Abstract
Remodeling of mitochondria is a dynamic process coordinated by fusion and fission of the inner and outer membranes of the organelle, mediated by a set of conserved proteins. In metazoans, the molecular mechanism behind mitochondrial morphology has been recruited to govern novel functions, such as development, calcium signaling, and apoptosis, which suggests that novel mechanisms should exist to regulate the conserved membrane fusion/fission machinery. Here we show that phosphorylation and cleavage of the vertebrate-specific Pβ domain of the mammalian presenilin-associated rhomboid-like (PARL) protease can influence mitochondrial morphology. Phosphorylation of three residues embedded in this domain, Ser-65, Thr-69, and Ser-70, impair a cleavage at position Ser77–Ala78 that is required to initiate PARL-induced mitochondrial fragmentation. Our findings reveal that PARL phosphorylation and cleavage impact mitochondrial dynamics, providing a blueprint to study the molecular evolution of mitochondrial morphology.
Keywords: protein evolution, protein phosphorylation, rhomboids, mitochondrial dynamics, intramebrane proteolysis
Mitochondrial biogenesis is an essential cellular process governed by a small set of proteins with membrane pro-fusion and pro-fission activities which are conserved in all eukaryotes (1–3). During metazoan evolution, this process has been recruited to coordinate novel mitochondrial functions, such as apoptosis (4–6), thereby suggesting the emergence, in higher eukaryotes, of novel mechanisms of regulation of the fusion and fission machinery of the organelle. Formal, mechanistic evidence supporting this hypothesis is, however, still missing.
Recently, rhomboid proteases have been implicated in the regulation of mitochondrial membrane remodeling. Studies in Saccharomyces cerevisiae demonstrated that PCP1P is required to cleave Mgm1p, an intermembrane space dynamin family member that participates in mitochondrial fusion events (7, 8). The yeast PCP1P protein belongs to a subfamily of mitochondrial rhomboid proteases typified by presenilin-associated rhomboid-like (PARL) protein (9, 10), the human ortholog of PCP1P (8). Despite their functional and structural conservation, PCP1P and PARL have unrelated N-terminal domains. The N-terminal region of PARL shows no detectable similarity to any other available protein sequences. This region of PARL, designated Pβ (spanning amino acids 40–100), is vertebrate-specific, as indicated by the notable conservation among mammals and, to a lesser extent, other vertebrates, but not between vertebrates and insects (11). Although the function of the Pβ domain remains unknown, its biological relevance is evident from its sequence conservation. Indeed, in the four available mammalian PARL sequences, 58 of the 62 residues of the Pβ domain are invariant, and there are no insertions or deletions (11), which suggests that at least during mammalian evolution, the N-terminal region of PARL was subject to strong purifying selection, which can be explained by functional constraints. In unconstrained sequences evolving neutrally, very few, if any, invariant residues would be expected to survive the ≈100 million years of evolution separating mammalian orders (12, 13). This analysis suggests that emergence of the Pβ domain at the outset of vertebrate evolution may be associated with the appearance of a new mechanism of regulation of PARL. We have recently shown that this part of the PARL molecule undergoes two consecutive cleavage events, termed α and β. The proximal α-cleavage is a constitutive processing associated with the protein import in the mitochondria, whereas the distal β-cleavage is regulated through a mechanism of proteolysis requiring PARL activity supplied in trans (11). Whether this cleavage occurs in vivo is unknown. In addition, its mechanism of regulation and its functional significance remain unexplored.
Results
Human PARL Is Subjected to β-Cleavage in Vivo.
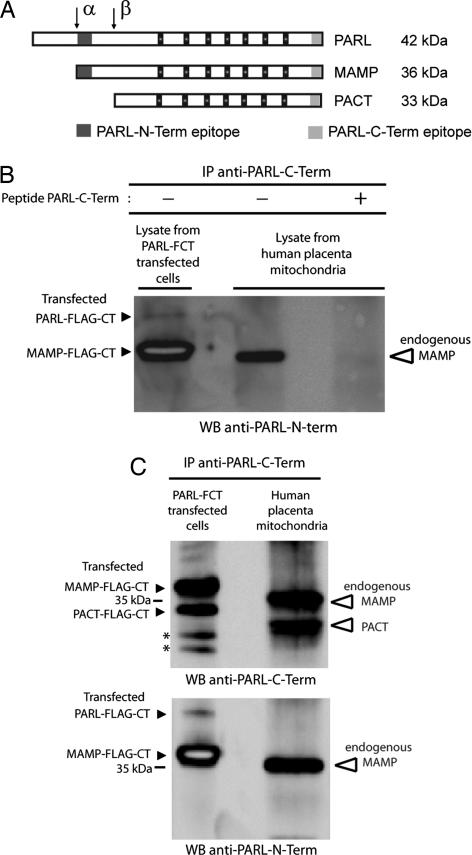
PARL transfected in HEK 293 cells is cleaved at position Ser77–Ala78, which maps within the vertebrate-specific Pβ domain (11), suggesting that, in vivo, the rhomboid protease may undergo the same processing. To address this possibility, we generated a polyclonal antibody against a peptide spanning the C terminus of PARL (anti-PARL-C-Term; Fig. 1A). This specific antibody (Fig. 1B) was used to immunoprecipitate endogenous PARL from lysates of mitochondria isolated from human placenta. Using antibodies recognizing the N-terminal and C-terminal regions of PARL (Fig. 1A), we then examined the cleavage of the endogenous protein relative to the transfected PARL-FCT by epitope mapping. Data showed two bands whose mobility is, as expected, slightly higher than that of the FLAG-tagged positive control (Fig. 1C Upper). Although both forms are immunoreactive against anti-PARL-C-Term, only the slower migrating band was positive to anti-PARL-N-Term, indicating that the corresponding epitope was absent in the faster migrating band (Fig. 1C). These data strongly suggest that endogenous PARL N terminus undergoes β-cleavage, indicating that this processing may mechanistically coordinate the function of the rhomboid protease in vivo.
