Abstract
One of the fundamental interactions in cell biology is the binding of cell receptors to adhesion ligands, and many aspects of cell behavior are believed to be regulated by the number of these bonds that form. Unfortunately, a lack of methods to quantify bond formation, especially for cells in 3D cultures or tissues, has precluded direct probing of this assumption. We now demonstrate that a FRET technique can be used to quantify the number of bonds formed between cellular receptors and synthetic adhesion oligopeptides coupled to an artificial extracellular matrix. Similar quantitative relations were found between bond number and the proliferation and differentiation of MC3T3-E1 preosteoblasts and C2C12 myoblasts, although the relation was distinct for each cell type. This approach to understanding 3D cell–extracellular matrix interactions will allow one to both predict cell behavior and to use bond number as a fundamental design criteria for synthetic extracellular matrices.
Keywords: fluorescence resonance energy transfer, myoblast, osteoblast, proliferation, RGD peptides
Cell–extracellular matrix (ECM) binding interactions, via cell surface receptors such as integrins (1, 2), regulate a wide array of developmental, pathological, and regenerative processes (3–5). Both the number of these bonds that form and the specific receptor–ligand pairing may mediate this signaling (6, 7). Although identification of the pairing may be readily accomplished and clearly regulates intracellular signaling and gene expression (2, 4, 7, 8), tools to quantify bond formation, especially for cells within a 3D environment, are lacking. Delineating this latter relation may be particularly important both in understanding the cellular behavior in different ECM and in developing a strategy to design synthetic ECM or cell-instructive materials, because biomaterials are frequently functionalized with molecules containing a receptor-binding domain to direct cell fate (5, 9). The ligand molecular structure, overall density, and spatial distribution at the micrometer and nanometer scale may all influence cell adhesion (e.g., focal adhesion formation), signaling pathways, cellular proliferation, apoptosis, migration, differentiation (9–14), and tissue formation (15, 16). It is unclear whether a subset or all of these variables alter the cellular response by simply altering bond number, but this assumption underlies many aspects of current biomaterials design. One can analyze receptor–ligand bonding with radioactive molecules (17), fluorescent molecules (18), and chemical assays (19), but these methods have been limited to analyzing cells suspended in medium or adherent to 2D substrates. Encapsulation of cells in 3D better reflects in vivo cell adhesion to ECM (20) and leads to distinct patterns of gene expression (21, 22) but is not amenable to the application of existing methods to quantify bond formation.
In this study a FRET technique is demonstrated to allow one to quantify the number of receptor–ligand bonds formed between cells and adhesion peptides coupled in a 3D gel matrix. FRET techniques have been widely used to probe molecular interactions at the nanometer scale in the past (23–25). For this study a biomimetic cell adhesion substrate was designed by covalently linking synthetic oligopeptides containing the Arg-Gly-Asp sequence [i.e., (Gly)4-Arg-Gly-Asp-Ala-Ser-Ser-Lys-Tyr(G4RGDASSKY)] to polymer molecules. These oligopeptides allow these polymer chains, which are originally inert to cells, to induce cell adhesion to 2D substrates and 3D matrices via specific interactions and to regulate cell phenotype (5, 9, 15, 26). Both bone-forming (MC3T3-E1 preosteoblasts) and muscle-forming (C2C12 myoblasts) cells, which provide useful models for tissue engineering studies, were used in these studies to broadly examine the relation between bond formation and proliferation and differentiation (27, 28), because both processes are critical to both natural and engineered tissue morphogenesis (29, 30). Furthermore, both processes in these cell types are known to depend on binding to RGD ligands or entire fibronectin molecules (15, 26).
Results
Calibration of FRET Technology with Cell Suspensions.
