Abstract
Long interspersed element type 1 (L1) retrotransposons are ubiquitous mammalian mobile elements and potential tools for in vivo mutagenesis; however, native L1 elements are relatively inactive in mice when introduced as transgenes. We have previously described a synthetic L1 element, ORFeus, containing two synonymously recoded ORFs relative to mouse L1. It is significantly more active for retrotransposition in cell culture than all native L1 elements tested. To study its activity in vivo, we developed a transgenic mouse model in which ORFeus expression was controlled by a constitutive heterologous promoter, and we established definitive evidence for ORFeus retrotransposition activity both in germ line and somatic tissues. Germ line retrotransposition frequencies resulting in 0.33 insertions per animal are seen among progeny of ORFeus donor element heterozygotes derived from a single founder, representing a >20-fold increase over native L1 elements. We observe somatic transposition events in 100% of the ORFeus donor-containing animals, and an average of 17 different insertions are easily recovered from each animal; modeling suggests that the number of somatic insertions per animal exceeds this number by perhaps several orders of magnitude. Nearly 200 insertions were precisely mapped, and their distribution in the mouse genome appears random relative to transcription units and guanine-cytosine content. The results suggest that ORFeus may be developed into useful tools for in vivo mutagenesis.
Keywords: gene trap, integration site preference, LINE-1, retrotransposon, transgenic mouse
Long interspersed elements (LINEs) are common components of mammalian genomes, and approximately one-fifth of the human and mouse genomes consist of LINE-1 elements (L1s) (1–3). The human genome contains >500,000 L1 copies, most of which are 5′ truncated (2, 4–7). Full-length L1s are ≈6 kb in length, containing an internal promoter in the 5′ UTR, two nonoverlapping ORFs (ORF1 and ORF2) and a 3′ UTR followed by a poly(A) tail (8–11). L1 ORF1 encodes a single-strand RNA-binding protein with nucleic acid chaperone activity in vitro (12–15); ORF2 encodes a protein with endonuclease and reverse transcriptase activities (16, 17); both proteins are required for retrotransposition in tissue culture (18). It is estimated that up to 100 L1s are active in the average diploid human genome (19), whereas ≈3,000 are potentially active in a diploid mouse genome (20). Human and mouse L1s are similar to each other but molecularly distinct (21). Nevertheless, human L1s can transpose in mouse cells and vice versa (18, 22). Such ex vivo experiments have extensively documented the ability of L1 to retrotranspose to new genomic locations, resulting in a wide spectrum of mutations ranging from innocuous neutral insertions to large rearrangements and deletions (23–25). In contrast, the activity of L1 in vivo has not been widely studied. Two human L1 isolates, L1RP (26) and L1LRE3 (27), are among the most active native L1s isolated thus far, but their transposition frequencies were less than satisfactory when introduced into mice: ≈1.5% of progeny animals had one germ line insertion event with L1RP (28, 29), and only ≈50% of donor-containing progeny animals had detectable somatic insertions with the more active L1LRE3 (30). The low activity of native L1s in mice hinders our current understanding of regulatory mechanisms of L1 propagation at the systems level, and it makes the use of L1 for in vivo mutagenesis impractical.
Retrotransposition frequency may in part be limited by the effect of an elongation defect affecting ORF1 and ORF2 transcription (31). To circumvent this defect, we designed a synthetic L1 in which both ORFs were recoded synonymously to mouse L1 by using the most favored codons in highly expressed mammalian genes. This element, termed ORFeus (after the similarly named legendary Greek God who revitalized his spouse), is significantly more active for retrotransposition in cultured cells than all native L1 elements tested (32). We report introduction of ORFeus into the germ line and characterization of ORFeus retrotransposition in mice.
Results
Construction of Transgenic L1 Mice.