Fig. 1.
Cleavage of PARL Pβ domain in vivo. (A) Schematic representation of the α- and β-cleaved forms of PARL, MAMP, and the PARL C-terminal product of β-cleavage (PACT). The locations of the epitopes recognized by the anti-PARL-N-Term and anti-PARL-C-Term antibodies are indicated. Small black squares depict the seven transmembrane helixes of PARL. (B) Specificity of the anti-PARL-C-Term antibody. The anti-PARL-C-Term antibody specifically immunoprecipitates (IP) PARL-FLAG-CTerminus (PARL-FCT) from HEK 293-transfected cells as well as endogenous PARL from mitochondrial lysates of human placenta. The specificity was addressed by preadsorbing the antisera with the synthetic peptide PARL-C-Term used to generate the corresponding antibody. WB, Western blotting. (C) Endogenous PARL is subjected to N-terminal β-cleavage as observed for transfected PARL. Transfected PARL-FCT and endogenous PARL were immunoprecipitated with anti-PARL-C-Term and subjected to epitope mapping. (Upper) Both the transfected and endogenous PARL are present in two forms, MAMP and PACT, which migrate according to the predicted molecular mass shown in A. The lower band labeled as PACT-FLAG-CT corresponds to the product of β-cleavage at residue Ser77–Arg78 (11). Note the slight difference in mobility between transfected and endogenous PACT, which is caused by the presence/absence of the FLAG tag. (Lower) Transfected and endogenous PACT lack the N-Term epitope because of N-terminal β-cleavage. Asterisks indicate nonspecific cleavages.
Endogenous and Transfected PARL Are Hyperphosphorylated at the Vertebrate-Specific Pβ Domain.
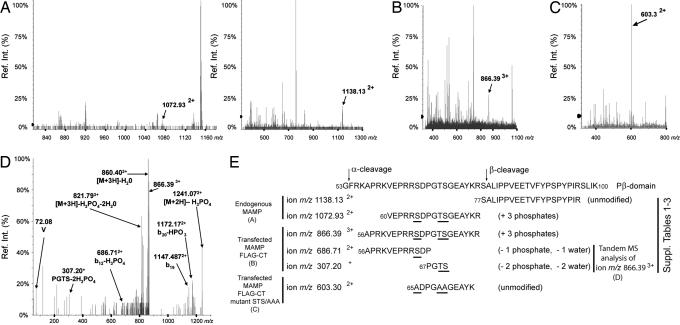
To investigate the mechanism of regulation of PARL β-cleavage in vivo, we conducted mass spectrometric studies to identify posttranslational modifications on the Pβ domain (14, 15). PARL was immunoprecipitated from lysates obtained from 250 mg of solubilized human placenta mitochondria, it was digested, and the peptides were subjected to LC/MS analysis. Data showed that >35% of the entire protein was covered (Table 1, which is published as supporting information on the PNAS web site), with two molecular ions spanning nearly the entire Pβ domain of the mitochondrial mature form of PARL, MAMP (Fig. 1A). Ion m/z 1072.932+ corresponded to a triple-phosphorylated 60VEPRRSDPGTSGEAYKR76 peptide, which maps between the α- and β-cleavage sites; ion m/z 1138.132+ corresponded instead to an unmodified peptide spanning the β-cleavage site and its distal region (77SALIPPVEETVFYPSPYPIR96; Fig. 2A and E).
Fig. 2.