The first studies involved calibrating the FRET technology by correlating energy transfer to peptide bond formation assessed by using 125I-G4RGDASSKY peptides coupled to the polymer in solution. Polymer chains that present 125I-labeled G4RGDASSKY oligopeptides (RGD peptides) (Fig. 1a) were added to cell suspensions at various concentrations, and the degree of substitution for RGD peptides on a single polymer molecule was also varied from 1 to 20 peptides per polymer chain. The number of 125I-G4RGDASSKY bound to cells increased in both cases in an approximately linear fashion as the total number of RGD peptides (NRGD/cell) was increased (Fig. 2a), as expected. The degree of FRET, represented by DFRET, between the RGD peptides coupled to polymers and the cell receptors was assessed in parallel first with polymer chains that present a single G4RGDASSKY oligopeptide labeled with rhodamine (acceptor) (Fig. 1b) and cells labeled with the fluorescein (donor) (Fig. 1c). Fluorescein molecules adjacent to receptors were presumed to exclusively participate in the energy transfer to labeled RGD peptides that bind to these receptors (31). DFRET was quantified with the emission intensity of fluorescein in the absence (Γfluorescein,0) and presence (Γfluorescein) of rhodamine-G4RGDASSKY-polymer following Eq. 1 (23):
Mixing fluorescein-labeled cells with rhodamine-G4RGDASSKY-polymer decreased the emission intensity of fluorescein [maximized at wavelength (λ) of 520 nm], which indicates energy transfer between the fluorescein and rhodamine probes (Fig. 2 b–d). The minimal increase in the intensity of rhodamine emission in these conditions is attributed to bleed of the donor emission at 580 nm that obscures the acceptor fluorescence at low levels of energy transfer (see Fig. 3c for acceptor fluorescence at higher energy transfer). In contrast, both mixing of fluorescein-labeled cells with rhodamine-G4RGEASSKY-polymer (non-receptor binding peptide) (diamond in Fig. 2c) and exposure of cells to a high concentration of free RGD peptides before mixing with the G4RGDASSKY-polymer, to block the cell receptors (red curve in Fig. 2d and Fig. 5, which is published as supporting information on the PNAS web site), greatly inhibited energy transfer. These results confirm that changes in the donor emission are the result of specific peptide–receptor interactions and rule out an influence of bleaching between fluorescein molecules in the system. The inverse system (cell membranes stained with donor and peptide stained with acceptor) showed similar results (results not shown). The difference of DFRET between the experimental conditions is larger than that caused by instrumental error and the biological variation, which are typically 0.02–0.05 (32). Energy transfer increased with the number of polymer chains presenting peptides (Fig. 2 c and d), until a saturating peptide concentration, likely because of a saturation of cellular receptors. This response of energy transfer to NRGD/cell was different from the measurement using 125I peptides, especially when multivalent polymer chains (containing multiple peptides) were used. Therefore, the energy transfer measured only using monovalent polymer chains (containing a single 125I peptide) was calibrated to the number of RGD peptides bound to cells (Fig. 2e). This calibration curve was used in all subsequent studies of cells immobilized within 3D peptide-presenting gels.
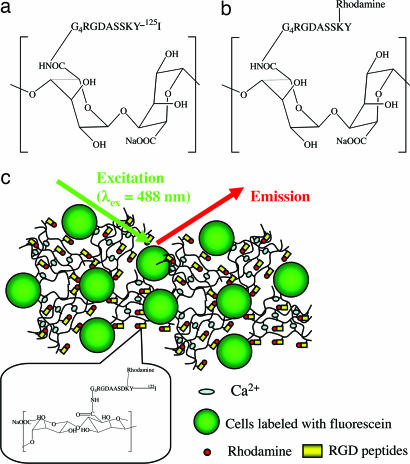
Fig. 1.
G4RGDASSKY oligopeptides (RGD peptide) were covalently bound to alginate molecules with varying degrees of substitution. 125I was linked to the tyrosine (Y) residue for the radioactive assay (a), and rhodamine was linked to the lysine (K) residue for the FRET experiments (b). In FRET experiments, cell membranes were stained with fluorescein (c). The fluorescent cells were mixed with various numbers of rhodamine-G4RGDASSSKY-polymer molecules. Gels were formed by cross-linking polymer chains with Ca2+ ions (c).
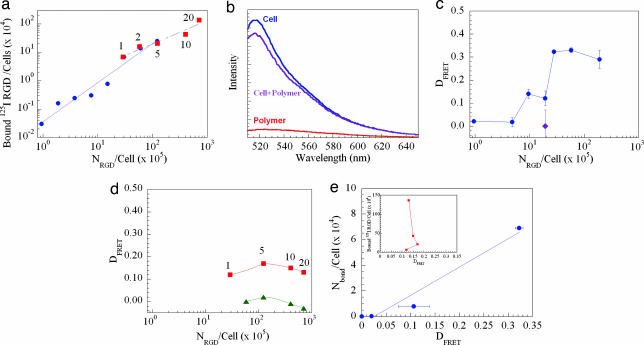
Fig. 2.
