Abstract
Because current methods of imaging prostate cancer are inadequate, biopsies cannot be effectively guided and treatment cannot be effectively planned and targeted. Therefore, our research is aimed at ultrasonically characterizing cancerous prostate tissue so that we can image it more effectively and thereby provide improved means of detecting, treating and monitoring prostate cancer. We base our characterization methods on spectrum analysis of radio frequency (rf) echo signals combined with clinical variables such as prostate-specific antigen (PSA). Tissue typing using these parameters is performed by artificial neural networks. We employedand evaluated different approaches to data partitioning into training, validation, and test sets and different neural network configuration options. In this manner, we sought to determine what neural network configuration is optimal for these data and also to assess possible bias that might exist due to correlations among different data entries among the data for a given patient. The classification efficacy of each neural network configuration and data-partitioning method was measured using relative-operating-characteristic (ROC) methods. Neural network classification based on spectral parameters combined with clinical data generally produced ROC-curve areas of 0.80 compared to curve areas of 0.64 for conventional transrectal ultrasound imaging combined with clinical data. We then used the optimal neural network configuration to generate lookup tables that translate local spectral parameter values and global clinical-variable values into pixel values in tissue-type images (TTIs). TTIs continue to show can cerous regions successfully, and may prove to be particularly useful clinically in combination with other ultrasonic and nonultrasonic methods, e.g., magnetic-resonance spectroscopy.
Keywords: Biopsy guidance, neural networks, prostate cancer, radiation therapy, spectrum analysis, therapy targeting, ultrasonic imaging
I. INTRODUCTION
Prostate cancer currently is detected using ultrasound-guided needle biopsies. Transrectal ultrasound is able to define prostate anatomy, i.e., prostate borders and internal regions, but it cannot reliably depict cancerous foci in the gland. Therefore, biopsy needles are directed into the gland simply with respect to gland regions, such as the left and right base, mid and apex, but blindly with respect to occult tumor foci that may be present. In early stages of disease, when cancerous foci are small and widely separated but easily treatable, biopsy needles frequently miss existing lesions resulting in false-negative determinations. Similarly, CT, MRI, PET and other current clinical imaging modalities are ineffective in imaging cancers of the prostate, even though many of them can show the prostate itself with excellent definition.1 As a consequence, current imaging methods cannot reliably provide information that is essential for effectively guiding biopsies, planning treatment and monitoring treated or ‘watched’ cancers.
In our previous analyses of repeat-biopsy data, we showed that the sensitivity of current transrectal ultrasound-guided procedures appears to be less than about 50%; therefore, half the actual cancers presenting for biopsy may be missed.2 Our current results, described below, confirm previously published results;2-7 they suggest that significant (e.g., 50%) improvements in sensitivity may be possible using ultrasonic tissue-type images (TTIs) to guide biopsies. Similarly, improvements in outcomes and reduced deleterious side effects may be achieved if TTIs can show cancerous and, equally important, noncancerous regions with higher confidence than current imaging methods provide. For example, imaging that reliably shows cancer-free regions of the prostate possibly could improve outcomes in nerve-sparing surgical treatments and could minimize damage to the rectum, bladder and urethra in radiation treatments.
This article describes new work and presents new results that we have obtained since our previously published research; these latest results confirm and validate our prior studies. The findings we describe in this article are derived from a demographically-different population of patients at a different medical center using a different ultrasonic scanner, computer, software and data-acquisition hardware to acquire radiofrequency (rf) echo-signal data. Furthermore, different software was used for subsequent, off-line spectrum analysis and also for neural network classification studies.