Phosphorylation of PARL Pβ domain at Ser-65, Thr-69, and Ser-70 in vivo and in vitro. (A) Phosphorylation of endogenous PARL. LC/MS analysis of PARL from mitochondria purified from human placenta is shown. The protein was immunoprecipitated with the anti-PARL-C-Term antibody (Fig. 1), digested, and subjected to LC/MS analysis. (Left) Molecular ion m/z 1072.932+, which corresponds to a triple-phosphorylated 60VEPRRSDPGTSGEAYKR76 peptide mapping between PARL α- and β-cleavage sites (Fig. 1A) (11). (Right) Ion m/z 1138.132+, corresponding to the unmodified 77SALIPPVEETVFYPSPYPIR96 peptide, which also maps on the vertebrate-specific Pβ domain of PARL. More than 35% of the mature form of PARL (MAMP; Fig. 1A) could be found through this analysis; the complete list of the ions is shown in Table 1. The identity of each peptide was determined manually and with a Bayesian reconstruction algorithm as well as searching against both theoretical peptide and fragmentation data from the PARL sequence. (B) Phosphorylation of transfected PARL-FCT. LC/MS analysis of transfected PARL-FCT purified from HEK 293 cells is shown. MAMP-FLAG-CTerminus (MAMP-FLAG-CT) was immunoprecipitated with anti-FLAG monoclonal antibody, purified by gel electrophoresis, digested, and analyzed by LC/MS analysis. The triple-phosphorylated 56APRKVEPRRSDPGTSGEAYKR76 peptide, ion m/z 866.393+, is indicated. More than 51% of MAMP sequence could be found through this analysis; the complete list of ions is shown in Table 2. (C) PARL mutant S65A/T69A/S70A is not phosphorylated. LC/MS analysis of transfected PARL-FCT mutant AAA purified from HEK 293 cells is presented. The data show ion m/z 603.32+, corresponding to the unphosphorylated 65ADPGAAGEAYK75 peptide. Note that no phosphorylated peptides encompassing the Pβ domain of this mutant protein were found (data not shown). (D) Tandem MS analysis of phosphorylated PARL. Ion m/z 866.393+ was fragmented to detect peptides that, through the loss of phosphate group(s) and/or water (−H3PO4), finely map phosphorylation at Ser-65, Thr-69, and Ser-70. The N-terminal ion m/z 686.712+ (56APRKVEPRRSDP–H3PO4) and m/z 307.2+ (67PGTS–2H3PO4) are shown in (E). The complete list of molecular ions is shown in Table 3. The identity and phosphorylation state of each peptide were determined by both manual interpretation of the spectra and a Mascot search of all of the enhanced product ion scans. (E) Schematic representation summarizing the results showing phosphorylation of endogenous and transfected PARL at residue Ser-65, Thr-69, and Ser-70.
To investigate whether transfected PARL is also phosphorylated, we overexpressed PARL-FCT in HEK 293 cells. The protein was immunoprecipitated with anti-FLAG to isolate the transfected protein. It was unlikely that endogenous PARL was copurified during this step because coimmunoprecipitation studies with PARL constructs harboring different tags did not reveal homodimers or oligomers (data not shown). The α-cleaved form of PARL, MAMP (Fig. 1A), was then isolated by gel electrophoresis, digested, and subjected to LC/MS analysis. More than 51% of the protein was covered (Table 2, which is published as supporting information on the PNAS web site). Within this peptide data set we also observed a triple-phosphorylated 56APRKVEPRRSDPGTSGEAYKR76 peptide, molecular ion m/z 866.393+ (Fig. 2B), which overlaps with most of the triple- phosphorylated m/z 1072.932+ identified during the analysis of the endogenous PARL. Molecular ion m/z 1138.132+, 77SALIPPVEETVFYPSPYPIR96, was also found (Table 2), indicating that sample preparation and analyses were performed under comparable experimental conditions. Similar results were also obtained from PARL-FCT isolated from transfected HeLa cells (data not shown).
To refine these results, we subjected ion m/z 866.393+ to tandem MS analysis. Data showed a series of three water and phosphoric acid losses as the primary detected fragments (Fig. 2 D and E and Table 3, which is published as supporting information on the PNAS web site), consistent with the fragmentation pattern of a peptide with phosphorylated Ser and Thr, rather than Tyr residues. Additionally, the nonphosphorylated y3 ion and the Y immonium ion but not their corresponding phosphorylated species were detected, indicating lack of phosphorylation at Tyr-74 of PARL. Furthermore, the N-terminal ion b12–H3PO4, an internal ion series (GTSG–2H3PO4, PGTS–2H3PO4, DPGTSG–2H3PO4), and the C-terminal ion y18–H3PO4 indicate phosphorylation at Ser-65, Thr-69, and Ser-70 (Fig. 2 D and E and Table 3). This conclusion was further supported by the lack of phosphorylated peptides in the data set obtained from LC/MS analysis of a transfected PARL mutant bearing Ala substitutions at these residues (Fig. 2 C and E and data not shown). A monophosphorylated peptide spanning Ser-65, Thr-69, and Ser-70 was detected in vivo as well as in vitro (Table 1); however, its relative abundance was very low compared with the triple- phosphorylated form (data not shown), indicating that most of the α-cleaved form of PARL is phosphorylated. We conclude that the vertebrate-specific Pβ domain of endogenous and transfected PARL is phosphorylated at residues Ser-65, Thr-69, and Ser-70, and, by implication, that this modification has the same function in vivo and in vitro.
The Phosphorylated Pβ Domain Is Exposed to the Matrix.
