic subgroups of H9N2 viruses (13).
Neuraminidase.
The most notable difference between the amino acid sequences of the N2 neuraminidases of the two human H9N2 viruses and the sequences of N2s of other avian H9 and human H3 viruses is a deletion of two amino acids, residues 38 and 39, in the stalk of the protein. The same deletion in the N2 of Qa/HK/G1/97 further emphasizes the close relationship between the quail and human isolates. The patterns of glycosylation sites, at asparagine 61, 69 or 70, 86, 146, 200, 234, and 402, of the various N2s were similar; the only difference between the avian/human H9N2 and human H3N2 viruses is the presence of an extra potential glycosylation site at Asn-329 in N2s of the latter viruses. One of the six amino acid differences between the N2 of Qa/HK/G1/99 and the N2s of the two human H9N2 viruses results in the absence of glycosylation at residue 402 of the quail virus protein which may contribute to observed differences in antigenicity (Table 2).
Discussion
The two cases of human infection were shown to be caused by similar H9N2 viruses, closely related to a quail virus Qa/HK/G1/97, which represents one of three genetically distinguishable subgroups of H9N2 viruses which cocirculated with H5N1 viruses in live poultry markets in Hong Kong in late 1997. It is evident, therefore, that they had not acquired (by genetic reassortment) any genes from another source including contemporary human viruses and that infection most likely resulted from direct transmission from infected birds, although the actual source has not been identified. Although Qa/HK/G1/97 was the only example of the genetic subgroup, the two human isolates indicated that similar viruses have continued to circulate, as subsequently confirmed by studies of avian viruses (K. Shortridge, unpublished results). The antigenic differences between the human and swine H9N2 isolates indicate that those swine viruses were not intermediates in avian to human transmission.
The most notable feature of these human H9N2 viruses is that they possess a set of internal genes similar to those of the H5N1 viruses which caused 18 cases of human infection in 1997. From studies of avian viruses, Guan et al. (13) concluded that the H5N1 viruses were reassortants which derived their internal genes from a Qa/HK/G1/97-like virus, although the origin of the genes has yet to be identified. Whether the commonality between the internal genes of the H9N2 and H5N1 viruses is an important factor in their ability to infect humans, and which, if any, of the genes are necessary, has yet to be established.
Various studies have shown that internal virus proteins, such as NP and PB2, are important in determining the host range of influenza A viruses (29, 30) and comparisons of the sequences of proteins of human and avian viruses have indicated certain amino acid residues which are typical of human rather than avian viruses. With a few exceptions, however, the majority of amino acid residues characteristic of the proteins of the H9N2 and H5N1 human viruses did not correspond to “human-like” residues and thus provide few clues as to why this particular complement of genes might enable these avian viruses to transmit to humans. Of particular interest is PB1, because the novel subtype viruses which emerged in 1957 (H2N2) and 1968 (H3N2) each acquired a novel “avian” PB1 in addition to HA and NA (H2N2 only) by reassortment of the preceding human viruses with avian viruses (31). None of the amino acids characteristic of PB1s of the human H9N2 and H5N1 viruses, including the extra penultimate Gly at the C terminus, were typical of the PB1 proteins of H1, H2, and H3 human viruses; neither did Asn-375 correspond to Ser-375 of human viruses, suggested to be important in influencing host specificity (31). Although the high proportion of coding changes in M2 sequences is consistent with strong selection for replication in a new host, it is not evident what advantage the amino acid changes might have for human infection.
Receptor specificity of the HA is important in determining host range and changes in preference for SAα2,3Gal to SAα2,6Gal moieties have been observed to accompany establishment of avian viruses (or their HAs) in human and porcine hosts. The H5N1 viruses which transmitted from chickens to humans in Hong Kong in 1997 were shown to retain specificity for SAα2,3Gal (32). The presence of an additional carbohydrate attached to Asn-158, close to the receptor-binding site, of the HAs of some of those viruses was shown to reduce binding affinity. It was suggested that this may be important to compensate for low receptor-destroying activity of the neuraminidase. It may be that glycosylation of the two sites close to the receptor-binding site of the HAs of the human and quail H9N2 viruses influences receptor binding and increases host range. The combination of amino acids, His-183, Glu-190, and Leu-226 distinguishes the human and quail HAs from those of the other H9N2 viruses and is typical of early human H3 HAs. Which features of the receptor-binding site affect species specificity and contribute to human infection have yet to be determined.
An extensive deletion of 19 or 20 amino acids in the stalks of the NAs of H5N1 and H5N2 viruses, respectively, has been correlated with the appearance of these viruses in chickens (32). In this respect, acquisition of the small deletion in the H9N2 neuraminidase may be related to changes in characteristics of the hemagglutinin and adaptation of these viruses in domestic poultry after their recent introduction from ducks (26).
The earlier isolation of H9N2 viruses in southern China (14) together with the preliminary results of serological studies (16) suggest that other human infections may have occurred. However, extensive surveillance in Hong Kong failed to identify other examples of this subtype among some 2,000 human A and B influenza viruses isolated during 1999. H9N2 viruses are prevalent in avian species in various parts of the world. Whether circulation of similar viruses in poultry in other Asian and European countries (ref. 33 and Y.P.L., J. Banks, D. Alexander, and A.H., unpublished results) has resulted in human infections is presently unknown and no cases of any severity have been reported. More extensive studies are required to establish the extent of human infection, and the source of infection and mechanism of transmission. It is of particular interest to determine whether these H9N2 viruses have a greater potential than the H5N1 viruses for human-to-human spread and the ability to become established in the human population.
