Abstract
Although apomixis has been quoted as a technology with the potential to deliver benefits similar in scale to those achieved with the Green Revolution, very little is currently known of the genetic mechanisms that control this trait in plants. To address this issue, we developed Hieracium, a genus of daisies native to Eurasia and North America, as a genetic model to study apomixis. In a molecular mapping study, we defined the number of genetic loci involved in apomixis, and we explored dominance and linkage relationships between these loci. To avoid difficulties often encountered with inheritance studies of apomicts, we based our mapping effort on the use of deletion mutagenesis, coupled with amplified fragment length polymorphism (AFLP) as a genomic fingerprinting tool. The results indicate that apomixis in Hieracium caespitosum is controlled at two principal loci, one of which regulates events associated with the avoidance of meiosis (apomeiosis) and the other, an unlinked locus that controls events associated with the avoidance of fertilization (parthenogenesis). AFLP bands identified as central to both loci were isolated, sequenced, and used to develop sequence-characterized amplified region (SCAR) markers. The validity of the AFLP markers was verified by using a segregating population generated by hybridization. The validity of the SCAR markers was verified by their pattern of presence/absence in specific mutants. The mutants, markers, and genetic data derived from this work are now being used to isolate genes controlling apomixis in this system.
Keywords: amplified fragment length polymorphism (AFLP), meiosis, parthenogenesis
Apomixis is the asexual formation of seed. It is a process that results in the formation of genetically uniform populations (1, 2) and also in unique patterns of speciation (3, 4). Approximately 400 flowering plant taxa are recorded as apomictic, including members of 35 diverse plant families (5, 6). Very few crop species, however, are known to be apomictic. Among those that are, most are tropical tree species, such as citrus and mango, or tropical forage grasses, such as Brachiaria and Paspalum.
It is widely reported that apomixis holds the promise of providing significant benefits to agriculture and to overall global welfare if it could be installed into seed-propagated crops in an inducible format (7–11). For rice alone, an economic analysis conducted on the scenario of free access and relatively modest adoption rates of apomixis predicted an improvement in welfare in excess of 4 billion U.S. dollars per annum (12). Despite this recognized potential, very little is known about the genetic and developmental processes that underlie the expression of apomixis, in part because of the absence of apomixis in classic model species. Some aspects of apomixis are under study in Arabidopsis, using mutagenesis to explore possible mechanisms for converting this obligate sexual species into an apomict (13). Most notably, this work has highlighted the critical role played by chromatin-remodeling factors and other epigenetic factors in the specification of early embryo and endosperm development (14–19). Other researchers are developing apomictic species into model systems to study the trait in its native form. Several models are emerging, including Panicum (20, 21), Pennisetum (22), Paspalum (23), Tripsacum (24–26), Brachiaria (27), Poa (28, 29), Taraxacum (30, 31), Hypericum (32), and Erigeron (33).
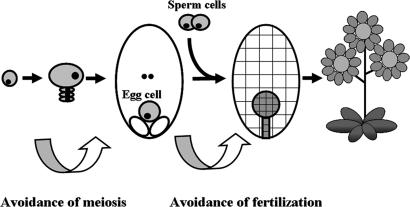
One of the best characterized systems is Hieracium, a genus of daisies native to Eurasia and North America (34). In Hieracium, apomixis occurs by apospory, a developmental process characterized by three distinct deviations from sexual reproduction (Fig. 1). In the first instance, a cell type develops within the ovule that initiates embryo sac formation without first proceeding through meiosis. This process is known as apomeiosis, and the cell type is called an aposporous initial. Aposporous initials typically develop near the time of meiosis at sites adjacent to the meiotic apparatus. They then divide and enlarge in apparent competition with meiotic products during early ovule development (35). Ultimately, their development results in the formation of one or more unreduced (2n) embryo sacs (35). Tucker et al. (36) monitored gene expression in the unreduced embryo sacs of an apomictic accession of Hieracium and in the reduced embryo sacs of a sexual accession. After initiation, the two were seen to be very similar. This finding is in agreement with the generally held belief that apomixis represents a modified form of sexual reproduction (13), as illustrated in Fig. 1. The second major deviation from sexual reproduction occurs at the level of egg-cell fate. Within each unreduced embryo sac, an egg cell develops. In common with meiotically derived egg cells, they may be fertilized by a suitable sperm cell, resulting in the formation of a zygote. Most commonly, however, the unreduced eggs of apomicts divide spontaneously, directly initiating the processes of embryogenesis. The spontaneous formation of an embryo is common to all apomictic systems, and it is also recorded in many animal systems (37). In both plants and animals, this process is known as parthenogenesis. Finally, the endosperm of Hieracium develops spontaneously without requiring the fertilization of the polar nuclei, a phenomenon referred to as autonomous endospermy.