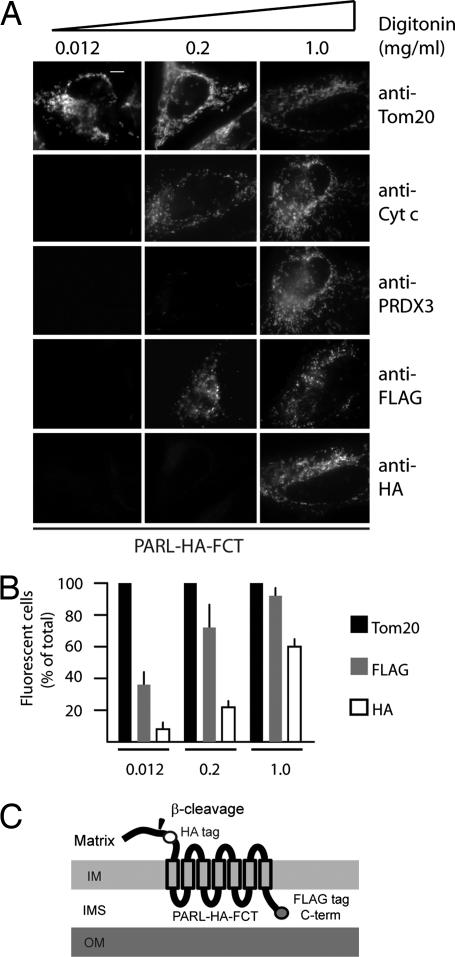
To determine the localization of the phosphorylated, vertebrate-specific Pβ domain of PARL, we investigated the topology of the protein. To this end, we used a PARL construct with a hemagglutinin (HA) tag inserted at its N terminus, at position 91, and a FLAG tag at its C terminus (PARL-HA-FCT; Fig. 3C). We permeabilized PARL-HA-FCT-transfected HeLa cells with increasing amounts of digitonin, and we performed immunofluorescence with antibodies against either the FLAG or HA tag. At low concentrations of digitonin, only the plasma membrane was permeabilized, allowing the outer membrane receptor Tom20 to be efficiently recognized by its antibody in ≈100% of cells (Fig. 3 A and B). As the digitonin concentration increased, first, 70% of cells were efficiently labeled with the FLAG along with the intermembrane space marker cytochrome c. In contrast, the HA epitope was efficiently labeled (≈60% of cells) only on treatment with the highest concentrations of digitonin, which appeared concomitantly with the matrix marker peroxidoreductase 3/sp-22 (16). These data indicate that the C-terminal domain of PARL is located in the intermembrane space, whereas the N-terminal phosphorylated Pβ domain is exposed in the mitochondrial matrix, thereby correcting our previous examination of PARL topology using immunogold-labeled EM sections (11).
Fig. 3.
Localization of PARL Pβ domain in the matrix. (A) HeLa cells were transfected with a construct expressing PARL-HA-FCT (see scheme in C), fixed, and permeabilized with the indicated concentrations of digitonin. For each condition, cells were coimmunostained with anti-FLAG or anti-HA, and anti-Tom20 (to label the outer membrane), or anti-cytochrome c (to label the intermembrane space), or anti-peroxiredoxin 3 (to label the matrix). (B) Quantitation of the experiments shown in A. (C) Scheme summarizing the topology of PARL.
Phosphorylation of the Pβ Domain Regulates β-Cleavage.
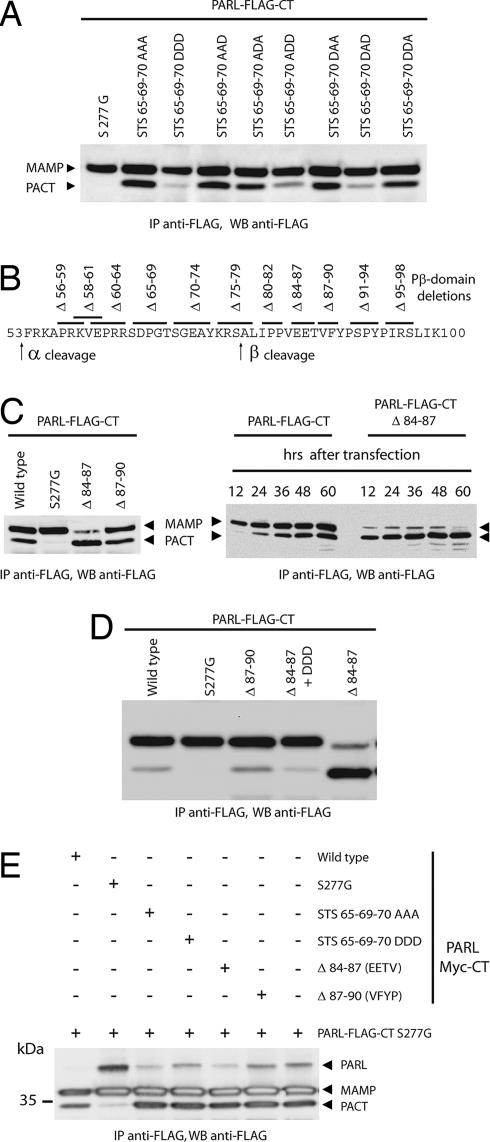
Given the proximity of the phosphorylated residues to the β-cleavage site, we investigated whether this modification could have a regulatory function on the cleavage itself. We therefore mutated residues Ser-65, Thr-69, and Ser-70 to alanine, to abolish phosphorylation, and to aspartic acid, which is commonly used to mimic phosphorylation by introducing a negative charge (17, 18). We then tested the effect of these mutations on β-cleavage, and we found that Asp substitutions of all three phosphorylated amino acids led to strongly reduced levels of β-cleavage (Fig. 4A). Impaired β-cleavage was also observed with two double Asp mutants, S65D/T69A/S70D and S65A/T69D/S70D (Fig. 4A) but not with single substitutions (data not shown), suggesting an additive inhibitory effect of each phosphorylated amino acid on β-cleavage. To demonstrate further the inhibitory effect of Asp substitutions on β-cleavage, we performed a large-scale mutagenesis screening to identify PARL mutants with constitutive β-cleavage. A mutant carrying the 84EETV87 deletion in the Pβ domain, Δ84–87, showed dramatically increased β-cleavage (Fig. 4 B and C). However, in this mutant, Asp substitutions in all three phosphorylated residues dominantly reestablished block of β-cleavage (Fig. 4D). Notably, none of the PARL mutants analyzed in this work with deletions and/or substitutions on the Pβ domain had compromised protease activity, as indicated by their ability to cleave in trans a catalytically dead (S277G) PARL protease (Fig. 4E and data not shown). Therefore, lack of β-cleavage of the mutant mimicking constitutive phosphorylation of PARL, DDD, is not the result of loss of proteolytic activity. This observation also implies that the import and insertion were not compromised, as further confirmed in digitonin-permeabilization experiments (data not shown). We conclude that the stable phosphorylation of residues Ser-65, Thr-69, and Ser-70 inhibits PARL β-cleavage.
