Abstract
Agricultural productivity and sustainability are continually challenged by emerging and indigenous pathogens. Currently, many pathogens can be combated only with biocides or environmentally dangerous fumigants. Here, we report a rapid and pathogen-specific strategy to reduce infection by organisms that target plant roots. Combinatorially selected defense peptides, previously shown to effect premature encystment of Phytophthora capsici zoospores, were fused to maize cytokinin oxidase/dehydrogenase as a display scaffold. When expressed in tomato roots, the peptide-scaffold constructs were secreted and accumulated to sufficient concentrations in the rhizosphere to induce zoospore encystment and thereby deter taxis to the root surface. Pathogen infection was significantly inhibited in roots expressing bioactive peptides fused to the maize cytokinin oxidase/dehydrogenase scaffold. This peptide-delivery technology is broadly applicable for rapid development of plant defense attributes against plant pathogens.
Keywords: disease resistance, Phytophthora
Agricultural crops are under constant challenge from emerging pathogens and indigenous pathogens that are recalcitrant to control (1–4). Often, host plant resistance determinants for these pathogens are not readily available in germplasm collections. Development of appropriate genetic resources for incorporation into resistance breeding efforts can take many years. Although genomic profiling of pathogens may provide targets for the development of specific anti-infective agents, these profiles are lacking for many indigenous and emerging pathogens.
Although a great deal has been learned in recent years about the contributions of resistance genes to host plant defense responses (5, 6), there has been limited success in developing resistant germplasm on the basis of this information. The complexities of plant perception of pathogen signals and the multifaceted nature of subsequent plant responses have made applications of new knowledge difficult. Given the limitations in availability of natural resistance and application of resistance genes, we have focused on a general and rapidly attainable strategy for direct disruption of pathogen development and pathogenesis.
The basis of this strategy is the identification of small peptides that function as inhibitory ligand mimics for pathogen cell-surface factors involved in either host signal reception or growth and development. Peptides that we have selected as ligand mimics have come from combinatorial libraries and are typically species- or strain-specific (7, 8). These defense peptides differ from numerous naturally occurring antimicrobial peptides that function via disruption of cell membranes and that affect a broad range of prokaryotic and some eukaryotic microorganisms (9, 10).
Three component steps of our plant defense strategy include (i) selection of peptides, from combinatorial libraries, that show high affinity for members of a plant cell-surface receptor population, (ii) assessment of impacts of affinity-selected peptides on pathogen development, and, finally, (iii) delivery of bioactive peptides as defense factors in susceptible tissues of host plants. We recently implemented this strategy against the indigenous oomycetous plant pathogen, Phytophthora capsici (Kingdom Stramenophila), which causes root disease in solanaceous crops such as tomato, melon, pepper, or squash. There is little genetic resistance available to thwart these diseases.
Ph. capsici infects plant roots, particularly at the zone of cell elongation located behind the root apical meristem, as motile zoospores that are chemotactic toward exudates from susceptible root tissues (11, 12). Once adjacent to the root surface, zoospores encyst and produce germ tubes that are also chemotactic in their growth toward a root surface (13). After root penetration and colonization, oospores are produced, and these persist in soil for many years without loss of the ability to produce infectious zoospores.
We previously addressed the first two component steps toward defense peptide deployment with a focus on the zoospore stage of Ph. capsici (7). We selected peptides from a combinatorial phage-display library (14) and chose from the affinity-selected peptide population those peptides that induced premature zoospore encystment. Approximately 50% of peptides with binding affinity induced significant premature encystment and thus could potentially disrupt the chemotactic potential of these zoospores.
Here, we report achievement of the final step in defense peptide implementation. We show that antipathogen peptides can be delivered as fusions with the naturally occurring protein, maize cytokinin oxidase/dehydrogenase (ZmCKX1) (15). ZmCKX1 is an attractive scaffold candidate for peptide display and delivery in plants. It is synthesized with an N-terminal signal peptide sequence that directs secretion from plant cells to the intercellular space (apoplast) and the rhizosphere, where contact with zoospores or invasive germ tubes is possible. Additionally, as predicted by recent 3D structural analysis (16), the solvent-exposed C terminus of ZmCKX1 provides an opportune position for peptide display.