Generation of a calibration curve relating the degree of energy transfer (DFRET) to the number of 125I-RGD peptides on polymer chains bound to cells (MC3T3-E1 preosteoblasts). The number of RGD peptides bound to cells suspended in serum-free medium was measured by using 125I-G4RGDASSKY-polymer (a). The overall number of RGD peptide (NRGD) was varied either by altering the number of polymers with one RGD peptide per chain in the solution (●) or by varying the degree of substitution of the polymer chains (■). Mixing cells containing fluorescein (donor) with rhodamine-G4RGDASSKY-polymer decreased the emission intensity of the donor, which was maximized at a wavelength (λ) of 520 nm (b). The DFRET increased both as the number of polymer chains containing a single RGD peptide was increased in the solution (c) and as the degree of substitution was raised (d). The ♦ in c indicates the DFRET measured with rhodamine-G4RGEASSKY-polymer. Curves –■– and –▴– in d represent the DFRET measured with unblocked cells and cells blocked with free RGD peptides before adding cells to RGD-coupled polymer chains, respectively. DFRET was calibrated to the number of RGD peptides bound per cell (Nbond/cell), assessed with the 125I-G4RGDASSKY-polymer (R2 = 0.90), over the linear response region of DFRET (e). Inset in e shows that DFRET measured by using multivalent RGD peptides is not correlated to the number of 125I-labeled peptides bound to cells. Numbers in a and d represent the substitution degree. In the FRET measurements, mixtures of cells and polymers were excited at a λ of 488 nm by using a fluorometer. Data points and error bars in c and e represent the mean and SD from four independent experiments.
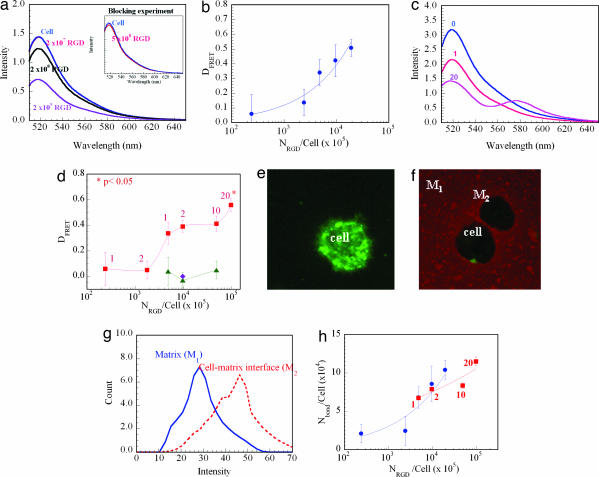
Fig. 3.
Quantification of the number of receptor–ligand bonds (Nbond) formed by cells (preosteoblasts) in the 3D gel matrices. The overall number of RGD peptides (NRGD) was first varied by increasing the number of rhodamine-G4RGDASSKY-polymer chains (each presenting one RGD peptide per chain) (a and b). These polymer chains were mixed with unmodified polymer at different volume ratios. Increasing the NRGD/cell decreased the emission intensity of fluorescein from the cell membrane (a). The numbers in a represent NRGD/cell. Blocking cell receptors with free RGD peptides before encapsulating cells in the gel matrix limited the decrease in the emission intensity of fluorescein (Inset in a). DFRET was related to the NRGD/cell in the gel matrix following a power law (R2 = 0.92) (b). Alternatively, the NRGD was varied with the degree of substitution while keeping the ratio between G4RGDASSKY-polymer and unmodified polymer constant at 1:4 (c and d). Increasing the degree of substitution decreased the emission intensity of fluorescein (c). The numbers in c represent the number of RGD peptides conjugated to each single polymer chain. The increase in DFRET was also related to NRGD following a power law. The difference in the values between the 1 NRGD/cell and 20 NRGD/cell in d were statistically significant (P < 0.05). Besides, curves –■– and –▴– in d represent the FRET measurements with normal cells and cells blocked with soluble RGD peptides, respectively, before their encapsulation. ♦ in d represents the condition in which cells were encapsulated in gels of 1:4 rhodamine-G4RGEASSKY-polymer/unmodified polymer. The FRET between cells and RGD peptides in the gel matrix was also visualized by using a confocal microscope. The strong color intensity of fluorescein staining in the cell membrane in the unmodified gel (e) was greatly decreased as cells were encapsulated in the gel presenting rhodamine-G4RGDASSKY-polymer chains (f). The color intensity of rhodamine at the interface between cells and the gel matrix was also increased as illustrated with microscopic images (f) and quantification using image analysis software (g). e corresponds to the fluorescence emitted between 500 and 540 nm, and f was prepared by overlaying fluorescence emitted between 500 and 540 nm and fluorescence emitted between 580 and 620 nm. By using the calibration curve (Fig. 2e), the Nbond/cell in the gel was calculated for experiments by using both approaches to modify the peptides available per cell (h). In h, curves –●– and –■– represent the situations in which Nbond/cell was varied with the number of polymer chains containing one RGD peptide/chain and the degree of substitution, respectively. Data points represent the mean and SD from four independent experiments.
