The first verse of “John Barleycorn,” a ballad Robert Burns based on an old song of the same name, concludes with “John Barleycorn should die.” Happily, John Barleycorn does not long lie interred with “clods upon his head.” Rather, he sprouts to life, produces a head “arm'd wi'pointed spears,” and ultimately fills the “nut brown bowl” and “brandy glass” with liquid joy. The field of barley genetics is also alive and well, as is evident from the report of Rostoks et al. (1) in this issue of PNAS. This article presents compelling evidence that molecular plant breeding, based on naturally occurring allelic diversity, will be a powerful tool for the improvement of self-pollinated crops.
The evidence is based on estimates of genomewide rates of linkage disequilibrium (LD) in barley, a self-pollinated crop plant. LD is the nonrandom association of alleles at two or more loci. Barley, the fourth most important cereal crop in the world, is a model system for inbreeding species, particularly the Triticeae. Knowing the extent of LD is a prerequisite for detecting relationships between genotypes and phenotypes in sample populations. Various analysis methods, generally referred to as association genetics, are widely used for mapping disease genes in humans, and there is increasing interest in their use for gene discovery in plants (2).
Barley has long played an important role in human affairs. It was one of the first crop plants to be domesticated. The gladiators of ancient Rome were known as the barley men (hordeii). Barley was a subject of the Reinheitsgebot, a pioneering food purity law. The U.S. Food and Drug Administration recently approved the claim that diets containing certain barley foods reduce the risk of coronary heart disease. Barley and the human have in common a large genome size. In other respects, their genetics are quite different. Barley is hermaphroditic: self-pollination and homozygosity are the norms. This inbreeding simplifies and facilitates many genetic analyses and has allowed barley researchers to be early adopters of molecular tools. The many reference mapping populations, abundant linkage data, and comprehensive quantitative trait locus information generated over the last decade will provide a comparative framework for interpreting the results of association genetics.
One of the reasons for interest in association genetics is the prospect of analyzing large germplasm arrays, rather than the few genotypes that can be sampled as parents of mapping populations. The principal technical barriers to implementing association mapping in barley were a paucity of gene sequences and limited capability of efficiently detecting differences in these sequences. Consequently, there were no reliable estimates of genomewide rates of LD. Rostoks et al. (1) overcame the technical constraints by using the large collection of publicly available ESTs and a high-throughput SNP detection platform. An SNP is a single nucleotide difference in the aligned DNA sequences of two or more individuals. Thanks to a cooperative international effort, there are ≈460,000 barley ESTs in GenBank. Assembly of overlapping ESTs from multiple varieties to form unigenes (harvest.ucr.edu) allowed for in silico detection of SNPs in the aligned sequences from multiple varieties. The Illumina GoldenGate Assay provided the throughput for detection of SNPs.
Surveys of germplasm arrays will establish a global perspective on barley varieties.
In humans and in outcrossing plants such as maize, LD occurs at very short (kilobase) distances and may allow for the detection of SNPs that are the functional determinants of phenotypes (3, 4). Rostoks et al. (1) show that in barley, LD can occur at great distances (up to 60 cM). They ascribe this extensive LD to the “artificial outcrossing” accomplished by plant breeders using shared genetic resources. These controlled crosses are the crucial factor that has allowed for sufficient genetic recombination to remove spurious relationships between alleles at different loci but at the same time has preserved extensive nonrecombinant regions (haplotype blocks). Even 1 cM encompasses an enormous physical distance (5) and may contain many genes, a challenge to geneticists sifting for candidate genes in these haplotype blocks. The gene cloner's bane is the breeder's delight because meiotic recombination is the principal source of genetic diversity in breeding programs. Some haplotype blocks will contain “prepackaged” combinations of favorable alleles. The bottom line is that breeders will be able to make some marker-assisted selection (MAS) decisions using SNPs in the genes that determine target traits and many more MAS decisions based on SNPs in genes linked to the determinants of target phenotypes.