Our research seeks to de velop TTI methods capable of reliably identifying and characterizing cancerous prostate tissue and thereby able to improve the effectiveness of biopsy guidance, therapy targeting, and treatment monitoring. While the research described here utilizes artificial neural networks to characterize prostate tissue based on spectrum analysis parameters and clinical variables such as prostate-specific antigen (PSA), age, race, etc., we recognize that other ultrasonic methods and also other imaging modalities can provide additional, independent information that potentially could be combined with ultrasonic spectrum-analysis in TTIs to markedly improve the efficacy of TTIs in depicting prostate cancer. The ultrasonic methods we plan to investigate in the future include perfusion imaging that utilizes ultrasonic contrast agents to assess normal and abnormal vasculature and elasticity imaging to depict relatively stiff regions of the gland. In addition, magnetic-resonance spectroscopy offers some exciting possibilities for sensing local chemical changes associated with prostate cancer. We also note that investigations by Scheipers et al have applied multifeature approaches to characterizing prostate cancer; their methods emphasize analysis of texture features in B-mode ultrasonic images, but they also have employed rf data and confirmed our successful application of ultrasonic spectral parameters in neural-network-based classification methods.8
II. METHODS
In collaboration with several medical centers (including the Memorial Sloan-Kettering Cancer Center in New York City, the Veterans Affairs Medical Center in Washington, DC, and the Virginia Mason Medical Center in Seattle, WA) we have been acquiring rf echo-signal data and clinical variables, e.g., PSA, during biopsy examinations of the prostate. We computed the power spectra of rf signals site-matched to each biopsied region, and we trained neural networks with over 3,500 sets of ultra sonic, clinical and histological data acquired since the inception of our study. Intercept and midband spectral parameters were computed in accordance with the theoretical frame work of Lizzi et al.9-11 In every analyzed case, biopsy histology results served as the gold standard for training and evaluating classifiers. The new findings described here are derived from data obtained at the Veterans Affairs Medical Center in Washington, DC.
Data acquisition
Rf data were acquired during transrectal ultrasound-guided biopsy examinations by digitizing the radiofrequency (rf) echo signals accessed within a Hitachi (Twinsburg, OH) EUB 525 scanner using a EUP V53W end-fire curved-array probe with a nominal center frequency of 7.5 MHz. Digitization was performed by a GaGe (Lashine, QC, Canada) Compuscope 1250-1M data-acquisition board sampling at a rate of 50 MS/s. Each scan plane consisted of 155 digitized scan vectors, and each vector comprised 3,600, 12-bit samples.
Clinical data such as the value of each patient's level of prostate-specific antigen (PSA), age, race, ethnicity, etc., were entered into the rf data-file header. A key entry into the header was the level of suspicion (LOS) for cancer assigned to the biopsied region of each scan by the examining urologist. This LOS assignment was based on the appearance of the conventional B-mode image at the biopsy site combined with any other information available to the urologist, e.g., PSA value or results of the patient's digital rectal examination (rectal palpation examination). The LOS served as our essential baseline for assessing the relative performance of our classification methods.
Patient recruitment
Patients for the study described here were recruited from the pool of patients undergoing biopsy at the Washington, DC, Veterans Affairs Medical Center. Our latest data set consisted of ultrasonic, clinical and histology data from 705 biopsies administered to 67 patients; the racial composition of this patient population was predominantly Black or of African descent. (In comparison, our previously reported results were based on a predominantly white patient population.) Cancer was detected by the biopsies in 26 (38.8%) of these 67 patients and in 119 (16.9%) of the 705 biopsy cores. The incidence of positive biopsies and the fraction of patients with detected cancer were slightly higher in this data set than we typically find in our studies; e.g., in our prior data sets, the incidence of positive biopsies tended to be close to 30% and the fraction of patients with detected cancers tended to be between 10% and 11%.
Data analysis and maintenance
Rf echo-signal data were analyzed using the methods described by Lizzi et al.9-11 Custom software was developed using Lab VIEW (National Instruments, Austin, TX). This software computed spectral-parameter values over the entire digitized scan. A sliding Hamming window was applied to the rf data of each scan vector; the window width was 64 samples (∼ 0.96 mm) and the step between window locations was only 8 samples (∼ 0.12 mm), so considerable overlap existed between adjacent windows. The power spectrum, i.e., the squared magnitude of the Fourier transform of the windowed rf data, was computed from the windowed rf signals and expressed as decibels (dB) relative to the corresponding spectrum of a perfectly reflecting, planar calibration target, e.g., a glass plate. (The calibration target was placed in the ultrasound beam with its surface in the center of the focal zone and orientated to be exactly perpendicular to the beam axis, i.e., parallel to the planar wave fronts that are present at the focus.) Spectra were computed over an effective frequency range of 3.5 to 8.0 MHz at every window position along each of the 155 vectors in the digitized scan. (Actual tissue spectra showed that the signal-to-noise ratio was consistently adequate over the frequency band extending from 3.5 MHz to 8.0 MHz.) The slope and intercept values were computed using a standard linear regression approach and at each window location, slope was corrected for the assumed attenuation of 0.5 dB/MHz-cm. Then the average values of slope, intercept and midband parameters were computed over a region of interest (ROI) site matched to the biopsy region.