We hypothesized that peptides displayed as part of this stable scaffold would be secreted in sufficient quantity and stability to interact with an invading pathogen and protect roots from infection. Defense peptides were secreted into the rhizosphere of transgenic tomato roots at concentrations sufficient to induce zoospore encystment, thus halting chemotaxis and inhibiting root infection.
Results
In Vitro Confirmation of Scaffold-Peptide Bioactivity.
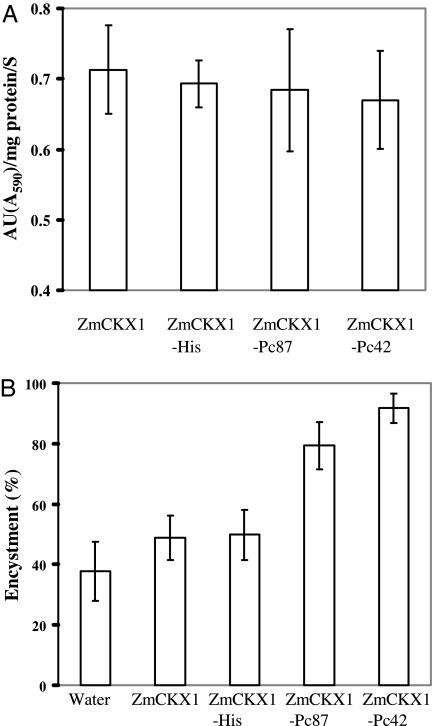
We assessed the functionality of purified scaffold peptides after expression and secretion by Pichia pastoris. We confirmed transformation of selected yeast colonies by PCR (data not shown). All scaffold peptides were secreted into the Pi. pastoris growth medium and accumulated to sufficient concentrations for detection of ZmCKX1 activity when evaluated by dichloroindophenol reduction assay (Fig. 1A). Enzymatic activities were similar among scaffold peptides and ranged from 0.67 activity units (AU) per mg−1·s−1 for ZmCKX1 alone to 0.71 AU per mg−1·s−1 for ZmCKX1-Pc42. We concluded that fusion of peptides to the C terminus of ZmCKX1 did not disrupt either the secretion of scaffold peptides or the structural aspects of ZmCKX1 required for normal enzymatic function.
Fig. 1.
Enzymatic activity of ZmCKX1 peptide constructs and induction of zoospore encystment. (A) Enzymatic activity (AU) of constructs secreted from Pi. pastoris. Secreted ZmCKX1 peptides were concentrated and purified before activity measurement. (B) Premature encystment of Ph. capsici zoospores induced by ZmCKX1 peptides. A 20-μl droplet containing ≈500 zoospores was incubated with Pi. pastoris-expressed scaffold peptides (final concentration of 0.5 μg/ul) for 90 min at room temperature. The number of zoospores encysted was counted and expressed as a percentage. The error bars indicate standard deviations (n = 6).
The fused peptides significantly affected zoospore encystment in supernatants from recombinant cultures of Pi. pastoris. ZmCKX1-Pc87 and ZmCKX1-Pc42 fusions induced encystment of 80–90% of challenged zoospores (Fig. 1B). In contrast, neither the ZmCKX1-His fusion nor native recombinant ZmCKX1 induced zoospore encystment above background levels in water. In control treatments, zoospores swam for 3 or more hours. Results of these in vitro assays confirmed that the scaffold effectively displayed peptides for interaction with zoospores.
In Planta Expression and Bioactivity of Scaffold Peptides.
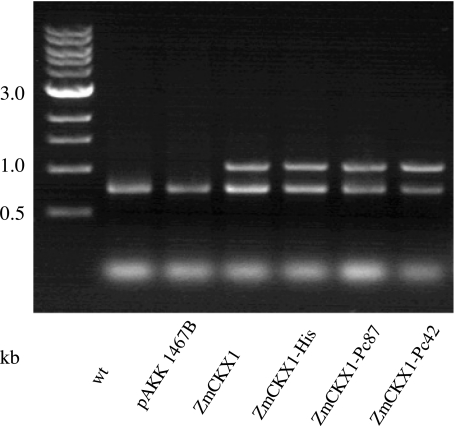
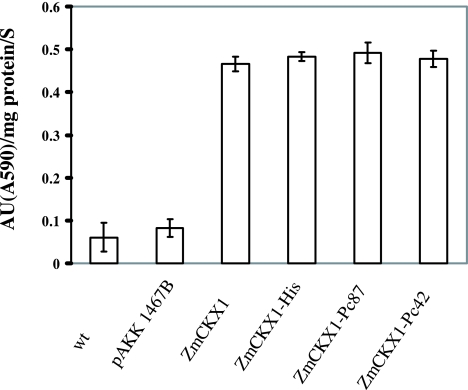
We next assessed the functionality of scaffold peptides in planta. All transgenic hairy root cultures expressed active ZmCKX1, as detected by RT-PCR (Fig. 2). The enzymatic activity of ZmCKX1 was approximately six to eight times greater in transgenic roots than in nontransformed roots or roots transformed with the pAKK 1467B vector alone (Fig. 3). The in planta functionality of ZmCKX1 corroborated conclusions drawn from experiments in Pichia that fusion of peptides to the C terminus did not disrupt secretion or enzymatic activity of the scaffold molecule.