Measurement of Number of Receptor–Ligand Bonds in a 3D Gel Matrix.
The FRET between cells and RGD peptides in a 3D gel matrix was next examined to quantify bond formation. The gel matrix was prepared by cross-linking a mixture of rhodamine-G4RGDASSKY-polymer and unmodified polymer (Fig. 1c). A higher number of peptides was needed in 3D culture to obtain linear binding responses, and this likely is related to the greatly diminished mobility of polymer chains after cross-linking, and thus access of peptides to the encapsulated cells. In this study the substitution degree of RGD peptides was kept constant at 1 and the NRGD/cell in the gel matrix was varied by altering the ratio between the rhodamine-G4RGDASSKY-polymer and unmodified polymer at a given polymer concentration of 1% (vol/vol). The emission intensity of fluorescein from the cell membrane was significantly decreased as the NRGD/cell was increased (Fig. 3a). However, encapsulating cells in gels presenting RGE peptides (diamond in Fig. 3d) and exposing cells to a high concentration of free oligopeptides before encapsulating them in the gel (Inset in Fig. 3a and red curve in Fig. 3d) greatly inhibited the energy transfer, irrespective of the NRGD/cell, again confirming the specificity of the receptor–ligand bonding measured with FRET. DFRET was increased with the NRGD/cell in the gel matrix, following a power law (Fig. 3b). Increasing the degree of substitution also decreased the intensity of donor emission (Fig. 3c) and subsequently increased the DFRET (Fig. 3d), although the dependency of DFRET on NRGD/cell was smaller than found when NRGD/cell was increased by using monovalent polymer containing a single peptide per chain. At a substitution degree of 20, the intensity of the acceptor emission was readily appreciated. FRET at the interface between cells and the gel matrix was also confirmed with microscopy, and a decreased intensity of the donor emission on the cell membrane was noted (Fig. 3e) along with an increased intensity of acceptor emission in the polymer interfacial region (Fig. 3 f and g).
The number of receptor–ligand bonds per cell (Nbond/cell) for cells within these gels was calculated from the DFRET by using the calibration curve (Fig. 2e). The Nbond/cell increased up to 1 × 105 as the NRGD/cell was increased (Fig. 3h), and the dependency of Nbond/cell on the NRGD/cell was reduced as the degree of substitution for RGD peptide was raised [i.e., Nbond ∝ (NRGD)α; α was decreased from 0.5 to 0.2 as one alters NRGD by increasing the degree of substitution]. Similar studies were performed with C2C12 myoblasts, with similar results (data not shown).
Correlation of the Number of Receptor–Ligand Bonds to Cell Growth and Differentiation.
The relevance of bond number to cell function was next assessed by analyzing the proliferation and differentiation of MC3T3 preosteoblasts and C2C12 myoblasts. For this study the DFRET was again measured after incubating cell-encapsulating gels in the serum-containing cell culture medium (same culture conditions as cell proliferation/differentiation studies). The presence of serum decreased the DFRET, corresponding to a decrease in bond formation, but DFRET increased with NRGD/cell in the same manner as gels incubated in serum-free medium (Fig. 4a).
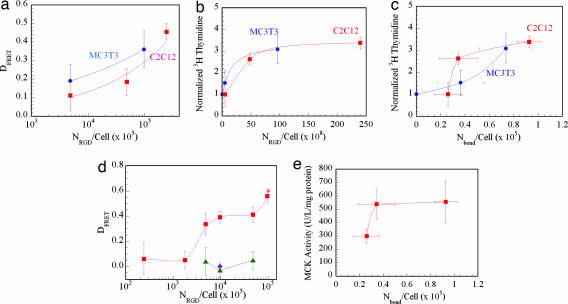
Fig. 4.
Cell proliferation and differentiation were regulated with the number of receptor–ligand bonds (Nbond). The larger NRGD in the gel matrix increased the degree of energy transfer (DFRET) between cells and RGD peptides (a) and subsequently the Nbond/cell for both MC3T3 preosteoblasts and C2C12 myoblasts. Passage through the cell cycle was increased with the overall number of RGD peptides (NRGD/cell) for both cell types as measured with the amount of [3H]thymidine incorporated into cells (b). The amount of [3H]thymidine, normalized to the lowest proliferation value at that condition, was also related to Nbond/cell for both cell types (c). Differentiation rates, measured with the level of osteocalcin secreted from preosteoblasts (d) and the level of MCK activity of myoblasts (e), were also related to Nbond/cell. Data points and error bars represent the mean and SD calculated from four independent experiments.