Fig. 4.
Substitutions mimicking phosphorylation at Ser-65, Thr-69, and Ser-70 inhibit PARL β-cleavage. (A) Constructs expressing the indicated mutant PARL protein were transfected in either HEK 293 or HeLa cells. The effect of mutations abolishing (Ala) or mimicking (Asp) phosphorylation (17, 18) is monitored by the amount of β-cleaved form of PARL detected, PACT (Fig. 1A). Note that Asp but not Ala substitutions block β-cleavage. IP, immunoprecipitation; WB, Western blotting. (B) Scheme of the deletions (Δ) within PARL Pβ domain that have been tested in this work. (C) The Δ84–87 mutant is constitutively cleaved at the β-cleavage site. (D) Asp substitutions at positions Ser-65, Thr-69, and Ser-70 in the Δ84–87 dominantly reestablish the block of β-cleavage. (E) Ala and Asp substitutions at the phosphorylated Ser-65, Thr-69, and Ser-70 residues do not affect PARL protease activity. HEK 293 cells were cotransfected with a catalytically dead PARL protein (PARL-Myc-CT S277G) and the indicated FLAG-tagged mutant, whose enzymatic activity is monitored by its ability to cleave the inactive PARL in trans and produce PACT (11).
β-Cleavage Mediates PARL Activity in Mitochondrial Morphology.
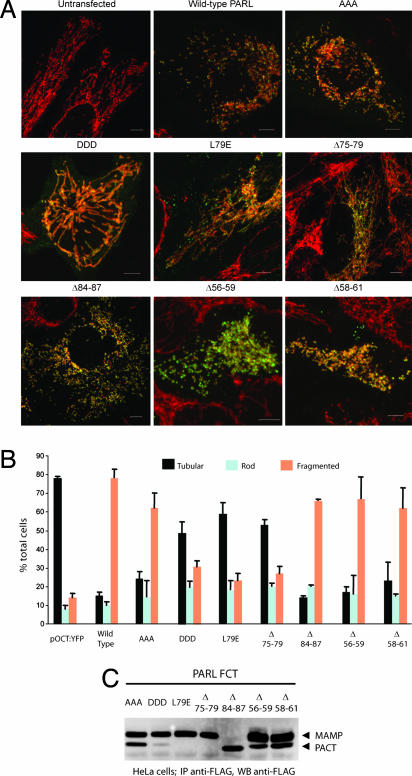
We next examined the role of β-cleavage and phosphorylation on mitochondrial morphology. Transient expression of wild-type PARL-FCT in HeLa cells resulted in a dramatic increase in mitochondrial fragmentation (Fig. 5A and B). Similar results were observed with the S65A/T69A/S70A mutant and the constitutively β-cleaved PARL protein, Δ84–87. By contrast, similar levels of expression of the S65D/T69D/S70D mutant (Fig. 5C and data not shown) did not result in significant mitochondrial fragmentation (Fig. 5), suggesting that transient expression of the nonphosphorylated, β-cleaved form of PARL leads to fragmented mitochondria. To investigate this observation further, we transfected mutants in which β-cleavage was abolished by removing (PARLΔ75–79) or mutating (PARLL79E) the cleavage site. Expression of either protein did not induce mitochondrial fragmentation (Fig. 5 A and B), further indicating that β-cleavage is required to initiate this process.
Fig. 5.
Cleavage of the vertebrate-specific Pβ domain is required to mediate PARL activity in mitochondrial morphology. (A) HeLa cells were transfected with the indicated PARL-FCT constructs, fixed, permeabilized, and stained with monoclonal anti-FLAG (green) and polyclonal anti-Tom20 (red). Images were taken with the Olympus FV1000 confocal microscope. (Scale bars, 5 μm.) (B) The mitochondrial morphologies of the wild-type and PARL mutants shown in A were quantified from three independent experiments, counting 100 cells per experiment. (C) Cells in A express similar levels of transfected proteins. Anti-FLAG immunoprecipitation (IP) and Western blot (WB) analysis of the FLAG-tagged PARL constructs used in A are shown. Equal amounts of immunoprecipitated protein were loaded. Note that in separate experiments, we verified that the immunoprecipitation efficiently depleted all of the transfected protein, validating this comparison (data not shown).