The average values of spectral parameters were entered into a FoxPro (Microsoft, Redmond, WA) data base along with the gold-standard biopsy results, examination parameters, clinical variables (e.g., PSA value) and patient variables (e.g., age and race). Queries applied to the data base extracted results needed for classification tasks, e.g., intercept, midband and PSA values along with the LOS values serving as our baseline. As always, the reference for ‘truth’ was the biopsy histology.
Tissue classification
As described in previous publications, our classification studies utilized standard and custom software for implementing neural network configurations with multilayer perceptrons and radial-basis functions.4-7 The inputs for the neural networks were intercept value, midband value and PSA value, with biopsy results serving as the gold standard for true tissue type. We used NConnect (SPSS, Chicago, IL) and MATLAB (MathWorks, Natick, MA) to classify spectral parameter values and PSA for biopsy-proven prostate tissues. Our classification efforts sought only to segregate cancerous from noncancerous peripheral-zone tissue within the prostate. However, the sampled tissue included a wide range of cancerous tissues, e.g., degrees of differentiation and aggressiveness as estimated histologically from the Gleason scores of the biopsy specimens, and numerous noncancerous types of diseased or unhealthy prostate tissue, e.g.,inflammatory conditions, benign hyperplasia, atrophy, etc. (The Gleason method of grading of prostate cancer is now virtually universal among pathologists.12) Gleason scores ranging from 1 to 5 indicate the degree of de-differentiation and aggressiveness found in prostate cancer; 1 indicates retention of a high degree of differentiation and 5 indicates virtually complete loss of differentiation. The final Gleason score is the sum of the two most-common grades in a specimen, e.g., if grades 3 and 4 were the two most common grades in a given specimen, then the score for that specimen would be 7.
We implemented multilayer-perceptron and radial-basis-function neural networks with NConnect and we implemented a multilayer-perceptron neural network with MATLAB. In NConnect, we randomly determined which cases were as signed to the training validation, and test sets. We allocated 80% of the data to the training, 10% to the validation and 10% to the test set. NConnect runs were repeated 10 times using a different randomization seed (i.e., selected different cases for training, validation and test sets) each time. In MATLAB, we used a custom script to limit the test set to a single patient and to place all the biopsies for that patient in the test set; all other data were as signed to the training and validation sets with 90% of those data in the training set and 10% in the validation set. A separate MATLAB run was performed for each patient; accordingly, we termed this a ‘leave-one-patient-out’ approach. We varied the network parameters used in NConnect and MATLAB to determine empirically the most-effective neural network configuration, e.g., in the multilayer perceptrons, we varied the number of hidden layers, number of nodes in each layer, activation functions, learning rates, etc. As shown below, all methods produced comparable classifier performances, but we chose to base further studies on the MATLAB leave-one-patient-out neural network because it assured in dependence among the data in the training set versus those in the test and validation sets.
ROC analyses
For evaluation purposes, the scores of all runs of each neural network configuration were merged into a single set and analyzed as a whole. That is, for the 10, NConnect runs that had 70 or 71 randomly selected biopsies in the test set for each run, the scores for all 10 separate runs were combined to create a single set of 705 scores; the classification performance of the full set of 705 scores then was evaluated using ROC methods. Similarly, for the 67, single-patient runs that had 10 to 12 usable biopsies for each patient and so had 10 to 12 biopsies in each test set, the 67 sets of 10 to 12 scores per patient were combined into a single set of 705 scores for ROC evaluation.
As mentioned above, our baseline for evaluating the improved classification provided by our methods was the LOS for cancer that was determined by the examining urologist using the B-mode appearance of the biopsy site combined with other available clinical information, e.g., PSA level, rectal-palpation impressions, etc. To assess the relative tissue-typing efficacy of the artificial neural network classifier, we compared its classification performance to the LOS representing conventional classification using ROC (relative-operating-characteristic also termed receiver-operator-characteristic) analyses.13
ROC curves depict the trade-off between sensitivity and specificity by plotting true-positive fraction (i.e., sensitivity) versus false-positive fraction (i.e., 1 - specificity) as the thresh-old for discriminating actually positive from actually negative cases is varied. The area under the curve provides a measure of classifier performance: an area of 0.5 indicates entirely random classification while an area of 1.0 indicates perfect classification.13 Software offered by various authors (e.g., ROCkit by C. Metz of the University of Chicago and MedCalc by F. Schoonjans, University Hospital, Gent, Belgium) provide area estimates, standard errors in area estimates, 95% confidence intervals for area estimates, statistics regarding the likelihood that two ROC curves are infact different, etc. As discussed under Results below, we have found that differences in area estimates so greatly exceed the sum of the standard errors in those estimates that ample confidence of classifier performance difference is demonstrated.