Fig. 2.
Confirmation of ZmCKX1 peptide expression in transformed tomato roots. Total RNA was extracted from apical hairy root segments and amplified by RT-PCR. Amplification of an 800-bp fragment of a tomato actin gene was used as a control to determine the relative expression of ZmCKX1 peptide constructs (1,000 bp).
Fig. 3.
Enzymatic activity of ZmCKX1 peptides secreted from tomato roots. ZmCKX1 peptides secreted from 2 g of root tissue were concentrated before determination of enzymatic activity (AU).
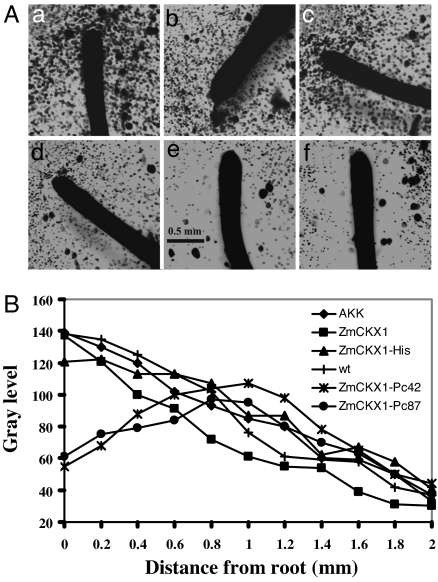
We tested the transformed roots for their influence on Ph. capsici zoospore chemotaxis. Roots transformed with scaffolds displaying bioactive peptides Pc87 and Pc42 induced spatial patterns of zoospore encystment that differed significantly from those induced by nontransformed roots or roots transformed with control scaffold constructs. Roots transformed with ZmCKX1-Pc87 or ZmCKX1-Pc42 induced “haloes” around the root cap and elongation regions that were devoid of encysted spores (Fig. 4A). In control roots, zoospores encysted immediately adjacent to the root surface and at greater distances.
Fig. 4.
Distribution of encysted Ph. capsici zoospores around the apical segments of tomato hairy roots. (A) Cyst distribution around nontransformed (wt) tomato hairy roots (a) and around hairy roots that expressed pAKK 1467B (b), ZmCKX1 (c), ZmCKX1-His (d), ZmCKX1-Pc87 (e), or ZmCKX1-Pc42 (f). Each root segment was incubated with 20,000 zoospores for 90 min before examination. (B) Density of encysted zoospores in relation to distance from the surface of roots that expressed ZmCKX1 peptides. Cyst density was estimated as the average gray-level value of pixels along 15 random transect in three images per hairy root treatment. Transects positioned perpendicularly to the root surface were 2 mm in length by 10 μm in width. Transect samples were made along the first 3 mm of root length from the root cap. Assessments of gray levels along transects were made by using MetaMorph software, Version 6.2r6 (Universal Imaging).
To assess the spatial distribution of encystment more precisely, we determined the cyst density in transect samples. In roots expressing Pc87 and Pc42, maximum cyst density, represented by maximum gray levels along sample transects, occurred at a distance of ≈1 mm from the root surface, coincident with the boundary of the cyst halo (Fig. 4B). Cyst density declined with increasing distance beyond the halo boundary. In contrast, maximum cyst density around control roots occurred at the root surface and declined monotonically with increasing distance.