Fig. 1.
Apomixis in Hieracium follows the developmental mechanism of apospory. Three critical deviations from sexual reproduction are apparent: an avoidance of meiosis (apomeiosis), an avoidance of fertilization before embryo formation (parthenogenesis), and an avoidance of fertilization before endosperm formation (autonomous endospermy).
Apomixis in Hieracium is reported to be genetically controlled (38–40). Intriguingly, Mendel studied inheritance in this genus (41), but apomixis would remain undescribed in these plants until the observations of Ostenfeld (42–44) and Rosenberg (45, 46; see also ref. 47). Several authors have reported that apomixis in Hieracium is conferred by the inheritance of dominant genetic elements (38–40), which is also widely reported for other apomicts as well (for reviews, see refs. 1 and 9). Ozias-Akins and colleagues (48–53) noted that the molecular mapping of apomixis loci in the grass genus Pennisetum was frustrated by an apparent repression of meiotic recombination around the site of an apospory-specific genomic region (ASGR). The size of this region of repressed recombination remains unclear, but it is estimated to be in excess of 50 megabases (52). Intriguingly, this region appears to be hemizygous in the apomicts studied because no similar region was found in sexual relatives (50). Furthermore, a very similar ASGR has also been described in the related apomict Cenchrus ciliaris (22, 51). Repressed recombination in association with elements of apomixis has also recorded for Paspalum (54, 55).
The observation that recombination is frequently repressed around loci associated with apomixis, together with the dominant inheritance of this trait in Hieracium, prompted the choice of deletion mutagenesis as a mechanism for mapping these elements in this system. This approach has several advantages over mapping that uses a segregating population: it utilizes near-isogenic mutants to define map positions; it is independent of difficulties associated with the suppression of cross-over at meiosis; and it simplifies marker validation because the mutants represent genomic subsets of the wild-type plant. Hieracium is also well suited for this approach because it can be regenerated easily in culture from very small tissue segments. This feature made it possible to break tissue chimerism and therefore to base the screen at the M1 level.
Results and Discussion
The Mutant Screen.
From an initial sample of ≈5,000 germinable seeds, ≈2,400 plants reached maturity after irradiation. The initial screen of chimeric M1 plants yielded 220 with potentially valuable mutant sectors, each of which was introduced into tissue culture for regeneration. Ninety regenerants were found to demonstrate deficiencies in apomeiosis and/or parthenogenesis, and 79 of these regenerants were identified as being sufficiently viable to be used for further study. Table 1 lists the mutant classes and the phenotypic characteristics of each. Mutants with a specific loss of the apomeiosis component of apomixis used reduced (meiotic) eggs to form progeny. Because these plants were tetraploids and they retained parthenogenesis, diploid seedlings were frequently recovered after the suppression of pollination (Table 1). In the apomixis literature, progeny of this type are called polyhaploids, and they are given the descriptive nomenclature n+0 to represent the formation of a seedling from an unreduced, unfertilized (1n) gamete (56). Following agreed conventions regarding the naming of loci identified by mutation in Arabidopsis, we suggest the designation loss of apomeiosis (LOA) for this locus in Hieracium. In the wild type, the dominant allele confers apomeiosis (LOA), whereas the recessive phenotype is reductional meiosis.
Table 1.
Mutant classes
| Mutant class | Progeny type(s) after bud decapitation* | Progeny type(s) after pollination | Locus | No. of mutants |
|---|---|---|---|---|
| Wild-type apomict | Unreduced maternal seedlings (2n+0) | Maternal seedlings (2n+0) | WT | NA |
| Loss of apomeiosis | Reduced polyhaploid seedlings (n+0) | Polyhaploids (n+0) | LOA | 24 |
| Loss of parthenogenesis | No seed forms | Unreduced hybrids (2n+n) | LOP | 30 |
| Combined class* | No seed forms | Reduced hybrids (n+n) | LOA + LOP | 25 |
| Loss of autonomous endospermy† | No seed forms | Maternal seedlings (2n+0) | Unidentified | 0 |
Nomenclature in parentheses follows the convention of Harlan and de Wet (56) in which the nuclear state of the egg is represented on the left of the addition sign and the nuclear state of the sperm is on the right. WT, wild-type; NA, not applicable.
*Many of the mutants in the combined class were severely compromised and/or had very large deletions that could not be used for mapping.
†Predicted mutant class that was not observed.