The proliferation of both cell types similarly increased with NRGD (Fig. 4b), irrespective of the valency of RGD peptides. Interestingly, cells encapsulated in gels that consist of polymers having a different valency but that lead to the similar Nbond/cell had a similar frequency of cell division (Fig. 6, which is published as supporting information on the PNAS web site). The dependency of cell growth on Nbond was specific for the cell type (Fig. 4c). The level of osteocalcin secretion, a marker of osteogenic differentiation (Fig. 4d), and the level of muscle creatine kinase (MCK) activity, a marker of myogenic differentiation (Fig. 4e), were also up-regulated as Nbond/cell was increased. Strikingly, both the dependency of cell proliferation and the dependency of cell differentiation on Nbond/cell were similar for each cell type.
Discussion
Altogether, these data demonstrate both a previously undescribed approach to first quantify receptor-adhesion ligand formation in 3D culture, and a quantitative relation between proliferation and differentiation and bond number. Specifically, it is demonstrated that the relationship between cell–ECM interactions and cellular phenotype is correlated with the number of receptor–ligand bonds. We speculate that the number of bonds quantitatively regulates the formation of focal adhesions and the rearrangement of cytoskeletal structures and subsequently alters signaling pathways that promote proliferation and cell differentiation (2, 4, 8, 9–13). This finding has significant impact for many areas of biology and will likely lead to many future studies probing the mechanism of these relations.
The FRET technique used in this study provides an accurate measurement for receptor–ligand bonds. Specifically, an advantage of FRET measurements of bond formation over 125I-based measurements, even in solution, is evidenced by the studies with polymer chains that present multiple peptides. The 125I-peptide measurements demonstrated a continuous increase in bond formation as the multivalency of the polymer was raised, whereas the FRET measurements indicated a saturation of binding. The 125I-peptide measurements will provide an inaccurate measurement for multivalent ligands, because if even one peptide on a polymer is bound to a cellular receptor the entire polymer chain will be associated with the cells and counted with this assay. This is avoided with the FRET-based analysis, because only ligands within 5–10 nm of a receptor will result in energy transfer (23).
Using the FRET technique, we demonstrate that the cellular microenvironment is able to modulate the number of receptor–ligand bonds. The smaller dependency of the bond number on the total ligand number with use of multivalent ligands (Fig. 3h) may be related to an increase in steric interference or unfavorable conformations of certain peptides. However, the effects of multivalent ligands on initiating receptor clustering (33), not monitored in this study, may lead to profound changes in cells even in the absence of changes in total bond number. In addition, the higher magnitude of energy transfer in gels than that observed in cell suspension likely results from an enhanced ability of cells to associate and perhaps cluster RGD peptides. Strikingly, for cells within the gels, the Nbond/cell is larger than the number of peptides theoretically available to each cell, if one assumes that cell receptors can only access peptides present in the nearest 10 nm of the gel as in previous calculations (6). This result suggests that cells either actively probe the gels and recruit peptides at distances >10 nm or expand their surface area (e.g., send extensions into surrounding gel) to access increasing numbers of peptides, in a manner similar to what has been observed for cells adhered on the surfaces of materials. Cells generate significant traction forces after adhesion (34, 35), and this allows them to rearrange adhesion ligands in a manner dependent on the mechanical properties of the adhesion substrate (36). This phenomenon may partially underlie the ability of cells in 3D culture in the current studies to bind to such high numbers of peptides. It is likely that the mechanical properties also influence cellular activity at least partially independent of changes in bond number (37), although this has not yet been directly evaluated.
This method, through appropriate modification, may be broadly applied to a range of cell types and adhesion ligands in a manner that allows the contribution of distinct cell receptors to total bond formation to be analyzed in vitro and even in vivo in a noninvasive manner. Furthermore, this method may be applied in the future to understand the relationship between bond formation and cell migration (38). This approach may also ultimately take the design of synthetic ECM, which currently is based on empirical testing of the relation between ligand presentation and cell response, to a predictive approach based on preestablished quantitative relations that tie fundamental biological interactions to cell behavior and greatly improve their utility in a variety of stem or progenitor cell-based therapies (39, 40).