Lookup tables
We generated lookup tables for translating spectral parameter and clinical variables to a relative likelihood for cancer using the optimal MATLAB multilayer-perceptron configuration, i.e., the combination of layers, nodes, etc., that gave the best ROC areas. The lookup table reduces computational time in generating TTIs by eliminating the need to run a neural network analysis in order to compute a score for cancer relative likelihood at every image pixel. Instead, when using an lookup table, spectral parameter values are computed at each pixel location and the computed parameter values along with patient's PSA level are referred to the lookup table, which then returns a cancer-likelihood score. Software for gray-scale imaging then translates the ‘raw’ score into a pixel value, e.g., a value of 0 for the lowest score in the set of raw scores to 255 for the highest score. The software also can present different score ranges as false colors to indicate different ranges in cancer likelihood, e.g., red above a certain threshold to indicate the greatest likelihood of cancer, orange between two thresholds to indicate a somewhat lesser likelihood, down to green to indicate a minimal likelihood. We routinely use five colors ranging from green through yellow-green, yellow, orange, and red overlaid on a midband image; alternatively, we use just orange and red to show areas of highest suspicion. The former false-color scheme might be used for guiding biopsies; the latter might be used for targeting radiation. This procedure is performed on a pixel-by-pixel basis either over the entire scan or over a user-specified subregion of the scan. Examples of a small subregion might be a limited biopsy area for use invalidating TTI performance by comparison with actual biopsy data or a some what larger sector window for use in guiding biopsies.
To date, we have used 64,000-element lookup tables consisting of 40 values of the midband parameter, 40 values of the intercept parameter, and 40 values of PSA. Values were selected to correspond to the predominant ranges of actual values in the data, e.g., −65 to −35 dBr for intercept, −65 to −47 dBr for midband, and 0 to 78 (or 39) ng/ml for PSA in our latest data set from the Washington, DC, Veterans Affairs Medical Center. For all three variables, the values were linearly incremented over these ranges; e.g., for PSA, the 40 values were incremented uniformly by 2 when the range was 0 to 78 and uniformly by 1 when the range was 0 to 39.
TTI self-consistency assessment
We compared TTIs to actual biopsy histology to further assess classification performance and assure self-consistency. Average score values were computed within regions of interest (ROIs) corresponding to actual biopsy locations, and the average score for each biopsy was compared to the biopsy histology. The average scores compared to the gold-standard histology determinations were evaluated using ROC methods.
III. RESULTS
ROC-curve areas for the analysis of our most-recent data set were greater for neural-network-based classification than for the B-mode-based LOS classification by more than 20% and, with small standard errors compared to the area differences, the area differences appear to be statistically quite significant. For this data set (67 patients with 705 biopsy specimens), our NConnect radial-basis-function ROC-curve area was 0.8100 ± 0.0249; our NConnect multilayer-perceptron area was 0.7986 ± 0.0248; and our MATLAB multilayer-perceptron area was 0.8030 ± 0.0213. In comparison, the LOS-based curve area was only 0.6473 ± 0.0290. Here, ± denotes the standard error in the estimate of the area under the curve.) The average of these neural-network-based-classifier ROC curve areas was approximately 0.804, which was approximately 24% greater than the LOS-based ROC curve area of 0.647. Further more, the area difference of approximately 0.157 greatly exceeds (by a factor of nearly 3) the sum of the standard errors in the area estimates, which we define here as the average of the three separate standard errors, approximately 0.024 plus the standard error for the LOS ROC curve, 0.029 and 0.024 + 0.029 = 0.053.