Mutants unable to perform the parthenogenesis component of apomixis did not form seed when pollination was prevented. However, they readily formed hybrid seed after pollination with the tetraploid tester plant A4Z (Table 1). Because apomeiosis remained intact in these plants, unreduced eggs were typically fertilized with diploid A4Z sperm cells, resulting in hexaploid-addition hybrids. In the apomixis literature, progeny of this type are given the descriptive nomenclature 2n+n to represent the fertilization of a 2n gamete with a reduced male gamete (56). We suggest the designation loss of parthenogenesis (LOP) for this locus in Hieracium. In the wild type, the dominant allele confers parthenogenesis (LOP), whereas the recessive phenotype is syngamy.
Another class of mutants was identified with deficiencies in both apomeiosis and parthenogenesis, which re-creates the sexual phenotype that is described by the nomenclature n+n (56). In many cases, these plants were weak, difficult to maintain, and displayed large deletions, making them of limited value for mapping. We also suspect that many of these plants represented mutants with embryo-lethal mutations that could be recovered by hybridization. Intriguingly, this type of mutation appears to have been identified principally in plants also demonstrating a loss-of-apomeiosis (loa) phenotype and not in plants that had only lost parthenogenesis (see Fig. 2). In loa mutants, the egg cell is reduced because of the reductional division of meiosis. Recessive, deleterious alleles causing embryo lethality are therefore more likely to be expressed in this background than in LOA plants that form unreduced, tetraploid egg cells. Mutants with the loa/LOP genotype form reduced eggs that can initiate parthenogenetic development (Table 2) to produce a polyhaploid (n+0) seedling. If they also carried a recessive embryo-lethal factor, however, they would fail to complete seed formation unless hybridization succeeded in complementing the lethality factor. At the level of detection for the mutant screen, therefore, these plants appeared initially to have the genotype loa/lop because reduced eggs formed and fertilization was required for seed development. We noticed, however, that they differed critically from this genotype in the ultimate quantity of seed that developed. Because loa/LOP plants are capable of parthenogenesis, hybridization is a rare event in this genotype. If these plants also carried an embryo-lethal factor requiring recovery through complementation, seed set after pollination would also be expected to be poor, which was observed (data not shown). In contrast, true loa/lop plants were seen to produce abundant seed after hybridization.
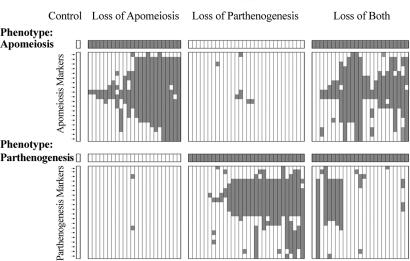
Fig. 2.
AFLP markers in a selection of 81 mutants in which apomeiosis and/or parthenogenesis was dysfunctional. Vertical columns represent mutants, horizontal rows represent markers, a light square represents a marker that is present, and a dark square represents a marker that is absent. Phenotypic data are shown in the bar at the top of each group. The controls were subjected to irradiation and regeneration, but they did not express a mutant phenotype with respect to apomixis.
Table 2.
Segregating population: P4 (sexual) = C4D (apomict)
| Segregant class | Progeny type(s) after bud decapitation | Progeny type(s) after pollination | Proposed genotype | No. of segregants |
|---|---|---|---|---|
| Apomict: apomeiotic, not requiring fertilization | Unreduced maternal seedlings (2n+0) | Maternal seedlings (2n+0) | AaaaPppp | 13 |
| Meiotic, not requiring fertilization | Reduced polyhaploid seedlings (n+0) | Polyhaploids (n+0) | aaaaPppp | 48 |
| Apomeiotic, requiring fertilization | No seed forms | Unreduced hybrids (2n+n) | Aaaapppp | 9 |
| Sexual: meiotic, requiring fertilization | No seed forms | Reduced hybrids (n+n) | aaaapppp | 22 |
Nomenclature in parentheses follows the convention of Harlan and de Wet (56) in which the nuclear state of the egg is represented on the left of the addition sign and the nuclear state of the sperm is on the right.
The mutant screen was also designed to detect a final expected mutant class (Table 1). In apomictic forms of Hieracium, the endosperm tissue forms spontaneously. In sexual forms, however, it will only form after fertilization (35). We had anticipated a mutant class in which the embryos arose asexually but the endosperm would need to be the product of fertilization. Many native apomicts utilize this mechanism (known as pseudogamy); however, for reasons that are unclear, it was not seen among the mutants recovered. Preliminary histological results indicate that lop mutants consistently demonstrated an inability to form either an embryo or an endosperm without fertilization (data not presented), indicating that the LOP locus may influence the formation of both tissues in this system.
Mapping.