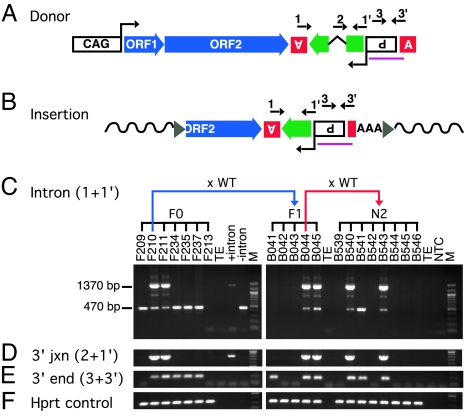
We constructed mouse transgenic lines containing ORFeus by pronuclear microinjection of fertilized eggs from B6/SJL F1 females. The ORFeus transgene is driven by a constitutive composite chicken β-actin promoter CAG (33), and it is marked by a retrotransposition indicator cassette (18, 34), in which a modified green fluorescent protein reporter gene (gfp) is disrupted by an intron (Fig. 1A). An “insertion” resulting from a retrotransposition event lacks the intron (Fig. 1B). We established a diagnostic PCR allowing ready distinction between the donor transgene and insertions (Fig. 1C; intron PCR using primers 1 and 1′ shown in Fig. 1 A and B). We initially PCR-screened tail DNA of potential founder animals for the donor transgene (Fig. 1C Left). Six of the screened 28 mice contained the intronless gfp signal (470-bp band), but only two of these mice (F210 and F211) contained the intron-containing gfp signal (1,370-bp band), suggesting the presence of the “donor element” in these two mice. The presence of the donor element was confirmed by further PCRs flanking the intron–exon junctions in the indicator cassette and throughout its length (Fig. 1 D–F and Fig. 5, which is published as supporting information on the PNAS web site) and verified by Southern blotting (Fig. 6, which is published as supporting information on the PNAS web site). Of two donor element-positive founders, only mouse F210 transmitted the donor element; line F210 was then expanded to examine ORFeus activity. Four other animals were “pseudofounders” bearing one or more transmissible new ORFeus insertions but lacking donor elements (Table 1, which is published as supporting information on the PNAS web site).
Fig. 1.
Construction and screening of founders with synthetic L1 (ORFeus) transgene. (A) The transgene construct consists of the following sequence elements from 5′ to 3′: (i) a composite CMV IE enhancer/modified chicken β-actin promoter, designated CAG (33); (ii) synthetic L1 ORF1, ORF2, and 5′ portion of 3′ UTR (32); (iii) herpes simplex virus thymidine kinase poly(A) signal (boxed inverted letter A) in antisense orientation to polyadenylate gfp mRNA; (iv) gfp (green block arrow), a modified version of the EGFP coding sequence [the gfp ORF is in antisense orientation relative to L1, and it is interrupted by intron 2 of the human γ-globin gene, which is in sense orientation relative to L1; gfp serves as a retrotransposition indicator gene (18, 34)]; (v) Rous sarcoma virus LTR promoter in antisense orientation relative to L1, which drives gfp transcription (boxed inverted P for promoter); (vi) β-globin poly(A) signal (boxed upright letter A). [numbered arrows above the diagram represent locations of genotyping PCR primers; the region used to generate Southern blotting probes is indicated (purple line)]. (B) Structure of a representative insertion. A typical insertion (i.e., a retrotransposition event derived from the donor transgene) is 5′ truncated, intronless, ends in a poly(A) tail (AAA), and is flanked by target-site duplications (gray triangles) and gDNA sequences (wavy solid lines). (C–E) PCR genotyping of founders (F0) and progeny (F1 and N2). PCRs were performed on mouse gDNA using primer pairs complementary to various transgene segments. Primers 1 and 1′ (intron) were used to amplify the indicator gene (C); primers 2 and 1′ (3′ junction) only amplify donor transgene (D); primers 3 and 3′ (3′ end) amplify the 3′ end of transgene sequence common to both donor and insertions (E); hprt primers were used as endogenous PCR controls (F). Mouse DNA samples are identified by corresponding mouse identifications: prefix F for founders and B for progeny from line F210. Kinship among mice is indicated at the top of the gel image. WT, wild-type C57BL/6J; TE, TE buffer; +intron, plasmid that carries indicator cassette with intron; −intron, plasmid with intronless indicator cassette; NTC, PCR mix only; M, 100-bp DNA ladder (New England Biolabs, Ipswich, MA).