Materials and Methods
Radiolabeling of G4RGDASSKY-Alginate.
(Gly)4-Arg-Gly-Asp-Ala-Ser-Ser-Lys-Tyr (G4RGDASSKY) oligopeptides (Commonwealth Biotechnology, Richmond, VA) were linked with 125I (PerkinElmer, Boston, MA). These iodinated oligopeptides were coupled to sodium alginate (FMC Biopolymer, Philadelphia, PA) by using a described carbodiimide chemistry (26). The molar ratio of oligopeptides to sugar residues was varied from 0.0015:1 to 0.03:1.
Fluorescent Labeling of G4RGDASSKY-Alginate.
The oligopeptides bound to alginate molecules were covalently linked to rhodamine succinimidyl ester (Invitrogen, Carlsbad, CA) following a described procedure (34). In certain experiments, G4RGEASSKY bound to alginate molecules were linked to rhodamine. The molar ratio between fluorophores and coupled oligopeptides was kept constant at 1:1. Polymers were reconstituted to 2% (wt/wt) solutions with serum-free αMEM (Invitrogen), after sterilization via filtering and freeze-drying.
Cell Culture.
Murine MC3T3-E1 preosteoblasts, a generous gift from Renny Franceschi (University of Michigan), were cultured in ascorbic acid-free αMEM supplemented with 10% FBS (Invitrogen) and 100 units/ml penicillin–streptomycin (PS) (Invitrogen). Cells with passage number between 14 and 20 were used in this study. C2C12 myoblasts (American Type Culture Collection, Manassas, VA) were cultured in high-glucose DMEM (Invitrogen) supplemented with 10% FBS and 100 units/ml PS. Cells with passage number between 3 and 12 were used in this study.
Measurement of the Number of RGD Peptides Bound to Cells Using 125I-G4RGDASSKY-Alginate.
Preosteoblasts were first suspended in the serum-free αMEM at a density of 20,000 cells per milliliter. Then, cell suspensions were mixed with 125I-G4RGDASSKY-alginate solution at varied polymer concentrations. The number of RGD peptides linked to a single alginate chain varied from 1 to 20. After incubating the cell–polymer mixture at 37°C for 20 min, 125I-G4RGDASSKY-alginate molecules unbound to cells were removed after centrifugation at 1,200 rpm (Eppendorf 5810R) for 5 min. Then, cells were resuspended in the fresh αMEM. The activity of 125I was measured with a γ-counter. The number of 125I-G4RGDASSKY-alginate bound to cells (A1) was calculated by using a calibration curve that relates the number of 125I-G4RGDASSKY to the radioactivity. Next, the number of nonspecific binding between cells and 125I-G4RGDASSKY-polymer (A2) was determined by measuring the radioactivity after the incubation of cells in high concentration of free RGD peptides. This process leads to the substitution of the 125I-G4RGDASSKY-polymer bound to cells via the receptor–ligand bond with free RGD peptides. Then, the number of specific bonds between RGD peptides and cell receptors was assessed by subtracting A2 from A1.
Measurement of FRET Between Rhodamine-G4RGDASSKY-Alginate and Cells Suspended in the Medium.
Cell membranes were stained with fluorescein by incubating cells with 5-hexadecanoylaminofluorescein (Invitrogen) in αMEM for 24 h. Then, fluorescently labeled cells (1 million cells per milliliter) were incubated with varied number of rhodamine-G4RGDASSKY-polymer in FBS-free αMEM for 20 min. The mixtures of cells and cell adhesion polymers were excited at a wavelength of 488 nm, and the resulting emission spectrum was collected by using a fluorometer (Fluoromax; Jobin Ivon, Edison, NJ) at 37°C. The degree of energy transfer was determined by comparing the peak height of emission curve from fluorescein, maximized at wavelength of 520 nm, in the presence and absence of rhodamine-G4RGDASSKY-polymer by using Eq. 1. In certain control experiments, cells were incubated with rhodamine-G4RGEASSKY-polymer. Alternatively, to block cell receptors, cells were preincubated with high concentrations of free RGD peptides before exposure to rhodamine-G4RGDASSKY-polymer. The degree of energy transfer was calibrated to the number of receptor–ligand bonds assessed with 125I-G4RGDASSKY-polymer.
Measurement of FRET Between Cells and Rhodamine-G4RGDASSKY-Alginate in the 3D Gel Matrix.