Cleavage of PARL at the β-site liberates the Pβ peptide, a 25-aa peptide that can target the nucleus when released to the cytosol (11). To investigate whether the liberated Pβ peptide is functionally implicated in the initiation of mitochondrial fragmentation, we transfected mutants in which we deleted parts of its sequence, Δ56–59 and Δ58–61 (Fig. 4B), and we analyzed the morphology of the mitochondria. Data showed that neither deletion impaired β-cleavage and mitochondrial fragmentation (Fig. 5), indicating that the function of the Pβ peptide is independent of the initiation of PARL-induced mitochondrial fragmentation.
Discussion
Considerable mechanistic and functional information on the mitochondrial rhomboid protease PCP1P in yeast is available (7, 8, 19, 20). On the other hand, the role of its mammalian ortholog PARL appears less clear. In higher organisms, dynamic changes in mitochondrial shape have been implicated in programmed cell death (4–6), a process that emerged late during metazoan evolution. Therefore, the machinery of mitochondrial fusion and fission is likely to be regulated by mechanisms additional to those for yeast. Our results implicate phosphorylation and cleavage of the Pβ domain of PARL in mitochondrial morphology. Because this domain is vertebrate-specific (11), this processing apparently is a regulatory mechanism that emerged during vertebrate evolution.
Recently, it has been shown that deletion of Parl in the mouse resulted in premature postnatal death (21), which correlated with reduced levels of cleaved OPA1. OPA1 has been shown to be involved in the regulation of the so-called “cristae remodeling” pathway of apoptosis (21, 22) and the regulation of mitochondrial fusion (23). Although PARL was shown to cleave OPA1, only minor changes in mitochondrial morphology were observed in Parl−/− fibroblasts (21), which indicates that PARL is not directly required for mitochondrial fusion (21). Therefore, the expression of a cleaved form of PARL appears to be a gain of function, leading to the fragmented morphology we have observed in this work. Because PARL-induced fragmentation depends on its phosphorylation and cleavage, the functional outcome of PARL expression on steady-state mitochondrial morphology is likely to be regulated by the abundance and activity of the yet-unidentified PARL kinase phosphatase, and protease. Thus, apparently disaccording results may arise from a complex set of regulators that could be expressed at different levels in different tissues and cells.
To date, the only reversible phosphorylation/dephosphorylation events known to occur within the mitochondrial intermembrane space or matrix compartment are limited to the E1 subunits of the pyruvate and branched-chain α-ketoacid dehydrogenase complexes (24, 25). The identification of the kinase/phosphatase couple that regulates PARL cleavage will be of critical importance to understand the regulation of mitochondrial morphology in different tissues. Moreover, the discovery of a role for phosphorylation in mitochondrial dynamics could also provide an explanation for the contrasting reports on the effect of proteins controlling mitochondrial dynamics. For example, expression of OPA1 increased mitochondrial fusion in mouse embryonic fibroblasts and in NIH 3T3 fibroblasts, whereas it resulted in dramatic fragmentation in COS-7 cells (23, 26, 27). Similarly, expression of DRP1 did not lead to fission in most cell types, but it was reported to fragment organelles in an inducible HeLa cell line (28). Finally, it is tempting to speculate that tissue selectivity of the clinical phenotype of dominant optic atrophy and Charcot–Marie–Tooth IIa, caused by mutations in OPA1 and MFN2 respectively, could similarly be a consequence of differential phosphorylation (6).
β-Cleavage liberates a 25-aa nuclear-targeted peptide termed Pβ peptide (11). Topology of PARL now shows that this peptide is generated in the matrix. A recent MS study has demonstrated the existence of a constant efflux of a large number of peptides from the mitochondria (29). These peptides, which originate from the cleavage of proteins localized mainly in the matrix and inner membrane, range in size from 6 to 27 aa, and they are extruded to the cytosol in an ATP- and temperature-dependent manner (29). Whether the Pβ peptide can be exported from the matrix to the cytosol remains to be demonstrated. However, the existence of specialized machinery for the export of matrix peptides of similar size supports this possibility. The fact that an integral Pβ sequence is not required for PARL-mediated mitochondrial fragmentation (Fig. 5A) is consistent with the possibility that the Pβ peptide mediates mitochondria-to-nucleus signaling (11).
Materials and Methods
Cell Lines and Antibodies.
HEK 293 and HeLa cells were purchased from American Type Culture Collection (Manassas, VA) and maintained under standard cell culture conditions. Cells were transfected at 40% confluence with FuGENE 6. The antibodies used in this work were: polyclonal anti-GFP (Clontech, Mountain View, CA), monoclonal anti-GFP (Invitrogen, Carlsbad, CA), monoclonal anti-cytochrome c and polyclonal anti-DsRed (PharMingen, San Diego, CA), monoclonal anti-HA (Covance, Denver, PA), mouse M2 monoclonal and rabbit polyclonal anti-FLAG. Polyclonal antibodies against the matrix marker peroxiredoxin 3 (30) were raised in rabbits against the recombinant human GST-PRDX3 protein following standard immunization protocols. Serum was tested for specificity by preadsorption with the antigen. Anti-PARL-N-Term antibody has been already described in ref. 11. Anti-PARL-C-Term was raised against a peptide spanning the last 12 aa of PARL conjugated to keyhole limpet hemocyanin, according to standard immunization protocols. Immunoprecipitations done with this antiserum were performed by covalently coupling this antiserum to protein A-agarose beads.