Figs. 2 and 3 illustrate our current model for marker order in the LOA and LOP loci. The two genomic regions identified align well with phenotypic data from the mutants (Figs. 2 and 3), indicating that both LOA and LOP have been correctly identified from regions of common marker loss. One mutant, γ-induced mutant 164 (γ164), demonstrated a combined loss-of-apomeiosis-and-parthenogenesis phenotype, yet it retained the most centrally located markers at the LOA locus (loa 300 and loa 343). Similarly, two mutants with a lop phenotype (γ133 and γ179) retained the most centrally positioned marker identified for this locus. We interpret these data as resulting from mutations that are either too small to be identified by the markers known to date or possibly because of positional changes, such as translocations and/or inversions that are not discernable by amplified fragment length polymorphisms (AFLP).
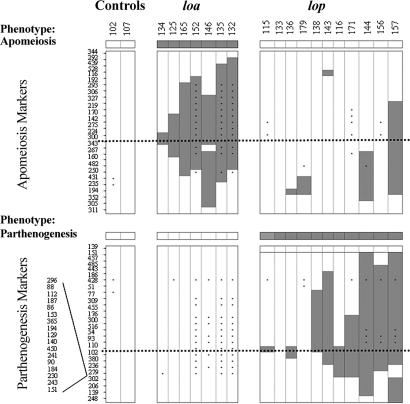
Fig. 3.
AFLP markers in an advanced selection of mutants displaying small deletions in either the LOA or LOP loci. Vertical columns represent mutants, horizontal rows represent markers, a light square represents a marker that is present, and a dark square represents a marker that is absent. Phenotypic data are shown in the bar at the top of each group. The control was subjected to irradiation and regeneration, but it did not express a mutant phenotype with respect to apomixis. Dotted lines indicate apparent midpoints for the consensus deletions, and therefore they are the most probable sites of critical loci. An asterisk represents a data point that was inferred from a previous experiment but not directly tested. A group of 17 lop markers was mapped to a site between markers lop 279 and lop 302. No further ordering is possible for this group because discriminating breakpoints were not available.
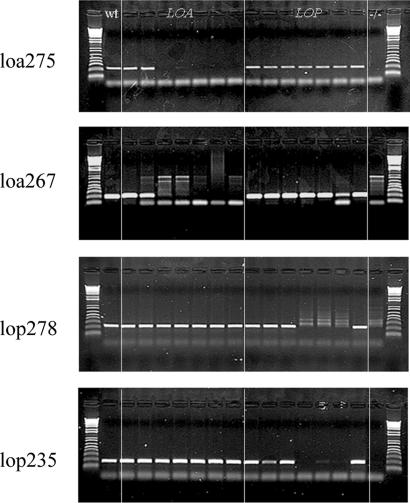
Seven AFLP bands were successfully converted into polymorphic sequence-characterized amplified region (SCAR) markers, including four markers for the LOA locus and three markers for the LOP locus. Fig. 4 illustrates the results for four of these markers, tested against a panel of seven loa mutants, seven lop mutants, one mutant demonstrating a loss of both characters, and a wild-type plant. In all but three cases, the SCAR marker results reflected those from the AFLP analysis. The exceptions were positive results for the SCARs loa 275 and loa 267 against mutant γ125, and lop 235 against mutant γ156 (Fig. 4). In all cases, the AFLP analysis had indicted a loss of the marker in the given mutant. For markers loa 267 and lop 235, AFLP predicted locations as just within the deletion regions of mutants γ125 and γ156, respectively. For loa 275, the predicted location was more central in mutant γ125. We suspect that some level of adjacent sequence duplication occurs at these sites, enabling the SCAR primers to amplify fragments, whereas the loss of critical restriction sites led to the loss of the AFLP markers.
Fig. 4.
Demonstration of SCAR marker use against the panel of mutants. In each gel lane 1 is a wild-type plant, lanes 2–8 are seven mutants in LOA (134, 125, 165, 152, 146, 135, and 132), lanes 9–15 are seven mutants in LOP (179, 138, 143, 116, 171, 144, and 156), and lane 16 is the dual mutant 168. The markers used are listed to the left of each gel.
The Segregating Population.