Cell membranes were stained with 5-hexadecanoylaminofluorescein in advance. Then, cells were mixed with a 1% (wt/wt) mixture of unmodified polymer and rhodamine-G4RGDASSKY-polymer. Subsequently, the cell–polymer mixtures were mixed with calcium sulfate (CaSO4; Sigma, St. Louis, MO) slurries to form gel matrices. Gel disks with 10-mm diameter and 1-mm thickness were punched after 10 min and incubated in the FBS-free αMEM at 37°C for 2 h. In certain experiments, gel disks were incubated in the αMEM supplemented with FBS for 4 h. The density of cells was kept constant at 10 million cells per milliliter. The number of RGD peptides in the gel matrix was varied by altering the volume ratio between the unmodified polymer and rhodamine-G4RGDASSKY-polymer (containing one peptide per polymer chain). In certain experiments, the substitution degree of RGD peptides varied from 1 to 20 while keeping the volume ratio between unmodified polymer and polymer with coupled RGD peptides constant. The emission spectrum from gel disks was collected by using a fluorometer with excitation of 488 nm. From the peak height of emission curve, the degree of energy transfer was quantified following Eq. 1. In control experiments, cells were encapsulated in the gel presenting rhodamine-G4RGEASSKY-polymer, or cells were preincubated with a high concentration of free RGD peptides before encapsulating them in the gel presenting rhodamine-G4RGDASSKY-polymer.
Cells stained with fluorescein were also visualized by using a laser scanning confocal microscope (Olympus, Center Valley, PA). Cells encapsulated in the gel matrix were excited at 488 nm by using a laser. The fluorescence emitted between 500 and 540 nm and the fluorescence emitted between 580 and 620 nm were collected through separate detector channels. Images were subjected to background subtraction by using National Institutes of Health image processing software (ImageJ 1.36b).
Cell Proliferation and Differentiation Assays.
Cells were encapsulated in the gel matrix with a varied number of RGD peptides. The density of cells was kept constant at 10 million cells per milliliter. The overall polymer concentration to form the gel matrix was kept constant at 1% (wt/wt), and the volume ratio between unmodified polymer and polymer to present RGD peptides was kept constant at 4:1 for MC3T3 preosteoblasts. For C2C12 myoblasts, the overall polymer concentration to form the gel matrix was kept constant at 2% (wt/wt). The volume ratio between unmodified polymer and polymer to present RGD peptides was kept constant at 0:1.
The cell culture medium supplemented with FBS and PS was exchanged every 2 days. Passage of cells through the cell cycle was examined by measuring [3H]thymidine incorporation at 7 days. Cells were incubated in the medium containing 3H-labeled thymidine (PerkinElmer) and collected by dissolving gels in 50 mM EDTA (Sigma)-PBS (Invitrogen). Cells were lysed with 12 M NaCl aqueous solution, and [3H]thymidine incorporation was quantified by using a scintillation counter (Amersham Pharmacia).
Osteogenic differentiation of MC3T3 preosteoblasts was evaluated from the level of osteocalcin secretion. Preosteoblast cells encapsulated in the gel matrix were incubated in the αMEM supplemented with 10% FBS, 100 units/ml PS, 50 μg/ml ascorbic acid (Sigma), 10 mM β-glycerophosphate (Sigma), and 0.1 μM dexamethasone (Sigma), while exchanging the medium every 2 days. Cell culture medium was collected after 1 and 2 weeks. The concentration of osteocalcin in the medium was analyzed with a mouse osteocalcin ELISA kit (Biomedical Technologies, Stoughton, MA). The osteocalcin concentration was normalized to the total protein concentration of lysed cells that had been encapsulated in gel matrices in parallel. Protein concentrations were measured by using protein assay solution (Bio-Rad, Hercules, CA).
Myogenic differentiation of C2C12 myoblasts was evaluated from the MCK activity. Cells encapsulated in the gel matrix were incubated in DMEM supplemented with 10% horse serum (Invitrogen) and 100 units/ml PS. After 1 week, cells isolated from the gel matrix were lysated by using a passive lysis buffer (Promega, Madison, WI). Then, cell lysates were analyzed by using a MCK assay kit solution (Pointe Scientific, Canton, MI), and the total MCK activity was normalized with protein concentration in the sample.
Supplementary Material
Acknowledgments
We thank Dr. Jeff Lichtman for allowing the use of the confocal microscope. This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research Grants R37 DE013033 and R01 DE13349 and U.S. Army Research Laboratories and Research Office Grant DAAD-19-03-1-0168.