As indicated in the Methods discussion above, we generated two lookup tables: one lookup t ble used PSA values of 0 to 79 ng/ml in increments of 2 ng/ml; the other used PSA values of 0 to 39 ng/ml in increments of 1 ng/ml. Examples of the lookup table for PSA values ranging from 0 to 79 ng/ml are shown in figure 1. The graph in figure 1A depicts the lookup table for PSA = 1 ng/ml; the graph in figure1 B depicts the lookup table for PSA = 8 ng/ml; the graph in figure 1C depicts the lookup table for PSA = 15 ng/ml; and the graph in figure 1D depicts the lookup table for PSA = 22 ng/ml. In each case, the vertical axis corresponds to the cancer-likelihood score; the horizontal axis on the lower left corresponds to midband value increasing from right to left; and the horizontal axis on the lower right corresponds to intercept value increasing from bottom to top; i.e., the lowest spectral-parameter values are at the nearest corner of the bottom plane and the highest values are at the farthest corner. The distribution of scores is markedly dependent on the PSA values. At lower PSA values, higher scores are apparent at lower values of midband and intercept; at higher PSA values, peak scores shift to higher spectral-parameter values and the surface contour becomes flatter. As PSA levels in crease further, this trend continues.
FIG. 1.
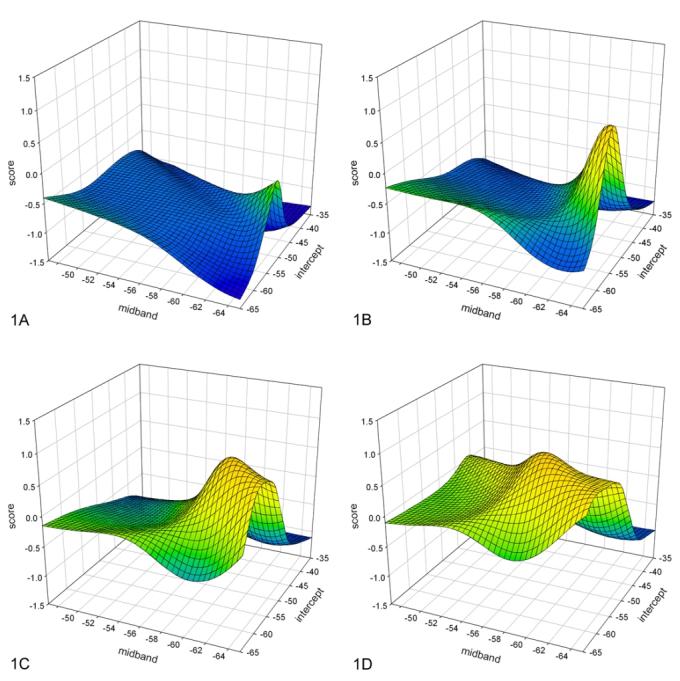
Lookup-table examples for different PSA values. A. Lookup table for PSA = 1; B. Lookup table for PSA = 8; C. Lookup table for PSA = 15; D. Lookup table for PSA = 22. The vertical axis is the score value (cancer likeli-hood) increasing toward the top; the lower axis is the midband value decreasing to the right; the right axis is the in tercept value decreasing toward the front. Note the distinct peak in score values at low midband and intercept values for low PSA values, and an increase in overall score value along with a shift of the peak scores to more-positive (higher) midband values as PSA value in creases.
We evaluated the lookup tables for self consistency by applying them directly to the parameter values originally computed in ROIs that were site matched to the biopsy regions and also by applying them to parameter values in ROIs that matched the biopsy regions in parameter images. We found that applying the lookup table with PSA values ranging from 0 to 78 ng/ml in steps of 2 to the original computed spectral-parameter values gave an ROC-curve area of 0.8456 ± 0.0205; applying it to parameter images gave an ROC-curve area of 0.8339 ± 0.0217. We then found that applying the lookup table with values ranging from 0 to 39 in steps of 1 to the original computed spectral-parameter values gave an ROC-curve area of 0.8736 ± 0.0191; applying it to parameter images gave an ROC-curve area of 0.8351 ± 0.0213.
Figure 2 shows TTIs limited to the biopsy region in two scan planes of the same patient. The biopsy in figure 2A was positive while the biopsy in figure 2B was negative. This is reflected in the red and orange coloring of figure 2A (indicating high scores for cancer likelihood) and green in figure 2B (indicating low scores for cancer likelihood). In biopsy guidance, larger TTI windows would be used, and in treatment planning, the TTI might apply to the entire scan. Examples of larger TTI windows biopsy-guidance windows are shown in figure 3 for the same patient; again, figure 3A shows the plane in which the cancerous biopsy was obtained while figure 3B shows the plane in which the noncancerous biopsy was obtained. (Spectrasonics, Inc., Wayne, PA, has developed a Hitachi-based prototype that is capable of generating TTIs in near real time and that is suitable for clinical guidance of biopsies. This prototype currently is in use by Dr. Christopher Porter at the Virginia Mason Medical Center for IRB-approved clinical research purposes.)