Constructs.
The vector used to express the human PARL protein in mammalian cells was pcDNA3. The PARL-FCT and PARL-HA-FCT constructs have been described in ref. 11. Mutants of these PARL constructs were obtained by site-directed mutagenesis. All mutations were confirmed by DNA sequencing.
PARL Cleavage Analysis.
Cells were transfected with the indicated PARL construct, grown for 24–36 h (unless otherwise indicated), washed with Dulbecco's PBS, and lysed in RIPA buffer containing a mixture of protease inhibitors and 1 mM sodium orthovanadate. Immunoprecipitations were performed with anti-FLAG-M2 monoclonal antibody or anti-PARL-C-Term at 4°C overnight as described in ref. 31. Immunocomplexes were washed four times with RIPA and denatured at 85°C for 4 min in Laemmli buffer. Samples were fractionated by SDS/PAGE on a 4–12% (wt/vol) 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)1,3-propanediol gel, blotted on PVDF membranes, immunodetected by Western blot analysis, and imaged by using the Versadoc system (Bio-Rad, Hercules, CA). For MS analysis, gels were stained with colloidal Coomassie blue. Stained bands were excised and stored at −70°C until ready for LC/MS analysis.
MS.
Samples were prepared as described in ref. 15. For capillary LC/MS, samples were analyzed by directed infusion of chromatographically separated components on an MDS Sciex QStar XL mass spectrometer (Applied Biosystems, Foster City, CA) interfaced with an Ultimate micropump (LC Packings, Sunnyvale, CA). Capillary columns (150 μm × 100 mm) were packed in-house with Majic C-18 reversed-phase packing material. A 150-min continuous gradient was used for separation with buffer A [2% ACN (vol/vol)/0.1% formic acid/0.01% TFA] followed by buffer B [10% isopropyl alcohol (vol/vol)/80% ACN (vol/vol)/0.1% formic acid/0.01% TFA]. Dried samples were resuspended in A buffer and loaded directly on the capillary column. Samples were sprayed at 4500V and MS along with tandem MS data were acquired on the fly by using the Analyst QS software (Applied Biosystems).
Data Analysis.
Resultant data were reconstructed manually and with a Bayesian reconstruction algorithm, and they were searched against both theoretical peptide and fragmentation data from the PARL sequence. Tandem data were used for web-based searches with Mascot (Matrix Science Ltd., London, U.K.). Matching tandem data were verified manually.
Mitochondrial Morphology Analysis.
Fluorescence imaging.
HeLa cells were transfected as described in ref. 32. To stain mitochondria with potentially sensitive dyes, cells were incubated with 50 nM MitoFluor Red 589 (Invitrogen) at 37°C for 15 min before imaging. For quantification of mitochondrial phenotypes, images were obtained with an Olympus IX70 microscope (Olympus Canada, Markham, ON, Canada) through a 100× objective U Plan Apochromat, NA 1.35–0.50 objective, excited at 500 nm (yellow fluorescent protein; YFP), 434 nm (cyan fluorescent protein; CFP), and 589 nm (MitoFluor Red 589) with the Polychrome IV monochrometer. The emitted light was filtered through a triple CFP/YFP/DsRed pass filter. Acquired images and multichannel overlaying were done with TillVision IV software (TILL Photonics, Pleasanton, CA). Quantification of mitochondrial morphology was done according to mitochondrial length to width ratio. When the ratio was >3:1, the mitochondria were classified as tubular and rod-shaped, and when the ratio was <3:1, mitochondria were classified as fragmented. Data were obtained from 100 cells from each condition, and standard deviations were calculated from three independent experiments. High-resolution images were obtained from samples transfected with PARL-FCT constructs and costained with monoclonal anti-FLAG and polyclonal anti-Tom20 by using an Olympus FV1000 confocal microscope (Olympus Canada). A 100X U Plan Apochromat objective NA 1.45 was used along with the argon laser to excite the 488 nm secondary Alexa 488 and the Red HeNe laser for Alexa 647 secondary antibody. The images shown are from 10–20 compressed Z stacks taken in 0.12-μm intervals to capture ≈2-μm sections of the cell by using Kalman averaging of two scans each (33).
Digitonin permeabilization.
Digitonin was recrystallized as described in ref. 34 and resuspended in PBS. Transfected cells were fixed in 4% paraformaldehyde and permeabilized with increasing concentrations of digitonin before standard immunofluorescence was performed with the indicated antibody as described previously. The data were quantified from 100 cells in three independent experiments (35).
Isolation of Mitochondria.
Fresh human placenta was obtained with appropriate permission, cut into fragments, and washed with PBS before decanting into 2 volumes per volume of mitochondrial isolation buffer (220 mM mannitol/68 mM sucrose/20 mM Hepes, pH 7.4/80 mM KCl/0.5 mM EGTA/2 mM MgOAc/protease inhibitors). The tissue was homogenized by using a Waring blender (Cole Palmer, Ansou, QC, Canada), and mitochondria were isolated by standard differential centrifugation. The mitochondrial pellet was resuspended in 100 ml of mitochondrial isolation buffer with 10% glycerol, then it was snap frozen and stored at −80°C.