Active Transposition in the Mouse Germ Line.
To assay ORFeus activity in mice, we backcrossed founder F210 to wild-type C57BL/6J mice, producing F1 progeny. These mice were themselves backcrossed, producing N2 progeny. All mice were genotyped by at least two PCR assays on genomic DNA (gDNA), including the intron and 3′ end primer pairs (Fig. 1 C–F Right). Nearly 500 N2 mice generated from such breeding were classified into three groups on the basis of PCR genotyping: group i, 50.4% (251 of 498) of these mice contained both the donor element and insertions (e.g., mice B540 and B543 in Fig. 1 C–F); group ii, 13.9% (69 of 498) had only insertions (e.g., mouse B541 in Fig. 1 C–F); and group iii, these mice were negative for both. Using the second genotyping PCR that targets the 3′ end of the ORFeus transgene sequence allowed detection of insertions as short as 200 bp in donorless animals. For example, F1 mouse B041 was negative for intron and 3′ junction PCRs (Fig. 1 C–D), but it was positive for the 3′ end PCR (Fig. 1E), suggesting an insertion(s) truncated at a nucleotide position between primers 2 and 3. The expression of the donor element was driven by a constitutive CAG promoter known to be active in both somatic and germ cells (35). Thus, insertions in donor element-containing mice (group i) consist of a mixture of germ line and somatic events (see the following section). In contrast, in donorless mice (group ii), insertions can only result from germ line transposition events that likely occurred during meiosis and segregated away from the donor element. To calculate the germ line insertion frequency, we divided the total number of group ii mice by the sum of groups ii and iii mice. This rather simplistic procedure (which underestimates the true frequency by ignoring multiple insertions and also does not discriminate new germ line insertions from preexisting insertions inherited from F1 donor parents) gave an overall germ line insertion frequency of 0.28 (69 of 247). Assuming that each insertion derives from an independent event, we refined our estimate of the mean number of insertions per progeny by extrapolating from the fraction of animals with no insertions; the frequency of insertions per animal is 0.33 according to the Poisson distribution (Supporting Methods and Table 2, which are published as supporting information on the PNAS web site).
To determine more accurately the germ line insertion frequency, we used Southern blot analysis on gDNA with a probe recognizing the donor element 3′ end (Fig. 2A). In a pilot experiment, we detected no discernable signals above background from group iii mice on blots. Therefore, only DNA samples positive in either of two genotyping reactions (groups i and ii mice) were subjected to a large-scale Southern blot analysis (109 PCR-positive mice from a population of 186 N2 mice, representing early litters among the total of 498 N2 animals genotyped by PCR). Representative blots for some N2 animals and their respective F1 parents are shown (Fig. 2B). As expected, all group i animals contained an intense ≈4.5-kb band corresponding to the donor element concatemer (e.g., animal B234). Donor-containing N2 animals also displayed additional bands that could reflect (a) junction fragments between the donor element and flanking genomic sequence; (b) preexisting insertions inherited from their F1 parents; or (c) putative new insertions. The insertions detected by blotting likely represent a mixture of germ line insertions and early somatic insertions that exist at near-single copy; signals from somatic insertions present at significantly less than single copy could be undetectable. Consistent with PCR genotyping results, group ii mice displayed no donor signal, but they had different numbers of bands of varied sizes (Fig. 2B, animals B400, B404, B340, B356, and B358). We cloned flanking genomic sequences of such insertions from group ii mice, and we tested germ line transmissibility by breeding three different group ii mice to wild-type mice. In all three cases, we detected transmission to the progeny of these insertions by insertion-specific PCRs (data not shown). Thus, these group ii animals provide definitive evidence that there are new transposition events in the germ line of N2 animals.