Plant phenotyping for this population also indicated the action of two loci, one associated with the inheritance of apomeiosis and the other with the inheritance of parthenogenesis, which is in agreement with reported observations in two other daisy genera, Taraxacum (30, 31) and Erigeron (33, 57). In a previous publication (38), we concluded that apomixis was inherited as a monogenic trait in Hieracium. It is now clear that this finding is not the case and that the earlier incorrect conclusion resulted from the screen used to measure apomixis at that time. That screen represented a score of parthenogenesis rather than of apomixis as a whole. For both the LOA and LOP loci, segregation distortion appeared to have acted during the formation of the hybrid population. For LOP, the dominant allele was inherited by 67% of the progeny, whereas the dominant allele for LOA was only inherited by 24% of the progeny. Roche et al. (51) noted a similar imbalance in the inheritance of apomixis in Pennisetum, and Grimanelli et al. (25) also reported it in Tripsacum. Nogler (58) noted that a dominant allele for apomixis in Ranunculus auricomus appeared to be gamete-lethal in homozygous form. In a previous study (38), we observed a similar effect in Hieracium. In this case, however, the effect appeared to result from a zygote-lethal mechanism. In either case, the effect need not be the direct result of apomixis genes but rather the product of linkage drag, where deleterious lethal alleles arise within regions of reduced recombination associated with the components of apomixis (1, 59). Matzk et al. (32) have proposed that this effect may be associated with the accumulation of transposable elements in these regions. Factors for apomeiosis (LOA) and parthenogenesis (LOP) segregated independently in this population. No significant linkage association was apparent among the segregant classes (Table 2).
The segregation patterns of two AFLP markers, one linked to the LOA locus (loa 300) and one linked to the LOP locus (lop 102), were determined for 37 members of the P4 × C4D hybrid population.
For parthenogenesis, the marker and trait demonstrated absolute cosegregation across all 37 of the plants tested. For apomeiosis, all 11 of the plants that scored positive for apomeiosis had inherited the loa 300 marker. Of the plants that scored negative for apomeiosis, 24 of 26 lacked the loa 300 marker, indicating that the loa 300 marker lies ≈5 cm from the LOA locus. We conclude that the markers used are closely linked to the LOA and LOP loci in Hieracium. Furthermore, no evidence was seen to indicate a role for any other major loci in the inheritance of apomixis in these plants, although modifier loci almost certainly influenced trait expression as previously suggested (60).
In summary, two major genomic regions in Hieracium caespitosum have been identified that collectively control apomixis at the level of the avoidance of meiosis and the avoidance of fertilization, respectively. The mutants and markers described in this work have become the basis of a gene-isolation strategy in Hieracium. A BAC library for C4D has been made, and it is now being used to isolate sequences corresponding to the LOA and LOP loci. Ultimately, it is our hope that this information will be used to install apomixis into target species and therefore to advance our goal of using this technology for the improvement of crop species to benefit global welfare.
Materials and Methods
Plant Material and Tissue Culture.
A tetraploid, apomictic accession of the species H. caespitosum (designated C4D), obtained from Dijon, France, was used for the mutagenesis and mapping studies reported in this work. Preliminary results indicated that this plant was simplex for dominant alleles associated with apomixis at both the LOA and LOP loci (Tables 1 and 2 and data not shown). Two other plant accessions were used as pollination partners: a tetraploid, apomictic accession of Hieracium aurantiacum, from Zurich, Switzerland (A4Z); and a tetraploid, sexual accession of Hieracium pilosella, from Caen, France (P4). All stock plants and mutants were maintained vegetatively in tissue culture and within a glasshouse. Flowering during winter months was encouraged by supplementary lighting to provide a 16-h photoperiod as described in ref. 38. To ensure the best possible representation of all progeny types, seedlings were germinated and raised to at least the third true-leaf stage in sterile culture as described in ref. 60.
Mutagenesis and Screening.
Asexually derived seed of the apomict C4D was collected from emasculated capitula (35) and mutated by using a 60Co source. A lethality curve was constructed, and a dose of 400 Gy (40 krad) of γ-irradiation was found to represent an approximate LD50 for fresh dry seed. Sufficient seed was treated at this level to ensure the survival of ≈2,500 individual seedlings. Assessment for loss of apomixis was initially made by visual detection of reduced seed set on sectors of M1 plants. This screen took advantage of the observation that even apomictic forms of Hieracium express sporophytic self-incompatibility (61). To generate stable nonchimeric mutants, tissue segments ≈2–4 mm in length, immediately subtending mutant seed heads, were harvested and used to regenerate plants as described by Bicknell (62).
Regenerants that continued to demonstrate a reduced-seed set phenotype were then assessed for the components of apomixis as described by Bicknell et al. (61). Briefly, for each mutant at least five immature capitula were severed above the plane of the developing ovaries, and the resulting progeny were assessed for ploidy by using a flow cytometer. Only plants capable of parthenogenetic development could set seed after this treatment because decapitation removes both anther and style tissue before anthesis and stigmatic receptivity. At least five capitula on each mutant were also cross-pollinated with the orange-flowered accession A4Z, and the progeny was assessed for ploidy and for morphology. In this manner, it was possible to assign each mutant into one of three classes: those that had lost apomeiosis, those that had lost parthenogenesis, and those without either (Table 1). To avoid any possibility of identity by decent within the mutant population, only one mutant sector was progressed from each chimeric M1 plant.
Segregating Population.