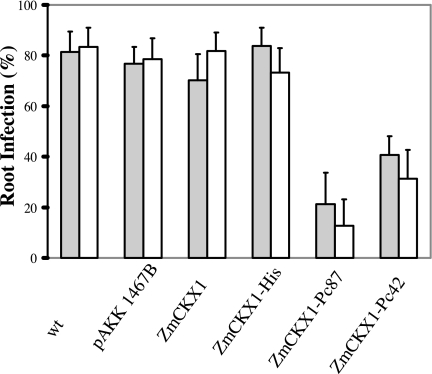
Although Pc42 and Pc87 effectively induce premature zoospore encystment, they do not inhibit germination of the cysts (7). Consequently, we assessed the impacts of ZmCKX1 peptides on root infection after encystment. The disruption of zoospore chemotactic movement to the root surface induced by ZmCKX1-Pc42 or ZmCKX1-Pc87 significantly reduced root infections caused by Ph. capsici 1794 or ATCC 15399. Fewer than 25% and 40% of the roots expressing these peptides were infected after 90 min of incubation in the presence of Ph. capsici zoospores, in contrast to roots expressing control scaffold-peptide constructs, where >70% of the root segments were infected (Fig. 5). The protective effects of ZmCKX1-Pc42 and ZmCKX1-Pc87 on zoospore chemotaxis and infection confirmed that these bioactive peptides serve as defense peptides when expressed in plant tissues.
Fig. 5.
Infection of tomato hairy roots by Ph. capsici 15399 (gray) or 1794 (white). Root infection was assessed after incubating roots with 500 zoospores of either isolate for 90 min, followed by rinsing for 45 min in running water, and culturing on corn meal agar containing 250 mg/liter ampicillin. Percentage of infection was assessed for nontransformed roots (wt) roots, roots that expressed pAKK 1467B alone, or roots that expressed ZmCKX1, ZmCKX1-His, ZmCKX1-Pc87, or ZmCKX1-Pc42. Percentage of infection represents the average from three experiments.
Discussion
This work demonstrates that peptides can be combinatorially selected to bind to pathogens and, when secreted by host plants, inhibit their development. These bioactive peptides reduced pathogen root infection when displayed by an appropriate protein scaffold.
A molecular scaffold for peptide display must have an appropriate structure to ensure that a bioactive peptide is available for interaction with a target, invasive microorganism. The scaffold must also be sufficiently stable to accumulate at functional concentrations within the plant apoplast and rhizosphere. The naturally occurring plant protein ZmCKX met these requirements. Our studies showed that this enzyme functions effectively as a peptide display scaffold.
ZmCKX1, originally characterized as an oxidase, was recently characterized more accurately to function as a dehydrogenase (17). ZmCKX1 is one of 12 members of the cytokinin oxidase/dehydrogenase family in maize. A search of genomic databases revealed only two ESTs from tomato with similarity to this gene family. ZmCKX1 degrades cytokinins and likely contributes to the maintenance of functional cytokinin concentrations (18, 19).
Morris et al. (15) suggested that the eight glycosylation sites in the ZmCKX1 molecule contribute to its ability to withstand extremes in temperature and pH. It is likely that glycosylation also contributes to ZmCKX1 peptide stability in the presence of plant proteases. Degradative proteases have been suggested as a limiting factor in attempts to express cecropin and other antimicrobial peptides in plants for protection against infection by the bacterial and fungal pathogens (20, 21).
Here, we have demonstrated that ZmCKX1 served as an effective scaffold for display of the combinatorially selected, bioactive peptides, Pc42 and Pc87, previously isolated from phage-display libraries for the ability to bind to Ph. capsici zoospores. Both scaffold-peptide constructs retained normal cytokinin-degradative activity, thus suggesting that ZmCKX1 structure was not significantly modified by fusion of either octapeptide to the exposed C terminus of the protein. The impacts of constitutively expressed ZmCKX1 peptides on cytokinin concentrations and development of tomato are unknown, but they are being studied in transgenic tomato plants that express the same scaffold-peptide constructs as used in these experiments (unpublished data). In addition, experiments are continuing to structurally modify ZmCKX1 and eliminate its enzymatic function, thus producing a neutral peptide-display scaffold.