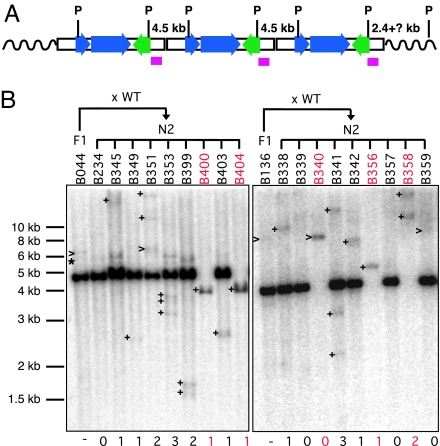
Fig. 2.
Estimating new insertion frequency by Southern blot analysis. (A) Schematic of transgene concatemer illustrating expected bands for PstI (P)-digested gDNA. Three copies of transgenes are shown for illustration; the actual copy number was estimated to be 8 to 10 by real-time PCR (data not shown). The probe position is indicated by a purple box. (B) Representative blots are shown for N2 progeny mice derived by backcrossing two F1 mice, B044 and B136, to wild-type C57BL/6J mice (WT), respectively. Numbers below each lane indicate the putative number of new insertions per individual mouse. >, bands comigrating with preexisting bands in F1 donor parents; +, new bands in N2 progeny absent from F1 donor parents; ∗, junctions fragments between donor concatemer and flanking genomic sequence. Group ii mice are highlighted in red. DNA migration positions are indicated (1-kb ladder; New England Biolabs).
The number of new bands in each N2 animal from the blotting experiments were tallied. We estimated the frequency of germ line retrotransposition by examining animals descended from ORFeus donor element heterozygotes but lacking the donor element (21 group ii mice and 77 group iii mice). Among these 98 N2 mice, the minimum new germ line insertion frequency was 0.27 (26 of 98), consistent with the PCR results (Table 2). These are minimum estimates because short insertions may not be detected; the signal strength might be too weak to be detected if the insertion is significantly shorter than the probe.
High-Level Somatic Transposition in Donor-Containing Mice.
We detected “intronless” products in 100% of donor-containing animals screened (Fig. 1C), although not all donor-containing animals displayed discrete insertion bands of high intensity on blots (Fig. 2B, animals B234, B339, and B357). This observation implies a high level of retrotransposition activity in somatic tissues albeit beyond the detection limit of blotting analysis. To evaluate the transposition activity better, we developed an inverse PCR (iPCR; refs. 36 and 37)-based insertion profiling approach (Fig. 7A, which is published as supporting information on the PNAS web site). The iPCR strategy allowed efficient amplification of multiple insertions in a single round of PCR of 35 cycles for products ranging from 0.1 to 3 kb long (Fig. 3A). All donor element-containing mouse samples (Fig. 3A, mouse B524) generated multiple bands as expected if the donor transgene continuously produced new insertions during mouse development and growth. Interestingly, when independent PCRs are performed with aliquots of the same ligation reaction, a completely or largely distinct banding pattern is generated, suggesting that the gDNA sample contains a complex population of low-abundance insertions. In contrast, significantly fewer bands (or in some cases, no bands) were detected in donorless group ii mouse samples (Fig. 3A, mouse B523). Potential background amplification from genotyping PCR-negative mice is minimal (Fig. 3A, mouse B530). To assess the level of nonspecific amplification from donor-containing gDNAs, we characterized 17 individual bands from two separate iPCRs on donor-containing mouse samples (Fig. 7 B and C). Twelve putative ORFeus insertions were recovered from 10 bands, and indeed the majority of these bands represent different insertions; sequencing analysis suggests that the remaining seven bands (most were <500 bp in size) were PCR artifacts caused by mispriming (Fig. 7B). Using this technique, we could recover up to 29 different insertions from a single tissue biopsy of a donor-containing mouse (see the following section). Mathematical modeling suggests that each donor-containing mouse could possibly have millions of unique somatic insertions throughout its body (see Supporting Methods). In addition, a similarly high level of somatic retrotransposition activities was demonstrated by iPCR from the independently derived ORFeus mouse founder animal F211 (Fig. 8, which is published as supporting information on the PNAS web site).