FIG. 2.
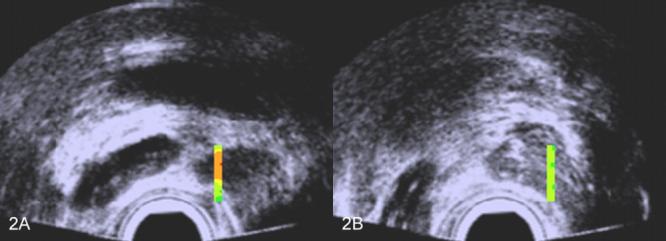
Scans with TTIs of one patient's prostate showing color encoding only in the actual biopsy region. A. TTI for which the corresponding biopsy was positive; B. TTI for which the corresponding biopsy was negative. The false colors are superimposed on a midband image: green and yellow-green indicate lower cancer likelihoods; orange and red indicate higher cancer likelihoods.
FIG. 3.
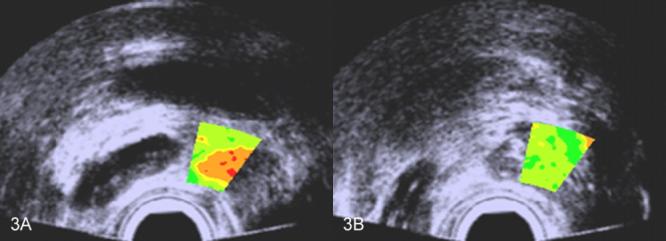
Scans with TTIs of one patient's prostate showing color encoding in a window that might be optimal for biopsy guidance. A. TTI for which the corresponding biopsy was positive; B. TTI for which the corresponding biopsy was negative. The false colors are superimposed on a midband image: green and yellow-green indicate lower cancer likelihoods; orange and red indicate higher cancer likelihoods.
V. DISCUSSION
Our initial, previously-described prostate-characterization studies were based on 1005 biopsy cores from patients at the Memorial Sloan-Kettering Cancer Center and had a cancer incidence of 10% in the cores. These earlier studies produced an ROC-curve area of 0.804 with a standard error in the estimate of 0.052 for NConnect multi-layer-perceptron neural networks with randomized assignments of cases to training, validation and test sets; and they produced an area of 0.662 with a standard error of 0.034 for the examining urologists' LOS assignments. Our most recent, currently-described studies were based on 705 biopsy cores from patients at the Washington, DC, Veterans Affairs Medical Center, and had a cancer incidence of 17% in the cores. These later studies produced ROC-curve areas very close to 0.800 with standard errors of essentially 0.024 for multilayer-perceptron and radial-basis-function neural networks implemented using two different software packages and two different schemes for assigning biopsies to training, validation and test sets; and they produced an area of 0.647 with a standard error of 0.029 for the examining urologists' LOS assignments. The initial studies utilized a classic, well-established urological ultrasound scanner that employed a single-element, mechanically-scanned transducer. Many urologists were involved in performing the sextant (three cores on each side of the gland) biopsies and assigning the LOSs. The latest studies utilized a modern scanner that employed an end-fire, curved, one-dimensional array probe. Only one urologist was involved in performing the double-sextant (six cores on each side of the gland) biopsies and assigning the LOSs. Despite the differences between many aspects of the two studies, the results are remarkably similar. In both, the ROC-curve area is approximately 23% higher for neural network classification than for conventional imaging combined with clinical information and the standard errors are sufficiently small to verify that the area differences are significant. Additional validity checks applied to the lookup tables and TTIs show encouraging consistency between the resulting TTIs and the classification scores produced by the neural networks.
V. CONCLUSIONS
ROC-curve areas for neural network classifiers in the latest data set are consistent with and strongly validate previously obtained results;2-7 they show remarkably higher classification performance than curve areas derived from conventional imaging combined with clinical gland evaluation. Lookup tables derived from neural networks to expedite imaging show score distributions as a function of midband and intercept parameter values that are consistent with current understandings of low echogenicity associated with some prostate cancers at low PSA values and evaluations of lookup tables themselves show consistency with the neural networks upon which they were based.