A subset of 92 seedlings was randomly selected from a population of H. pilosella × H. caespitosum F1 hybrids, providing a 90% probability of detecting a mapping distance of 2.5 cm or greater. All were assessed for the components of apomixis as described above.
Molecular Markers and Mapping.
Molecular markers were used to identify candidate genomic regions associated with the observed mutant classes. Because the mutants were derived from asexually generated seed and all subsequent operations retained the somatic genotype of the original M1 plant, the mutants were treated as near-isogenic lines. Furthermore, the deletion mutants were assumed to represent genomic subsets of the wild-type genotype. By using this feature, candidate genomic regions associated with apomixis were defined as regions of marker loss found to occur in at least three independent mutants. The marker class used for this work was secondary digest-amplified fragment length polymorphisms (SDAFLP) (63), which were separated by PAGE (16) and visualized by using either radioisotope exposure to film or by silver staining, or they were examined by using an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). SDAFLP was used because it is independent of template methylation.
Early results with simpler AFLP protocols (64) proved to be uninterpretable because the methylation status of many of the γ-irradiation-induced mutants was very different from that of the wild type (data not shown). Discerning marker bands were isolated from silver-stained polyacrylamide gels, amplified by PCR, and sequenced. For most of the central marker bands identified, minisequencing (65) was conducted to clarify band identification, enable the design of more specific AFLP primers, and facilitate band isolation.
SCAR markers were developed from the sequence data and tested against the mutants and segregant plants. A total of 256 AFLP primer combinations were tested against a panel of mutants, the wild type, and a control plant, which was isolated after shoot regeneration from an irradiated plant that had retained apomixis.
To expedite marker screening, a four-step process was conducted utilizing emerging mapping data to refine the mutant panel used to identify centrally positioned markers about commonly lost “consensus” genomic regions. In the first step, 79 mutants were tested by using eight AFLP primer combinations. This step provided 39 markers lost in more than 3 mutants. An analysis of these data led to the selection of 36 mutants that appeared to have discrete deletions about two distinct loci. This group included plants known to have lost either apomeiosis or parthenogenesis and a group of 13 in which neither apomeiosis nor parthenogenesis was expressed. The remaining plants typically demonstrated very large deletions, making them less suitable for mapping. In the second step, a further 20 primer combinations were tested on the 36 selected plants. After analysis of these data, the third step utilized an elite set of 8 mutants and 2 controls to test a final 228 primer combinations for markers close to the mid points of the consensus-deleted regions. This set included plants with small deletions and others with intermediate-sized deletions. In this way, it was possible to assign each marker to one of two classes: those more proximal than the most central breakpoint and those more distal. In the final step, 29 key mutants were used to order the most central markers.
The power of this multistep approach was that it allowed large numbers of primer combinations to be tested rapidly at low cost. The clear risk was that the early selection of mutants representing potential consensus regions could bias the results and possibly lead to the nonidentification of critical marker clusters. Because this effect would be particularly pronounced when the candidate mutant panel was constricted too rapidly, the multistep approach was taken to mitigate the risk.
Mapping was conducted empirically through two-dimensional tabulation. Matrices were constructed in which mutants were assigned to columns, and marker presence was recorded in rows (see Fig. 2). Marker order was discerned based on four principal assumptions: that each loss-of-function mutant had lost some, or all, of a locus responsible for its characteristic phenotype; that a marker close to the locus would be more frequently lost than one positioned more distally; that genomic regions are typically lost contiguously, and therefore adjacent markers would be more likely lost together than markers in nonadjacent positions; and that the most probable solution is the most parsimonious one.
Acknowledgments
We thank Suzanne Lambie, Pam Fletcher, and Dianne Hall for help with the plants; Michael Michelbart and Chris Winefield for helpful suggestions; and Philippa Barrell for editing assistance. This work was supported by New Zealand Foundation for Science and Technology Grant C02X0203.