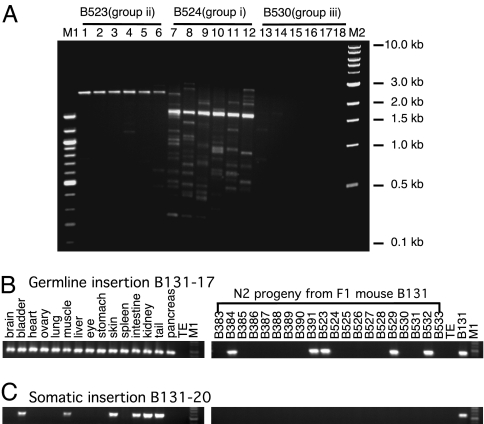
Fig. 3.
Insertion profiling by iPCR and insertion-specific PCR. (A) Insertion profiling by iPCR. Insertions were recovered by an iPCR technique (Fig. 7A). Six independent PCRs with the identical ligation mix for each mouse are shown. Samples were designated by respective mouse identification and PCR genotyping group. (B) Tissue distribution and inheritance pattern of germ line insertion B131-17. A panel of 15 different tissues from F1 mouse B131 (left) and tail biopsies from 20 of its N2 progeny were amplified by a primer specific to the flanking genomic sequence and an ORFeus primer. (C) Tissue mosaicism and lack of inheritance of a somatic insertion B131-20. M1, 100-bp DNA ladder; M2, 1-kb DNA ladder (New England Biolabs).
To map the genomic locations of individual insertions, we used iPCR to capture the 3′ integration junction at which the poly(A) stretch at the 3′ end of an insertion is joined to flanking genomic sequence, and we determined the DNA sequence. The sequencing data were subjected to a computerized annotation pipeline and subsequent manual inspection. To date, we have gathered a collection of 197 distinct insertions, derived from a total of 24 mice. Among these mice, 171 are mapped unambiguously to a specific genomic position (Table 3, which is published as supporting information on the PNAS web site), but the remaining 26 could not be assigned to specific chromosomes because they were either integrated in highly repetitive sequences, or the flanking sequences recovered were too short. Of note, 154 mappable insertions were from 9 donor-containing mice (an average of 17 insertions per animal); 15 mappable insertions were also recovered independently from ≥2 littermates or mice from different generations, providing evidence that there are germ line insertions in donor-containing mice (Table 3). We designed primers specific for individual insertions to characterize tissue mosaicism vs. germ line inheritance for insertions of interest. Examples of such experiments are shown (Fig. 3 B and C); insertion B131-17 was seen in all 15 tissues examined and in 5 of 20 its N2 progeny (Fig. 3B), suggesting that B131-17 is a germ line transposition event. In contrast, insertion B131-20 could only be detected in a subset of six tissues and in none of the 20 N2 progeny (Fig. 3C), confirming it as an early somatic event. All tissue samples and progeny were also subjected to the standard PCR genotyping procedures illustrated in Fig. 1 C–F (see also Fig. 9 A–D, which is published as supporting information on the PNAS web site).
Properties of de Novo Insertions and Chromosomal Distribution.