The extent of genetic reassortment involving H9N2 viruses, their ability to infect humans, and their continued circulation in domestic poultry and pigs in southern China emphasizes the potential for the emergence, by adaptation or genetic reassortment (possibly with circulating human H1N1 or H3N2 viruses), of a virus with the ability to establish itself in the human population and cause the next pandemic.
Acknowledgments
We thank Michael Bennett and José Saldanha for assistance with sequence analyses and Rosemary Sumray for preparation of the manuscript. All are affiliated with the National Institute for Medical Research. We thank Henrietta Hall, Yumi Matsuoka, and Xiyan Xu, all from the Centers for Disease Control and Prevention, for assistance in performing HI tests, sequencing, and sequence analyses, and Dr. Zhang Lijuan (University of Hong Kong) for technical support. These studies were supported in part by Public Health Research Grant AI29680 and Contract AI9537 from the National Institutes of Allergy and Infectious Diseases, Cancer Center Support Core Grant CA21765, and the American Lebanese Syrian-Associated Charities.
Abbreviations
- HA
hemagglutinin
- NA
neuraminidase
- NP
nucleoprotein
- Ck
chicken
- Dk
duck
- Qa
quail
- HK
Hong Kong
- SA
sialic acid
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ404735, AJ404736, AJ289871–AJ289874, AJ278646–AJ278649, and AJ404626–AJ404637).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160270697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160270697
References
- 1.Webster R G, Laver W G. Bull WHO. 1972;47:449–452. [PMC free article] [PubMed] [Google Scholar]
- 2.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster R G, Hinshaw V S, Bean W J, Van Wyke K L, Geraci J R, St. Aubin D J, Petursson G. Virology. 1981;113:712–724. doi: 10.1016/0042-6822(81)90200-2. [DOI] [PubMed] [Google Scholar]
- 4.Pensaert M, Ottis K, Vandeputte J, Kaplan M M, Backmann P A. Bull WHO. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 5.Guan Y, Shortridge K F, Krauss S, Li P H, Kawaoka Y, Webster R G. J Virol. 1996;70:8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholtissek C, Rhode W, von Hoyningen V, Rott R. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 8.Schafer J R, Kawaoka Y, Bean W J, Suss J, Senne D, Webster R G. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 9.Goldfield M, Bartley J D, Pizutti W, Black H C, Altman R, Halperin W E. J Infect Dis. 1977;136:S347–S355. doi: 10.1093/infdis/136.supplement_3.s347. [DOI] [PubMed] [Google Scholar]
- 10.Claas E, Osterhaus A, van Beek R, De Jong J, Rimmelzwaan G, Senne D, Krauss S, Shortridge K, Webster R G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 11.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Jing H, et al. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 12.Bender C, Hall H, Huang J, Klimov A, Cox N, Hay A, Gregory V, Cameron K, Lim W, Subbarao K. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- 13.Guan Y, Shortridge K F, Krauss S, Webster R G. Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y, Li J, Cheng X, Wang M, Zhou Y, Li X H, Cai F, Miao H L, Zhang H, Guo F, et al. Chin J Exp Clin Virol. 1999;13:105–108. [Google Scholar]
- 15.Anon Weekly Epidemiol Rec. 1999;14:111. [Google Scholar]
- 16.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L S, Lai R W M, Orr W K, Shortridge K F. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 17.Kendal A P, Pereira M S, Skehel J J. Concepts and Procedures for Laboratory-Based Influenza Surveillance, U.S. Department of Health and Human Services. Atlanta: Centers Dis. Control; 1982. [Google Scholar]
- 18.Belshe R B, Hall Smith M, Hall C B, Betts R, Hay A J. J Virol. 1988;62:1508–1512. doi: 10.1128/jvi.62.5.1508-1512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Gorman O T, Kawaoka Y, Bean W J, Webster R G. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay A J. In: Antiviral Drug Resistance. Richman D D, editor. Chichester, England: Wiley; 1996. pp. 43–58. [Google Scholar]
- 22.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Nature (London) 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 23.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 24.Webster R G, Rott R. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 25.Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk H-D, Garten W. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y J, Krauss S, Senne D A, Mo I P, Lo K S, Xiong X P, Norwood M, Shortridge K F, Webster R G, Guan Y. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 27.Deshpande K L, Fried V A, Ando M, Webster R G. Proc Natl Acad Sci USA. 1987;84:36–40. doi: 10.1073/pnas.84.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Nature (London) 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 29.Scholtissek C, Burger H, Kistner O, Shortridge K F. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 30.Subbarao E K, London W, Murphy B R. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaoka Y, Krauss S, Webster R G. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matrosovich M, Zhou N, Kawaoka Y, Webster R. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander D J. In: Proceedings of the Fourth International Symposium on Avian Influenza. Swayne D E, Slemons R D, editors. Tallahassee, FL: Rose Printing; 1998. pp. 9–13. [Google Scholar]