Supplementary Material
Acknowledgments
We thank Dr. Gordon Shore (McGill University, Montreal, QC, Canada) for the anti-Tom20 antibody and Dr. Jordan Fishman (21st Century Biochemicals) for assistance in the generation of antibodies. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada, the Canada Foundation for Innovation, and the Centre de Recherche Université Laval Robert-Giffard (to L.P.); grants from the Canadian Institutes of Health Research and the Canada Foundation for Innovation (to the H.M.M. laboratory); a Fonds de la Recherche en Santé du Québec Junior-2 Scholarship (to L.P.); and a Canadian Institutes of Health Research New Investigator Award (to H.M.M.).
Abbreviations
- LC/MS
liquid chromatography/mass spectrometry
- MAMP
mature mitochondrial PARL
- PACT
PARL C-terminal product (of β-cleavage)
- PARL
presenilin-associated rhomboid-like (protein)
- PARL-FCT
PARL-FLAG-CTerminus.
Footnotes
Conflict of interest statement: E.A.B. is an employee and stockholder of Century 21st Biochemicals. This company sells mass spectrometry analysis and antibody production services.
This article is a PNAS direct submission. W.N. is a guest editor invited by the Editorial Board.
References
- 1.Meeusen SL, Nunnari J. Curr Opin Cell Biol. 2005;17:389–394. doi: 10.1016/j.ceb.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto K, Shaw JM. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MP. Nat Cell Biol. 2003;5:497–499. doi: 10.1038/ncb0603-497b. [DOI] [PubMed] [Google Scholar]
- 4.Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. Curr Opin Cell Biol. 2003;15:706–716. doi: 10.1016/j.ceb.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Karbowski M, Youle RJ. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Chan DC. Hum Mol Genet. 2005;14(Suppl 2):R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- 7.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 8.McQuibban GA, Saurya S, Freeman M. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 9.Pellegrini L, Passer BJ, Canelles M, Lefterov I, Ganjei JK, Fowlkes BJ, Koonin EV, D'Adamio L. J Alzheimer's Dis. 2001;3:181–190. doi: 10.3233/jad-2001-3203. [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. Genome Biol. 2003;4:1–12. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sik A, Passer BJ, Koonin EV, Pellegrini L. J Biol Chem. 2004;279:15323–15329. doi: 10.1074/jbc.M313756200. [DOI] [PubMed] [Google Scholar]
- 12.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 13.Ogurtsov AY, Sunyaev S, Kondrashov AS. Genome Res. 2004;14:1610–1616. doi: 10.1101/gr.2450504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchers CH, Thapar R, Petrotchenko EV, Torres MP, Speir JP, Easterling M, Dominski Z, Marzluff WF. Proc Natl Acad Sci USA. 2006;103:3094–3099. doi: 10.1073/pnas.0511289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlman DH, Berg EA, O'Connor PB, Costello CE, Hu J. Proc Natl Acad Sci USA. 2005;102:9020–9025. doi: 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watabe S, Kohno H, Kouyama H, Hiroi T, Yago N, Nakazawa T. J Biochem (Tokyo) 1994;115:648–654. doi: 10.1093/oxfordjournals.jbchem.a124390. [DOI] [PubMed] [Google Scholar]
- 17.Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Proc Natl Acad Sci USA. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SY, Wenk MR, Kim Y, Nairn AC, De Camilli P. Proc Natl Acad Sci USA. 2004;101:546–551. doi: 10.1073/pnas.0307813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS. J Cell Biol. 2004;165:167–173. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sesaki H, Southard SM, Hobbs AE, Jensen RE. Biochem Biophys Res Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- 21.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, et al. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 23.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. Proc Natl Acad Sci USA. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris RA, Hawes JW, Popov KM, Zhao Y, Shimomura Y, Sato J, Jaskiewicz J, Hurley TD. Adv Enzyme Regul. 1997;37:271–293. doi: 10.1016/s0065-2571(96)00009-x. [DOI] [PubMed] [Google Scholar]
- 25.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- 26.Misaka T, Miyashita T, Kubo Y. J Biol Chem. 2002;277:15834–15842. doi: 10.1074/jbc.M109260200. [DOI] [PubMed] [Google Scholar]
- 27.Olichon A, Emorine LJ, Descoins E, Pelloquin L, Brichese L, Gas N, Guillou E, Delettre C, Valette A, Hamel CP, et al. FEBS Lett. 2002;523:171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- 28.Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T. J Biol Chem. 2005;280:2691–2699. doi: 10.1074/jbc.M410609200. [DOI] [PubMed] [Google Scholar]
- 30.Wonsey DR, Zeller KI, Dang CV. Proc Natl Acad Sci USA. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellegrini L, Passer BJ, Tabaton M, Ganjei JK, D'Adamio L. J Biol Chem. 1999;274:21011–21016. doi: 10.1074/jbc.274.30.21011. [DOI] [PubMed] [Google Scholar]
- 32.Harder Z, Zunino R, McBride H. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride HM. J Biol Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- 34.Kun E, Kirsten E, Piper WN. Methods Enzymol. 1979;55:115–118. doi: 10.1016/0076-6879(79)55016-2. [DOI] [PubMed] [Google Scholar]
- 35.Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.