In previous work (7), we found that zoospores induced to encyst by Pc42 or Pc87 were still able to germinate and grow. It was not known whether these germlings were still infective. Consequently, in the current experiments, it was important to examine the effects of scaffold peptides on root infection after chemotaxis was halted. We found that both ZmCKX1-Pc42 and ZmCKX1-Pc87 significantly reduced root infection relative to the infection levels seen when WT roots or roots transformed with nonbioactive peptides were challenged with the pathogen. Reductions in root infection induced by bioactive peptides may be even greater in soil or hydroponic culture where population densities of Ph. capsici zoospores are significantly lower than in these experiments. Furthermore, because roots are always elongating, the susceptible elongation region would continue to move forward in soil away from encysted zoospores. Using tomato plants stably transformed with ZmCKX1 peptides, we assessed rhizosphere accumulation of defense peptides and the validity of this root protection scenario in the presence of realistic population densities of Ph. capsici in soil (unpublished data).
The maximum size of peptides that can be displayed at the terminal position of ZmCKX1 is not certain. However, ZmCKX1 scaffolds created to display three copies of the peptides, Pc42 and Pc87, in tandem were equally effective at inducing zoospore encystment by in vitro assay (unpublished data).
Peptides Pc42 and Pc87 were shown previously to be species-specific in their zoospore encystment effect (7). Similarly, combinatorial peptides selected against the human bacterial pathogen, Haemophilus influenzae, were shown to be strain-specific in their effects on bacterial cell growth (8). Such specificity of action contrasts with the typically broad spectrum membrane disruption caused by antimicrobial peptides. The ability to select combinatorial peptides with this level of specificity in action is likely to be advantageous in designing plant defense strategies with minimal effect on nontarget beneficial microorganisms.
In addition to bioactive functions, affinity-selected peptides can be used to identify and characterize specific members of the population of cell-surface factors that interact with these ligand mimics. We expect that the identification of these surface factors, in combination with the increasing availability of genomic and proteomic information, will accelerate the pace at which specific bioactive peptides can be identified and deployed for plant defense.
The strategy for selective plant defense that we have developed did not require genomic or proteomic profiles of host–pathogen interactions. We were able to identify defense peptides by assessing the influence of affinity-selected peptides on aspects of development critical to a microorganism's success as a plant pathogen and by providing a means of stable delivery to sites of infection. In addition to selection of defense peptides that are effective against oomycetous and bacterial pathogens, we have recently selected peptides that inhibit the growth of Uromyces appendiculatus, the fungal pathogen that causes common bean rust (unpublished data). Our strategy could have broad utility in confronting indigenous and emerging oomycetous and fungal pathogens for which resistance is unknown and that lack genomic and proteomic profiles. These technologies will also enhance the ability to respond to new forms of resistance that develop within ever-changing pathogen populations in the field.
Materials and Methods
Fungal Species and Zoospore Production.
Ph. capsici isolates, 1794 and ATCC 15399, were grown on clarified 10% V8 vegetable juice agar (Campbell Soup, Camden, NJ) for 3–6 days in the dark. To induce sporangium formation, cultures were incubated 2 additional days at 25°C under fluorescent lights (12 h light/dark). To release zoospores from sporangia, cultures were flooded with sterile deionized water and incubated at room temperature for 30 min (7). To determine zoospore density, an aliquot of a zoospore suspension was vortexed for 30 s to induce encystment, and the number of cysts was determined with a hemocytometer.
Scaffold-Peptide Construction for Expression in Pi. pastoris.
A recombinant version of ZmCKX1 was used as a scaffold for peptide display. The 3′ end at the ZmCKX1-encoding gene was modified to include HindIII and XbaI restriction sites for insertion of sequences encoding the octapeptides, Pc42 (VSPNVHDG) or Pc87 (DRPSMSPT). These bioactive peptides were demonstrated previously to induce premature Ph. capsici zoospore encystment (7). A stop codon was located immediately after the peptide insertion site. The native N-terminal signal of ZmCKX1 was replaced with the α-factor propeptide of Saccharomyces cerevisiae to ensure efficient secretion of expressed ZmCKX1 peptides from Pi. pastoris (22).
A control construct included modified ZmCKX1 with a sequence insert encoding a His tag (HHHHHHG). This sequence was used to represent a nonselected, nonbioactive peptide. A second control treatment included nonmodified ZmCKX1.
Expression and Purification of ZmCKX1 Peptides from Pi. pastoris.
