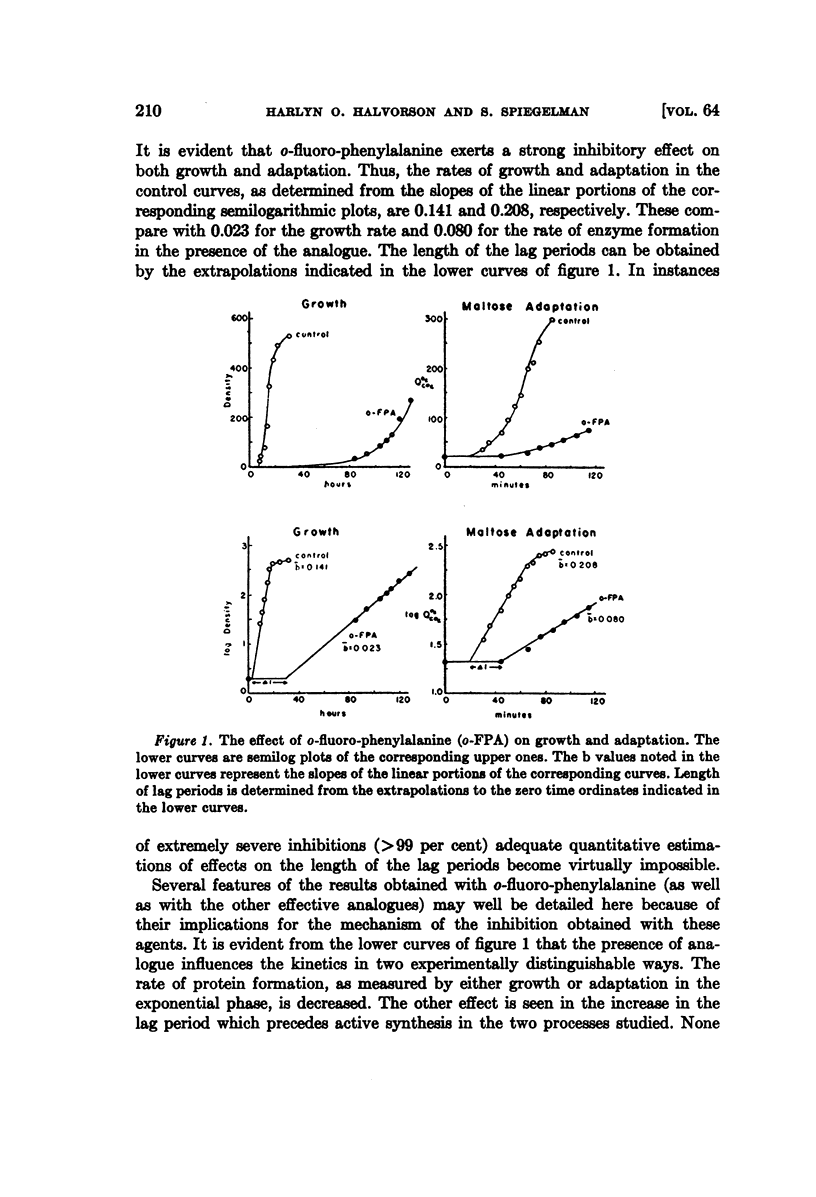
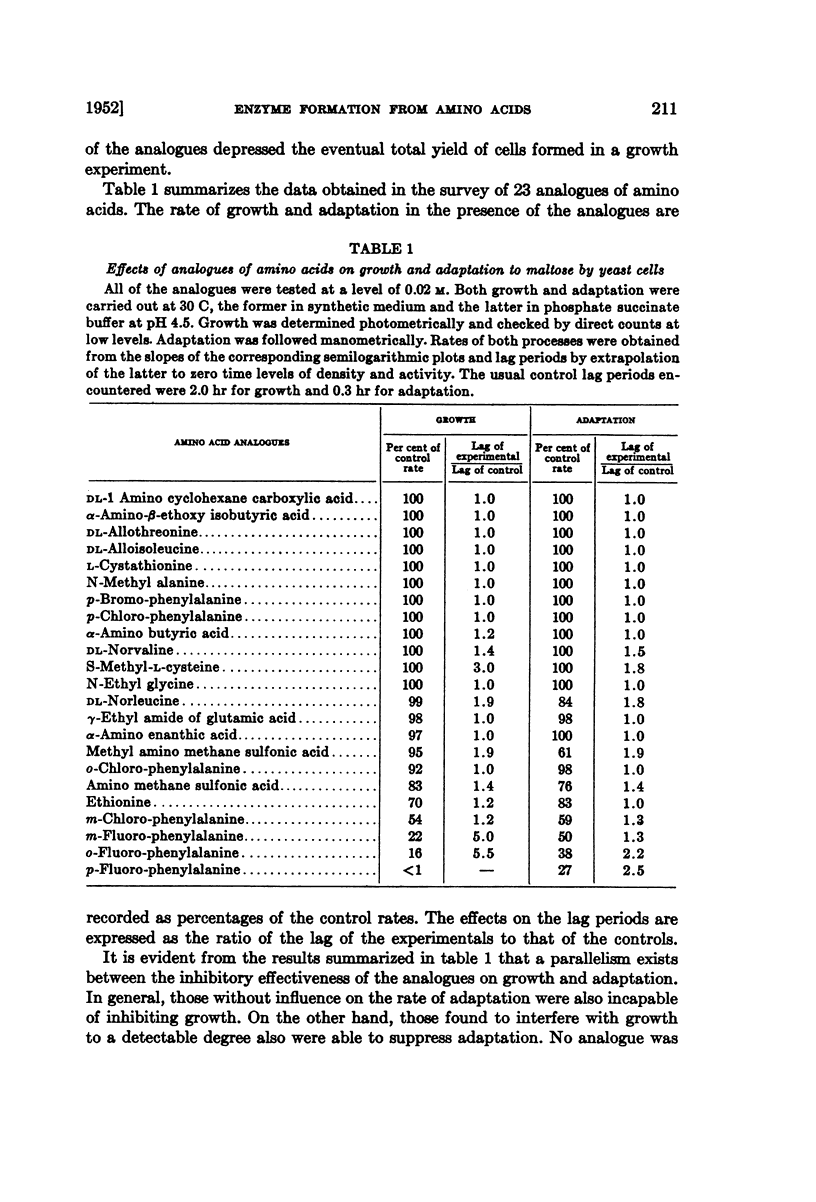
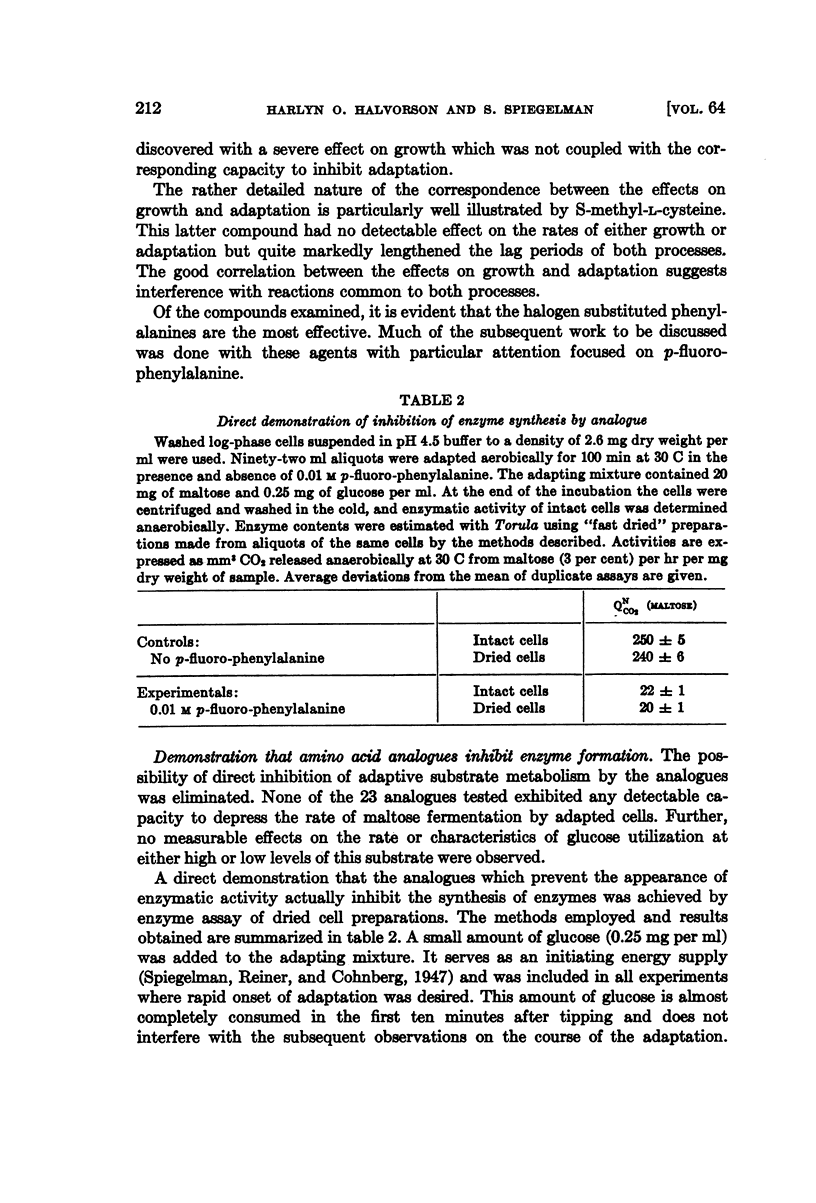
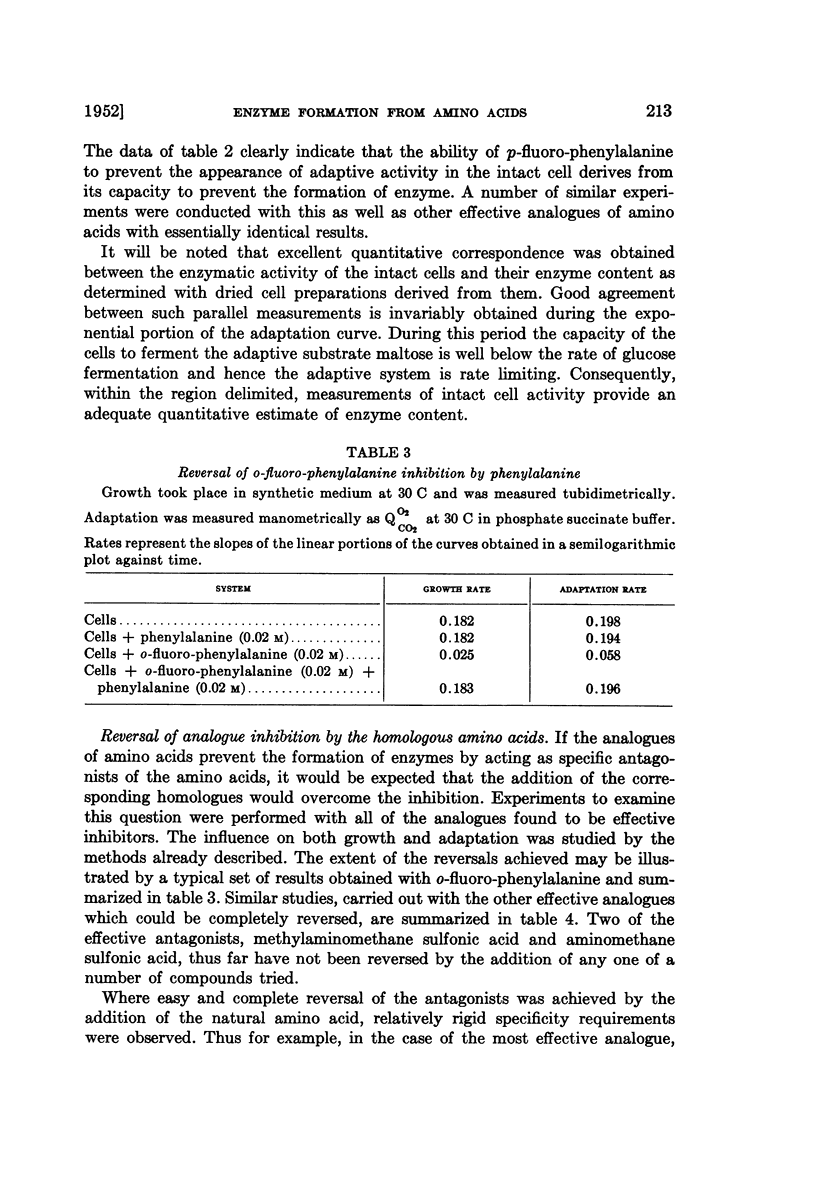
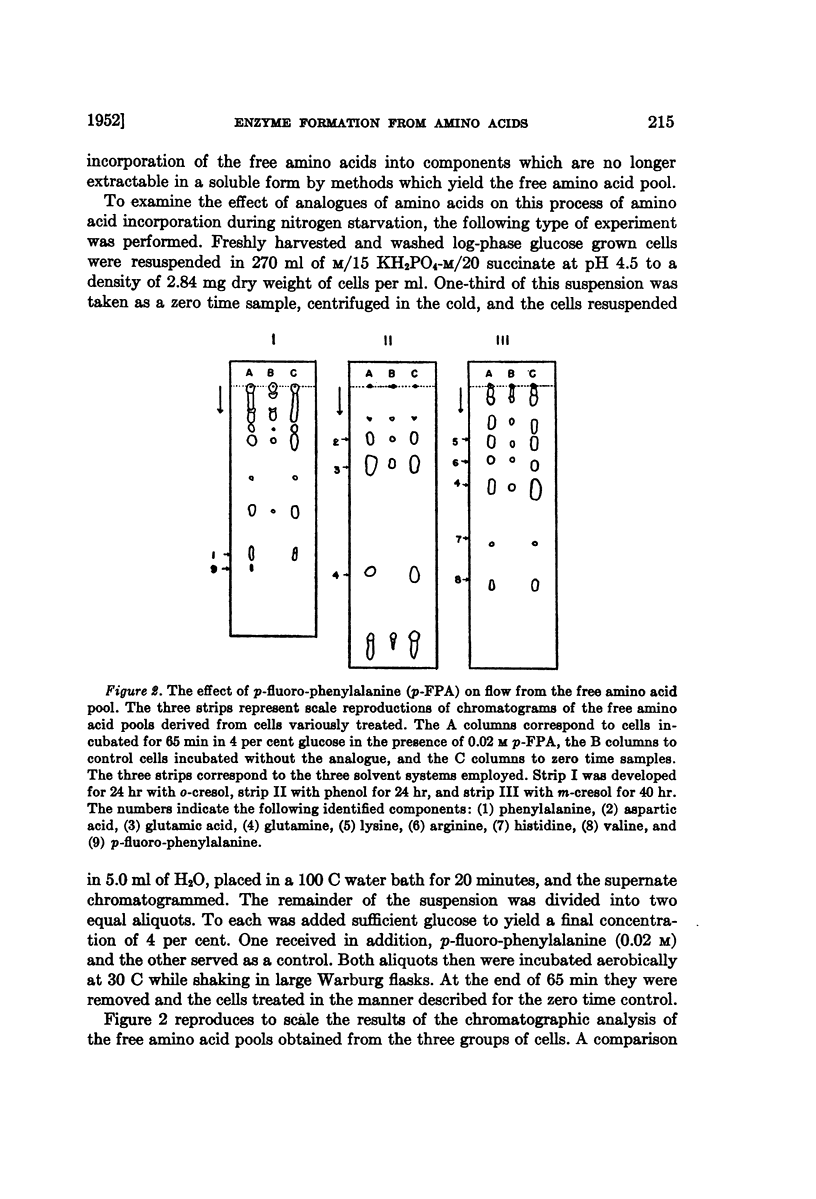
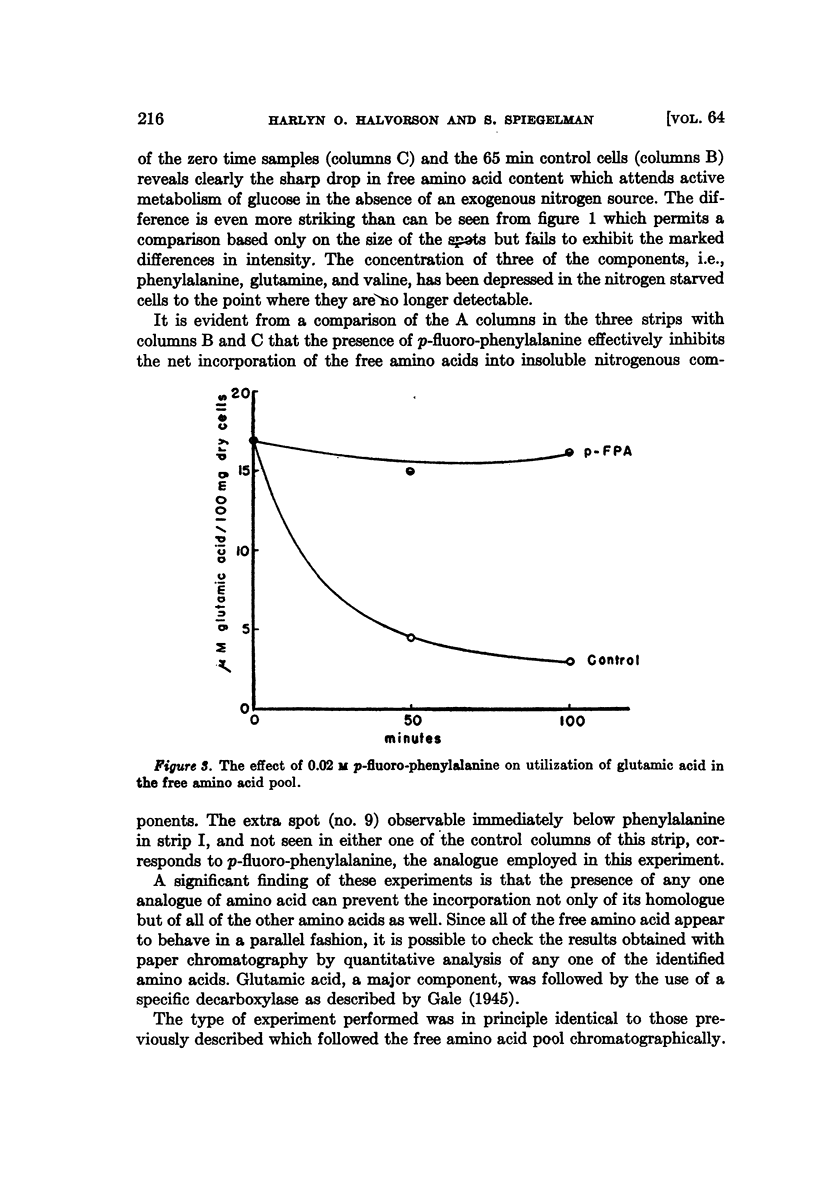
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANFINSEN C. B., STEINBERG D. Studies on the biosynthesis of ovalbumin. J Biol Chem. 1951 Apr;189(2):739–744. [PubMed] [Google Scholar]