The application of SNP detection to plant breeding lies at the core of integrated European (U.K. LINK) and U.S. projects. The U.S. component, the Barley Coordinated Agricultural Project (CAP) (www.barleycap.org), involves 29 scientists at 19 institutions and is funded by the U.S. Department of Agriculture/Cooperative State Research, Education and Extension Service. Over the next 4 years, ≈3,000 mapped SNPs will be assayed on ≈4,000 barley varieties and experimental lines submitted by 10 breeding programs. Approximately 40 traits will be measured on the same germplasm. This very large data set will be stored in the Germinate database, accessed through The Hordeum Toolbox web portal, and analyzed by using QTL Miner software developed specifically for this application. The SNPs reported by Rostoks et al. (1) are a launching point. In all, five oligonuceotide polymorphism arrays (OPAs) will be developed and applied by the U.K. LINK and Barley CAP projects.
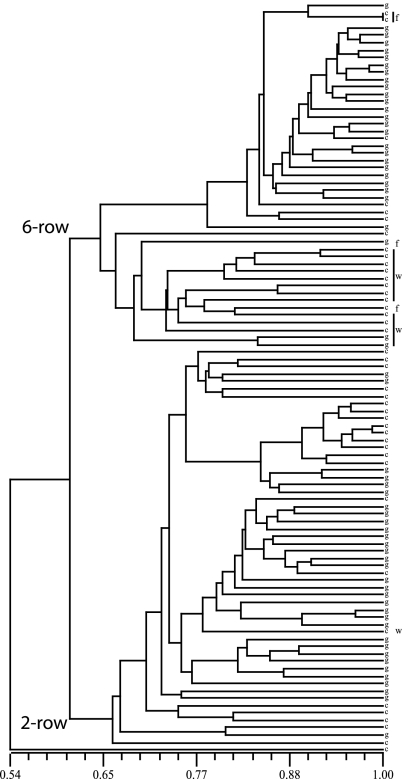
Surveys of many different germplasm arrays with these OPAs will establish a truly global perspective on the genetic relationships of barley varieties, experimental selections, genetic stocks, landraces, and ancestral species. Using 1,104 SNPs represented in pilot-OPA 2 and 102 accessions (most from North America), we generated the dendrogram shown in Fig. 1. Notable is the basal discriminating factor of six-row vs. two-row. These terms refer to the number of fertile florets per rachis node in a head of John Barleycorn. The sequence of Vrs1, the principal determinant of the number of fertile florets, is not yet published, but a prepublication search confirms that it is not represented in either of the two pilot-OPAs developed so far (Takao Komatsuda, personal communication). The extensive use of six-row spring barley for malting is peculiar to North America; in the rest of the world, spring two-row barley has long been preferred for brewing purposes. This example attests to the historical reluctance of plant breeders to mix the two germplasm pools.
Fig. 1.
UPGMA (Unweighted Pair Group Method with Arithmatic Mean) cluster analyses of 102 barley accessions genotyped with 1,104 SNPs. Genotypes are represented by their g/c SNP at VRN-H1. Inflorescence type, the basal discriminating factor, is noted as 2-row or 6-row. Winter (w) and facultative (f) accessions are indicated. Prepublication SNP data from the U.K. LINK and CAP projects were kindly provided by the OPA development team led by Timothy J. Close (University of California, Riverside, CA).
Rostoks et al. (1) identified winter vs. spring growth habit, rather than inflorescence type, as the primary determinant of structure in their data. Winter habit means that a period of low temperature (vernalization) is necessary to successfully transition from a vegetative to a reproductive state in a reasonable time frame. Spring and facultative habit accessions achieve the transition in a timely fashion, with or without vernalization. The epistatic interaction of alleles at the VRN-H1 and VRN-H2 loci determines vernalization sensitivity in barley (6). Winter and facultative accessions have a critical region in the first intron of VRN-H1 (putatively a binding site for a repressor encoded by VRN-H2) that is lacking in spring habit types (7). Functional allelic variation at VRN-H2 is caused by the presence or deletion of the gene (7, 8). This identification of growth habit as the cause of structure raises several points. The VRN-H2 gene is not represented in any of the OPAs developed so far: the spring habit varieties from which ESTs were obtained all have the deletion. Ideally, a broader representation of genes from all target germplasm will be achieved in future OPAs. Gene deletions as a source of allelic variation will need to be considered, and accounting for epistatic interactions will be essential.