ZmCKX1-peptide constructs were ligated into the Pi. pastoris cytoplasmic expression vector, pPZIC (Easy Select Pichia Expression Kit Version B; Invitrogen, Carlsbad, CA). Pi. pastoris strain X33 was transformed and manipulated further according to kit instructions. Typically, transformed yeast colonies were selected on Difco YPDS medium (Becton Dickinson, Sparks, MD) amended with 100 μg/ml zeocin. One confirmed transformant colony from each constructed ZmCKX1 peptide was inoculated into BMGY (Invitrogen) medium, grown overnight, resuspended in BMGY medium containing 0.5% (vol/vol) methanol, and grown at 30°C with vigorous aeration. Additional methanol was added (0.5%, vol/vol) at 24, 48, and 72 h postinoculation. At harvest, culture medium was centrifuged, and the supernatant was cleared by filtration through a 0.22-μm Stericup filter (Millipore, Billerica, MA). Recovered medium was concentrated through a Millipore NMWL:30,000 ultrafiltration membrane, and the medium was exchanged with TE buffer (10 mM Tris·Cl, pH 8.0/1 mM EDTA) in a stirred-cell filtration apparatus. Purification of protein to >95% electrophoretic homogeneity was achieved by size-exclusion chromatography on Superose 12 (Amersham Pharmacia Biotech, Piscataway, NJ) (18).
Enzymatic Activity of Native and Recombinant ZmCKX1.
Secreted ZmCKX1 activity was measured by a continuous dichloroindophenol (DCPIP) reduction assay (15, 18). In a microtiter well, 150 μl of a ZmCKX1-peptide construct was combined with 100 μl of reaction mixture consisting of 250 μM zeatin as substrate and 250 mM sodium phosphate (pH 7.0), 125 μM DCPIP, and 2.5 mM EDTA. A control treatment for each ZmCKX1-peptide construct included the same reaction mixture, but without zeatin. DCPIP reduction was measured at 590 nm at 10-s intervals for 10 min in a VersaMax (Molecular Devices, Sunnyvale, CA) microplate reader and quantified with Softmax Pro 4.3.1 software. Total protein in culture medium was assessed by Bradford assay.
Zoospore Encystment.
To assess zoospore encystment, a 20-μl droplet containing 500 zoospores was gently mixed with each ZmCKX1 peptide (final concentration 0.5 μg/μl) on a glass slide and incubated for 90 min at room temperature. Encystment was quantified over time by time-lapse photography (3-s exposure period). In captured images, encysted zoospores appeared as single points, and motile zoospores traced nonlinear tracks (7).
Scaffold-Peptide Constructs for in Planta Expression.
Scaffold-peptide constructs were created in the same manner as for Pichia expression, except that the native ZmCKX1 N-terminal signal sequence was retained. Scaffold peptides were inserted initially into the vector, pCGT 2998 (received from Christopher Taylor, Donald Danforth Plant Science Center, St. Louis, MO). Sequence region, TobRB7-5A, the tobacco root-specific promoter on vector pCGT 2998, was replaced by the sequence encoding the cauliflower mosaic virus 35S promoter, and the ZmCKX1-peptide fragment was substituted for the vector sequence between EcorI and Xba. Subsequently, the fragment containing the 35S promoter, ZmCKX1 peptide, and OCS terminator was excised from the vector with Sse8387I restriction enzyme and cloned into a transformation vector, pAKK 1467B (received from Christopher Taylor). This vector contained GFP driven by the constitutive superubiquitin promoter. The final constructs for plant root transformation included pAKK1476B containing nonmodified ZmCKX1, ZmCKX1-Pc42, ZmCKX1-Pc87, ZmCKX1-His, or AKK 1467B alone.
Tomato Transformation and Regeneration of Hairy Roots.
Root transformation was performed following the protocol of Collier et al. (23). Each binary scaffold-peptide construct was introduced into Agrobacterium rhizogenes strain K599. Bacterial cells were then grown in Difco (Franklin Lakes, NJ) LB broth, concentrated by centrifugation, and resuspended in GIBCO (Carlsbad, CA) 0.25× Murashige and Skoog basal medium (Invitrogen) at pH 5.8.