Current results match previously reported results from a different medical center and patient demographics, comparisons of TTIs with gold-standard histology show excellent correlations and provide very encouraging promise for improving the detection and management of prostate cancer, e.g., for guiding biopsies, planning dose-escalation and tissue-sparing options for therapy, and assessing the effects of treatment.
ACKNOWLEDGEMENTS
These studies were supported in part by NIH/NCI grant CA53561 awarded to Riverside Research Institute. This research would not have been possible without the invaluable participation and encouragement of the late William R. Fair.
REFERENCES
- 1.El-Gabry EA, Halpern EJ, Strup SE, Gomella LG. Imaging prostate cancer: current and future applications. Oncology. 2002;15:325–336. [PubMed] [Google Scholar]
- 2.Feleppa EJ, Ketterling JA, Kalisz, et al. Application of spectrum analysis and neural-network classification to imaging for targeting and monitoring treatment of prostate cancer. In: S. Schneider S, Levy M, McAvoy B, editors. Proc 2001 Ultrasonics Symp. IEEE; Piscataway, NJ: 2002. pp. 1269–1272. [Google Scholar]
- 3.Feleppa EJ, Ennis RD, Schiff PB, et al. Spectrum-analysis and neural networks for imaging to detect and treat prostate cancer. Ultrasonic Imaging. 2001;23:135–146. doi: 10.1177/016173460102300301. [DOI] [PubMed] [Google Scholar]
- 4.Feleppa EJ, Urban S, Kalisz, et al. Advances in tissue-type imaging (TTI) for detecting and evaluating prostate cancer. In: S. Schneider S, Yuhas D, editors. Proc 2002 Ultrasonics Symp. IEEE; Piscataway, NJ: 2003. pp. 1373–1377. [Google Scholar]
- 5.Feleppa EJ, Ketterling JA, Porter CR, et al. Ultrasonic tissue-type imaging (TTI) for planning treatment of prostate cancer. In: Walker W, Emelianov S, editors. Medical Imaging 2004: Ultrasonic Imaging and Signal Processing. Society of Photo-optical Instrumentation Engineers; Bellingam, WA: 2004. pp. 223–230. Proc SPIE 5373. [Google Scholar]
- 6.Feleppa EJ, Alam SK, Deng CX. Emerging ultrasound technologies for imaging early markers of disease. Disease Markers. 2004;18:249–268. doi: 10.1155/2002/167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lizzi FL, Feleppa EJ, Alam SK, Deng CX. Ultrasonic spectrum analysis for tissue evaluation. Pattern Recog Lett. 2003;24:637–658. [Google Scholar]
- 8.Scheipers U, Ermert H, Sommerfeld, et al. Ultrasonic multifeature tissue characterization for prostate diagnostics. Ultrasound Med Biol. 2003;29:1137–1149. doi: 10.1016/s0301-5629(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 9.Lizzi FL, Greenebaum M, Feleppa, et al. Theoretical framework for spectrum analysis in ultrasonic tissue characterization. J Acoust Soc Am. 1983;73:1366–1373. doi: 10.1121/1.389241. [DOI] [PubMed] [Google Scholar]
- 10.Lizzi FL, Ostromogilsky M, Feleppa, et al. Relationship of ultrasonic spectral parameters to features of tissue microstructure. IEEE Trans Ultrason Ferroelect Freq Contr. 1987;34:319–329. doi: 10.1109/t-uffc.1987.26950. [DOI] [PubMed] [Google Scholar]
- 11.Feleppa EJ, Lizzi FL, Coleman DJ, Yaremko MM. Diagnostic spectrum analysis in ophthalmology: a physical perspective. Ultrasound Med Biol. 1986;12:623–631. doi: 10.1016/0301-5629(86)90183-3. [DOI] [PubMed] [Google Scholar]
- 12.Egevad L, Granfors T, Karlberg L. Prognostic value of the Gleason score in prostate cancer. BJU Int. 2002;89:538–542. doi: 10.1046/j.1464-410x.2002.02669.x. [DOI] [PubMed] [Google Scholar]
- 13.Metz CE. ROC methodology in radiological imaging. Invest. Radiology. 1986;21:720–733. doi: 10.1097/00004424-198609000-00009. [DOI] [PubMed] [Google Scholar]