- DITTMER K. The structural bases of some amino acid antagonists and their microbiological properties. Ann N Y Acad Sci. 1950 Jul 7;52(8):1274–1301. doi: 10.1111/j.1749-6632.1950.tb54030.x. [DOI] [PubMed] [Google Scholar]
- Freeland J. C., Gale E. F. The amino-acid composition of certain bacteria and yeasts. Biochem J. 1947;41(1):135–138. doi: 10.1042/bj0410135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEIGER E. The role of the time factor in protein synthesis. Science. 1950 Jun 2;111(2892):594–599. doi: 10.1126/science.111.2892.594. [DOI] [PubMed] [Google Scholar]
- Gale E. F. Studies on bacterial amino-acid decarboxylases: 5. The use of specific decarboxylase preparations in the estimation of amino-acids and in protein analysis. Biochem J. 1945;39(1):46–52. doi: 10.1042/bj0390046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN H. P., DOUDOROFF M. The mutation of Pseudomonas putrefaciens to glucose utilization and its enzymatic basis. J Bacteriol. 1950 Jun;59(6):739–750. doi: 10.1128/jb.59.6.739-750.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilin D., Hartree E. F. The use of glucose oxidase (notatin) for the determination of glucose in biological material and for the study of glucose-producing systems by manometric methods. Biochem J. 1948;42(2):230–238. [PMC free article] [PubMed] [Google Scholar]
- POLLOCK M. R. Penicillinase adaptation in B. cereus; adaptive enzyme formation in the absence of free substrate. Br J Exp Pathol. 1950 Dec;31(6):739–753. [PMC free article] [PubMed] [Google Scholar]
- SLEEPER B. P., TSUCHIDA M., STANIER R. Y. The bacterial oxidation of aromatic compounds; the preparation of enzymatically active dried cells and the influence thereon of prior patterns of adaptation. J Bacteriol. 1950 Jan;59(1):129–133. doi: 10.1128/jb.59.1.129-133.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANIER R. Y. Enzymatic adaptation in bacteria. Annu Rev Microbiol. 1951;5:35–56. doi: 10.1146/annurev.mi.05.100151.000343. [DOI] [PubMed] [Google Scholar]