Tomato plants (Peto 343) were grown in vermiculite for 12–14 days. Roots were removed from each seedling by severing the shoot at the ground line. Each explant, with shoot and attached leaves, was inserted into a Fibrgro cube (Sarnia, Ontario, Canada) that had been saturated with transformed A. rhizogenes K599. Explants were maintained on the laboratory bench under high humidity. The regenerated root system represented a composite of transformed and WT tissues. The status of individual regenerated roots was evaluated by assessing GFP activity with a MZFLIII Stereoscope (Leica Microsystems, Bannockburn, IL) equipped with a Chroma (Rockingham, VT) filter 11000v3, Optronics (Chelmsford, MA) CCD camera, and MagnaFire acquisition software (Optronics).
Individual transgenic roots expressing GFP were excised and sterilized with a plant surface sterilization kit (Qbiogene, Irvine, CA). Roots were then placed on Whatman (Middlesex, U.K.) filter paper (8.2 cm) overlaid on Gamborg's B-5 medium (Sigma, St. Louis, MO) (pH 5.7) that was amended with 200 mg/liter Timentin (GlaxoSmithKline, Research Triangle Park, NC) and solidified with 0.8% Daishin agar (Brunschwig-Chemie, Amsterdam, The Netherlands). Cultures were incubated at 25°C in the dark to induce root elongation and branching. After incubation, 2-cm-long apical root segments were transferred to 15 ml of Gamborg's B-5 liquid medium. One milliliter of fresh medium was added every other day. Root cultures were incubated on a VWR (West Chester, PA) model 300 rotary shaker and agitated at 50 rpm for 10 days before challenge with Ph. capsici zoospores.
Confirmation of ZmCKX1 Peptide Expression in Regenerated Root Tissue.
Total RNA was isolated from hairy root tissue with a RNeasy plant mini-kit (Qiagen, Valencia, CA). Single-stranded cDNA was produced by reverse transcription with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) using appropriate primers (Table 1). PCR was carried out with TaqDNA polymerase (Invitrogen).
Table 1.
Primers for detection of ZmCKX1 peptide expression in tomato hairy roots
| Construct | Sense primer* | Antisense primer |
|---|---|---|
| Actin† | ACCCAATTGAGCACGGAATT | GCCAATGCAGTAATTTCTTTGC |
| wt‡ | CACGGCCCACAGATATCTAAT | CTAGACTAGTCAAAGCTTGTTG |
| AKK 1467B | Same | Same |
| CKX | Same | Same |
| CKX-Pc42 | Same | TCCATCATGTACATTAGGTGATAC |
| CKX-Pc87 | Same | TGTTGGTGACATTGATGGTCTATC |
| CKX-His | Same | TCCGTGGTGGTGGTGGTGGTG |
*All primer sequences are oriented 5′ to 3′.
†The tomato actin gene (GenBank accession no. U60480) was used as an internal reference.
‡Nontransformed roots.
Enzymatic Activity of ZmCKX1 Peptides Secreted from Transgenic Roots.
Two grams of root tissue from each transgenic root culture was harvested and incubated in 15 ml of Gamborg's B-5 medium (pH 5.7) at 25°C in the dark. After 12 h of incubation, the medium was passed through a Centricon YM-30 filter (Millipore) and concentrated to ≈300 μl by centrifugation. Concentrated medium was then exchanged twice with 10× volume of TE buffer (10 mM Tris·Cl, pH 8.0/1 mDM EDTA) and concentrated again. Finally, 300 μl of medium was cleared by filtration through a 0.22-um syringe filter (Fisher Scientific, Hampton, NH) before assessment of protein content and enzyme activity as described.
Assessment of Rhizosphere Zoospore Encystment.
Replicate apical root segments with intact meristems were removed from culture medium and placed individually on glass slides. Each terminal, nonwounded 1.5-cm segment was immersed in a water droplet (120 μl) containing 20,000 Ph. capsici zoospores. Zoospore accumulation and encystment around a root segment was visualized and quantified after 60–90 min of incubation by image capture and analysis using MetaMorph software, Version 6.2r6 (Universal Imaging, Downington, PA).
Patterns of zoospore encystment were assessed by quantifying cyst distribution along 2-mm transects positioned perpendicularly to the root surface at 15 randomly selected locations within 3 mm of the root cap along the elongation zone. In captured images, regions of visibly high cyst density were reflected by regions of high gray level. Cyst density (gray scale) was averaged across transects of three replicate root segments per scaffold peptide treatment.
Assessment of Root Infection.