Abbreviations
- AFLP
amplified fragment length polymorphism
- LOA or loa
loss of apomeiosis
- LOP or lop
loss of parthenogenesis
- SCAR
sequence-characterized amplified region
- SDAFLP
secondary digest-amplified fragment length polymorphism.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bicknell RA, Catanach AS. In: The Molecular Biology and Biotechnology of Flowering. Jordan BR, editor. UK: CABI, Wallingford; 2006. pp. 354–390. [Google Scholar]
- 2.Nogler GA. In: Embryology of Angiosperms. Johri BM, editor. Berlin: Springer; 1984. pp. 475–518. [Google Scholar]
- 3.Dickinson TA. Folia Geobot. 1998;33:327–332. [Google Scholar]
- 4.Horandl E. Folia Geobot. 1998;33:335–348. [Google Scholar]
- 5.Carman JG. Biol J Linn Soc. 1997;61:51–94. [Google Scholar]
- 6.Carman JG. In: Flowering of Apomixis: From Mechanisms to Genetic Engineering. Savidan Y, Carman JG, Dresselhaus T, editors. Mexico: CIMMYT, IRD, European Commission DG VI; 2001. pp. 95–110. [Google Scholar]
- 7.Hanna WW. Adv Agron. 1995;54:333–350. [Google Scholar]
- 8.Koltunow AM, Bicknell RA, Chaudhury AM. Plant Physiol. 1995;108:1345–1352. doi: 10.1104/pp.108.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savidan Y. Plant Breed Rev. 2000;18:13–86. [Google Scholar]
- 10.Savidan Y. Biofutures. 2000;198:38–43. [Google Scholar]
- 11.Toenniessen GH. In: Flowering of Apomixis: From Mechanisms to Genetic Engineering. Savidan Y, Carman JG, Dresselhaus T, editors. Mexico: CIMMYT, IRD, European Commission DG VI; 2001. pp. 1–7. [Google Scholar]
- 12.McMeniman S, Lubulwa G. Internal Report. Canberra: Australian Centre for International Agricultural Research; 1997. pp. 1–26. [Google Scholar]
- 13.Grossniklaus U. In: Flowering of Apomixis: From Mechanisms to Genetic Engineering. Savidan Y, Carman JG, Dresselhaus T, editors. Mexico: CIMMYT, IRD, European Commission DG VI; 2001. pp. 168–211. [Google Scholar]
- 14.Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES. Proc Natl Acad Sci USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossniklaus U, Vielle-Calzada J-P, Hoeppner MA, Gagliano WB. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 16.Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F. Development. 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohad N, Margossian L, Hsu Y-C, Williams C, Repetti P, Fischer RL. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorensen MB, Chaudhury AM, Robert H, Bancharel E, Berger F. Curr Biol. 2001;11:277–281. doi: 10.1016/s0960-9822(01)00072-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Miyazaki C, Kojima A, Saito A, Adachi T. J Plant Physiol. 1999;154:55–62. [Google Scholar]
- 21.Savidan Y. Theor Appl Genet. 1980;57:153–156. doi: 10.1007/BF00279706. [DOI] [PubMed] [Google Scholar]
- 22.Roche D, Cong PS, Chen ZB, Hanna WW, Gustine DL, Sherwood RT, Ozias-Akins P. Plant J. 1999;19:203–208. doi: 10.1046/j.1365-313x.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 23.Martinez EJ, Urbani MH, Quarin CL, Ortiz JPA. Hereditas. 2001;135:19–25. doi: 10.1111/j.1601-5223.2001.00019.x. [DOI] [PubMed] [Google Scholar]
- 24.Grimanelli D, Leblanc O, Espinosa E, Perotti E, Gonzalez de Leon D, Savidan Y. Heredity. 1998;80:33–39. doi: 10.1046/j.1365-2540.1998.00263.x. [DOI] [PubMed] [Google Scholar]
- 25.Grimanelli D, Leblanc O, Espinosa E, Perotti E, Gonzalez de Leon D, Savidan Y. Heredity. 1998;80:40–47. doi: 10.1046/j.1365-2540.1998.00264.x. [DOI] [PubMed] [Google Scholar]
- 26.Sokolov VA, Kindiger B, Khatypova IV. Genetika. 1998;34:499–506. [Google Scholar]
- 27.Pessino SC, Evans C, Ortiz JPA, Armstead I, Do Valle CB, Hayward MD. Hereditas. 1998;128:153–158. [Google Scholar]
- 28.Albertini E, Barcaccia G, Porceddu A, Sorbolini S, Falcinelli M. Mol Breed. 2001;7:293–300. [Google Scholar]
- 29.Albertini E, Porceddu A, Ferranti F, Reale L, Barcaccia G, Romano B, Falcinelli M. Sex Plant Reprod. 2001;14:213–217. doi: 10.1007/s00497-001-0116-2. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk PJ, van Baarlen P, de Jong JH. Sex Plant Reprod. 2003;16:71–76. [Google Scholar]