Because VRN-H1 was not represented in their OPA, Rostoks et al. (1) mapped this gene and found that it did not explain the structural separation of the winter and spring germplasm groups. This result could be caused by the failure to account for VRN-H2, a lack of rigorous growth habit phenotyping, or short distance LD decay, or (as Rostoks et al. argue) the cause of structure is linked to VRN-H1. The basis of functional allelic differences in VRN-H1 appears to be caused by the presence/absence of a putative VRN-H2 binding site and the length of adjacent sequence in the first intron (8). VRN-H1 is represented in the second pilot OPA as an SNP in the 3′ UTR. We found that this G/C SNP shows no relationship with growth habit in the North American germplasm set (Fig. 1), indicating that there is rapid LD decay within the gene. Growth habit does appear as an element of structure; most winter and facultative accessions group together. The exceptions are derived from winter × spring crosses, evidence that adventuresome plant breeders can break down structural barriers. Points to be made from this preliminary comparative analysis of two samples of germplasm are: LD in barley will not always be occurring at the centimorgan level, EST-based SNPs may not detect the functional basis of allelic differences, presumed causes of structure need to be verified, and causes of structure will vary with different samples of germplasm.
Rostoks et al. (1) did find a locus 27 cM away from VRN-H1 that showed a perfect relationship with growth habit. ABC14350 coincides with the predicted linkage map position of the Fr2 low-temperature tolerance quantitative trait locus and a CBF gene cluster (9). It is tempting to speculate that the winter vs. spring structural division is caused by allelic variation at one or more CBF genes involved in regulating the low-temperature tolerance pathway (10). Rostoks et al. do not, however, report whether any of the CBF gene family members mapping to this location are present in their OPA, and they did not directly map CBF genes. The prospect that differences in CBF genes may account for adaptation to winter vs. spring growing conditions is just one example of the many exciting biological questions that will be generated by these large-scale SNP detection projects.
The U.K. LINK and U.S. CAP projects will identify regions of the barley genome harboring genes that determine productivity, adaptation, and quality. Functional allelic variants will be found in a subset of candidate genes. This information will provide plant breeders with markers for identifying and manipulating haplotype blocks and for marker-assisted selection based on “perfect” markers. This information will also provide the John Barleycorn grist for geneticists to mill for years to come.
Footnotes
The authors declare no conflict of interest.
See companion article on page 18656.
References
- 1.Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, et al. Proc Natl Acad Sci USA. 2006;103:18656–18661. doi: 10.1073/pnas.0606133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta PK, Rustgi S, Kulwal PL. Plant Mol Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Schork NJ. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 4.Hirschhorn JN, Daly MJ. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 5.Kunzel G, Korzun L, Meister A. Genetics. 2000;154:397–412. doi: 10.1093/genetics/154.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi R, Yasuda S. In: Nilan RA, editor. Barley Genetics II: Proceedings of the Second International Barley Genetics Symposium; Pullman, WA: Washington State Univ Press; 1971. pp. 388–408. [Google Scholar]
- 7.von Zitzewitz J, Szu″cs P, Dubcovsky J, Yan LL, Pecchioni N, Francia E, Casas A, Chen THH, Hayes PM, Skinner JS. Plant Mol Biol. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- 8.Szu″cs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM. Mol Genet Genom. 2007 doi: 10.1007/s00438-006-0195-8. [DOI] [PubMed] [Google Scholar]
- 9.Skinner JS, Szu″cs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Theor Appl Genet. 2006;112:832–842. doi: 10.1007/s00122-005-0185-y. [DOI] [PubMed] [Google Scholar]
- 10.Stockinger EJ, Gilmour SJ, Thomashow MF. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]