Thirty apical root segments from cultures that expressed a specific scaffold peptide were immersed in 2 ml of water containing 5,000 zoospores. Only nonwounded tissues were immersed in the zoospore suspension. After 90 min of incubation, roots were rinsed under running water for 45 min to remove zoospores and cysts that had not germinated and penetrated root tissues. Roots were then plated on Difco cornmeal agar containing 250 mg/liter ampicillin. The percentage of root segments infected was determined after 2–3 days of incubation. Infection assays were performed in triplicate for each ZmCKX1 peptide construct.
Acknowledgments
We thank Christopher Taylor for providing vectors pCGT 2998 and pAKK 1467B. This work was supported in part by a grant from the U.S. Department of Agriculture–Illinois Missouri Biotechnology Alliance. F.J.S. was a grantee of the National Aeronautics and Space Administration Exobiology program during part of this work.
Abbreviations
- ZmCKX1
maize cytokinin oxidase/dehydrogenase
- AU
activity units.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
This article is a PNAS direct submission.
References
- 1.Bove JM. J Plant Pathol. 2006;88:7–37. [Google Scholar]
- 2.Pivonia S, Yang XB. Phytopathology. 2006;96:400–407. doi: 10.1094/PHYTO-96-0400. [DOI] [PubMed] [Google Scholar]
- 3.O'Donnell K, Kislter HC, Tacke BK, Casper HH. Proc Natl Acad Sci USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shubert TS, Rizvi SA, Sun X, Gottwald TR, Graham JH, Dixon WN. Plant Dis. 2001;85:340–356. doi: 10.1094/PDIS.2001.85.4.340. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Martin GB, Bogdanove AJ, Sessa G. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 7.Bishop-Hurley SL, Mounter SA, Laskey J, Morris RO, Elder J, Roop P, Rouse C, Schmidt FJ, English JT. Appl Environ Microbiol. 2002;68:3315–3320. doi: 10.1128/AEM.68.7.3315-3320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop-Hurley SL, Schmidt FJ, Smith AL. Antimicrob Agents Chemother. 2005;49:2972–2978. doi: 10.1128/AAC.49.7.2972-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 10.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 11.Erwin DC, Ribeiro OK. In: Phytophthora Diseases Worldwide. Erwin DC, Ribeiro OK, editors. St Paul, MN: Am Phytolpath Soc; 1996. pp. 1–7. [Google Scholar]
- 12.Warburton AJ, Deacon JW. Fungal Genet Biol. 1998;25:54–62. doi: 10.1006/fgbi.1998.1086. [DOI] [PubMed] [Google Scholar]
- 13.Morris P, Bone E, Tyler BM. Plant Physiol. 1998;117:1171–1178. doi: 10.1104/pp.117.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith GP, Petrenko VA. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 15.Morris RO, Bilyeu KD, Laskey JG, Cheikh NN. Biochem Biophys Res Commun. 1999;255:328–333. doi: 10.1006/bbrc.1999.0199. [DOI] [PubMed] [Google Scholar]
- 16.Malito E, Coda A, Bilyeu KD, Fraaije MW, Mattevil A. J Mol Biol. 2004;341:1237–1249. doi: 10.1016/j.jmb.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 17.Galuszka P, Frébort I, Šebela M, Sauer P, Jacobsen S, Peè P. Eur J Biochem. 2001;268:450–461. doi: 10.1046/j.1432-1033.2001.01910.x. [DOI] [PubMed] [Google Scholar]
- 18.Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ, Kramer MD, Morris RO. Plant Physiol. 2001;125:378–386. doi: 10.1104/pp.125.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mok DW, Mok MC. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 20.Oard SV, Enright FM. Plant Cell Rep. 2006;25:561–572. doi: 10.1007/s00299-005-0102-5. [DOI] [PubMed] [Google Scholar]
- 21.Owens LD, Heutte TM. Mol Plant–Microbe Interact. 1997;10:525–528. doi: 10.1094/MPMI.1997.10.4.525. [DOI] [PubMed] [Google Scholar]
- 22.Scorer CA, Buckholz RG, Clare JJ, Romanos MA. Gene. 1993;136:111–119. doi: 10.1016/0378-1119(93)90454-b. [DOI] [PubMed] [Google Scholar]
- 23.Collier R, Fuchs B, Walter N, Lutke WK, Taylor CG. Plant J. 2005;43:449–457. doi: 10.1111/j.1365-313X.2005.02454.x. [DOI] [PubMed] [Google Scholar]