- 31.Vijverberg K, van der Hulst R, Lindhout P, van Dijk PJ. Theor Appl Genet. 2004;108:725–732. doi: 10.1007/s00122-003-1474-y. [DOI] [PubMed] [Google Scholar]
- 32.Matzk F, Hammer K, Schubert I. Sex Plant Reprod. 2003;16:51–58. [Google Scholar]
- 33.Noyes RD, Rieseberg LH. Genetics. 2000;155:379–390. doi: 10.1093/genetics/155.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. Flora Europea. Vol 4. Cambridge, UK: Cambridge Univ Press; 1976. pp. 358–410. [Google Scholar]
- 35.Koltunow AM, Johnson SD, Bicknell RA. Sex Plant Reprod. 1998;11:213–230. [Google Scholar]
- 36.Tucker MR, Araujo A-CG, Paech NA, Hecht V, Schmidt EDL, Rossell J-B, de Vries SC, Koltunow AMG. Plant Cell. 2003;15:1524–1537. doi: 10.1105/tpc.011742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suomalainen E, Saura A, Lokki J. Cytology and Evolution in Parthenogenesis. Boca Raton, FL: CRC Press; 1987. [Google Scholar]
- 38.Bicknell RA, Borst NK, Koltunow AM. Heredity. 2000;84:228–237. doi: 10.1046/j.1365-2540.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- 39.Christoff M. Z Indukt Abstamm Vererb. 1942;80:103–125. [Google Scholar]
- 40.Gadella TWJ. Proc Kon Ned Akad V Wetensch; 1991. pp. 455–488. [Google Scholar]
- 41.Mendel G. Verh Naturforch Vereins Brünn. 1869;8:26–31. [Google Scholar]
- 42.Ostenfeld CH. Berichte Deutschen Botan Gesellschaft. 1904;22:537–541. [Google Scholar]
- 43.Ostenfeld CH. Bot Tidsskr. 1906;27:225–248. [Google Scholar]
- 44.Ostenfeld CH. Zeitschr Abst Vererb. 1910;3:241–285. [Google Scholar]
- 45.Rosenberg O. Berichte Deutschen Botan Gesellschaft. 1906;24:157–161. [Google Scholar]
- 46.Rosenberg O. Bot Tidsskr. 1907;28:143–170. [Google Scholar]
- 47.Nogler G. Genetics. 2006;172:1–6. doi: 10.1093/genetics/172.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goel S, Chen ZB, Conner JA, Akiyama Y, Hanna WW, Ozias-Akins P. Genetics. 2003;163:1069–1082. doi: 10.1093/genetics/163.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozias-Akins P, Lubbers EL, Hanna WW, McNay JW. Theor Appl Genet. 1993;85:632–638. doi: 10.1007/BF00220923. [DOI] [PubMed] [Google Scholar]
- 50.Ozias-Akins P, Roche D, Hanna WW. Proc Natl Acad Sci USA. 1998;95:5127–5132. doi: 10.1073/pnas.95.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roche D, Chen ZB, Hanna WW, Ozias-Akins P. Sex Plant Reprod. 2001;13:217–223. [Google Scholar]
- 52.Roche D, Conner JA, Budiman MA, Frisch D, Wing R, Hanna WW, Ozias-Akins P. Theor Appl Genet. 2002;104:804–812. doi: 10.1007/s00122-001-0795-y. [DOI] [PubMed] [Google Scholar]
- 53.Roche D, Hanna WW, Ozias-Akins P. Sex Plant Reprod. 2001;13:343–349. [Google Scholar]
- 54.Labombarda P, Busti A, Caceres ME, Pupilli F, Arcioni S. Genome. 2002;45:513–519. doi: 10.1139/g02-014. [DOI] [PubMed] [Google Scholar]
- 55.Pupilli F, Labombarda P, Caceres ME, Quarin CL, Arcioni S. Mol Breed. 2001;8:53–61. [Google Scholar]
- 56.Harlan JR, De Wet JMJ. Bot Rev. 1975;41:361–390. [Google Scholar]
- 57.Noyes RD. Int J Plant Sci. 2000;161:1–12. doi: 10.1086/314238. [DOI] [PubMed] [Google Scholar]
- 58.Nogler GA. Botan Helvet. 1984;94:411–422. [Google Scholar]
- 59.Bicknell RA, Koltunow AM. Plant Cell. 2004;16(Suppl):S228–S245. doi: 10.1105/tpc.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koltunow AM, Johnson SD, Bicknell RA. Sex Plant Reprod. 2000;12:253–266. [Google Scholar]
- 61.Bicknell RA, Lambie SC, Butler RC. Hereditas. 2003;138:11–20. doi: 10.1034/j.1601-5223.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- 62.Bicknell RA. Plant Cell Tissue Organ Cult. 1994;37:197–199. [Google Scholar]
- 63.Knox MR, Ellis THN. Mol Genet Genomics. 2001;265:497–507. doi: 10.1007/s004380000438. [DOI] [PubMed] [Google Scholar]
- 64.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brugmans B, van der Hulst RGM, Visser RGF, Lindhout P, van Eck HJ. Nucleic Acids Res. 2003;31:e55. doi: 10.1093/nar/gng055. [DOI] [PMC free article] [PubMed] [Google Scholar]