Abbreviations
- ECM
extracellular matrix
- MCK
muscle creatine kinase
- PS
penicillin–streptomycin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Adams JC, Watt FM. Development (Cambridge, UK) 1993;117:1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 2.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 3.Sakai T, Larsen M, Yamada KM. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 4.Clark EA, Brugge JS. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 5.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 6.Massia SP, Hubbell JA. J Cell Biol. 1991;114:1089–1110. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asthagiri AR, Nelson CM, Horwitz AF, Lauffenburger DA. J Biol Chem. 1999;274:27119–27127. doi: 10.1074/jbc.274.38.27119. [DOI] [PubMed] [Google Scholar]
- 8.Langholz O, Rockel D, Mauch C, Kozlowska E, Bank I, Krieg T, Eckes B. J Cell Biol. 1995;131:1903–1915. doi: 10.1083/jcb.131.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano Y, Mooney DJ. Adv Mater. 2004;16:17–25. [Google Scholar]
- 10.Ingber DE. Proc Natl Acad Sci USA. 1990;87:3579–3583. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen L, Mooney DJ, Vacanti JP, Ingber DE. Mol Biol Cell. 1994;5:967–975. doi: 10.1091/mbc.5.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 13.Lee KY, Alsberg E, Shiong S, Comisar WA, Linderman J, Ziff R, Mooney DJ. Nano Lett. 2004;4:1501–1506. doi: 10.1021/nl0493592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallant ND, Michael KE, Garcia AJ. Mol Biol Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsberg E, Anderson KW, Albeiruti A, Franceschi RT, Mooney DJ. J Dental Res. 2001;80:2025–2029. doi: 10.1177/00220345010800111501. [DOI] [PubMed] [Google Scholar]
- 16.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 17.Su Z, Liu GZ, Gupta S, Zhu ZH, Rusckowski M, Hnatowich DJ. Bioconjugate Chem. 2002;13:561–570. doi: 10.1021/bc0155566. [DOI] [PubMed] [Google Scholar]
- 18.Lee FH, Haskell C, Charo IF, Boetigger D. Biochemistry. 2004;43:7179–7186. doi: 10.1021/bi0362121. [DOI] [PubMed] [Google Scholar]
- 19.Keselowsky BG, Garcia AJ. Biomaterials. 2005;26:413–418. doi: 10.1016/j.biomaterials.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 20.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 21.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, Davis GE. J Cell Sci. 2001;114:2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 22.Birgersdotter A, Sandberg R, Ernberg I. Semin Cancer Biol. 2005;15:405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Kluwer Academic/Plenum; 1999. [Google Scholar]
- 24.Baneyx G, Baugh L, Vogel V. Proc Natl Acad Sci USA. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myong S, Rasnik N, Joo C, Lohman TM, Ha T. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 26.Rowley J, Mooney DJ. J Biomed Mater Res. 2002;60:217–223. doi: 10.1002/jbm.1287. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Christensen K, Chawla K, Xiao GZ, Krebsbach PH, Franceschi RT. J Bone Miner Res. 1999;14:893–903. doi: 10.1359/jbmr.1999.14.6.893. [DOI] [PubMed] [Google Scholar]
- 28.Garcia AJ, Vega MD, Boettiger D. Mol Biol Cell. 1999;10:789–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinman HK, Phip D, Hoffman MP. Curr Opin Biotechnol. 2003;14:526–532. doi: 10.1016/j.copbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Rosso F, Giordano A, Barbarisi M, Barbarisi A. J Cell Physiol. 2004;199:174–180. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- 31.Chigaev A, Buranda T, Dwyer DC, Prosnittz FR, Sklar LA. Biophys J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel SS, Thaler C, Koushik SV. Sci STKE. 2006;331:1. doi: 10.1126/stke.3312006re2. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto S, Akiyama SK, Yamada KM. Science. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- 34.Wang N, Butler JP, Ingber DE. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 35.Choquet D, Felsenfeld DP, Sheetz MP. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 36.Kong HJ, Polte T, Alsberg E, Mooney DJ. Proc Natl Acad Sci USA. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engler A, Bacakova L, Neewman C, Hategan A, Griffin M, Discher D. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palecek SP, Huttenlocher A, Horwitz AF, Lauffenburger DA. J Cell Sci. 1998;111:929–940. doi: 10.1242/jcs.111.7.929. [DOI] [PubMed] [Google Scholar]
- 39.Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 40.Orive G, Hernandez RM, Gascon R, Chang TNS, De Vos P, Hortelano G, Hunkeler D, Lacik I, Shapiro AMJ, Pedraz JL. Nat Med. 2003;9:104–107. doi: 10.1038/nm0103-104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.