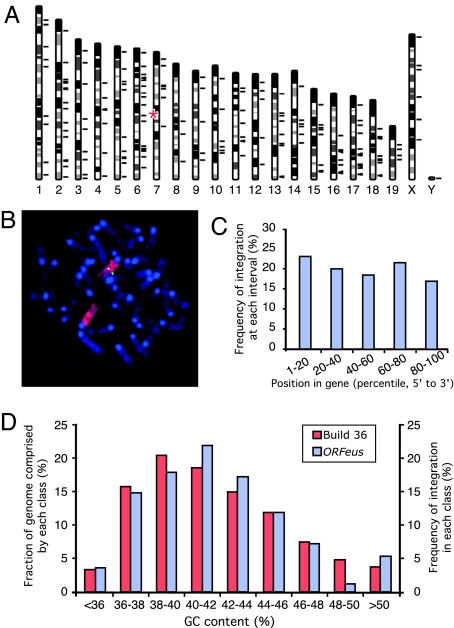
With the large number of insertions recovered from ORFeus line F210, we could survey the integration pattern of de novo synthetic L1 insertions in vivo. These insertions were located on all chromosomes including the Y chromosome (Fig. 4A), whereas the donor element transgene concatemer was mapped to chromosome 7 by fluorescent in situ hybridization (FISH) and whole chromosome painting (Fig. 4B and Supporting Methods). Of the new insertions, 27.6% (47 of 170) were mapped to RefSeq genes, and 28.0% of the mouse genome is covered by annotated RefSeqs. Thus, ORFeus appears to have no bias in its integration relative to transcription units (P = 0.99, χ2 test). Intragenic hits are distributed uniformly across the length of target genes, neither favoring nor avoiding 5′ or 3′ ends (Fig. 4C; P = 0.94, χ2 test); intergenic hits do not cluster near genes any more than expected, given the distribution of all intergenic distances in the build 36 database (P = 0.96, χ2 test). In addition, no bias toward AT-rich regions was detected (Fig. 4D; P = 0.93, χ2 test).
Fig. 4.
Chromosomal distribution of mapped insertions. (A) A total of 171 mappable insertions were charted to mouse genome build 36 (short black lines to the right of individual chromosomes). The approximate position of donor concatemer on chromosome 7 is marked (red asterisks). The Y chromosome insertion was mapped to multiple Y chromosome-specific BAC clones. (B) Mapping donor transgene location by FISH. Metaphase spreads of splenocytes from donor-containing mice were probed with fluorescently labeled full-length transgene cDNA probe (green) and subsequently with a whole-chromosome paint probe for chromosome 7 (red). Chromosomes were counterstained with DAPI (blue). (C) Distribution of insertions within annotated genes. The point of integration for every intragenic insertion (a total of 65) was transformed into a percentile value relative to the gene length (from 5′ to 3′) and binned into five-percentile intervals (x axis). Integration frequency was derived by dividing the number of intragenic insertions in each interval by the total number of intragenic insertions (y axis). (D) Distribution of ORFeus insertions relative to local GC content. GC content was determined for a 50-kb region centered at each insertion, and the frequency of integration was plotted as a function of the local GC content (right y axis); as a comparison, the mouse genome was binned as 50-kb segments by guanine-cytosine (GC) content (left y axis).
We have also obtained sequence information for the 5′ junction of 25 insertions (Table 4, which is published as supporting information on the PNAS web site). The 5′ junction information for 19 of these insertions was obtained simultaneously from the 3′ junction iPCR product, thus representing an unbiased sample of short insertions. The remaining 6 insertions were longer, and their 5′ junction was recovered by additional PCRs, each using an ORFeus-specific primer and a primer complementary to the 5′ flanking genomic sequence of the insertion. All 25 were 5′ truncated, and the length of these insertions ranged from 0.2 to 4.4 kb. All of these insertions contained a poly(A) tail at the predicted cleavage site of the polyadenylation signal of the donor elements. Most were flanked by target-site duplications ranging in size from 1 to 37 bp. Four were accompanied by 1- to 238-bp deletions at the integration site. The deduced consensus of first-strand nicking site for ORFeus was TTTT|AA, identical to that previously determined for L1 endonuclease (17, 38). These features are typical of the structure of native L1 elements in vivo and insertions isolated in tissue culture cells (23–25, 30, 39–41), with the exception on the spectrum of target site deletion sizes (see Discussion).
ORFeus-Mediated Gene Trapping in Mammalian Cells.
To date, we have aged a cohort of 60 donor-containing mice for >18 months without observing obvious fitness reductions despite the high level of retrotransposition activities in germ line and somatic tissues. This result is not surprising because the ORFeus transgene in this mouse line lacked potent gene-trapping elements. L1-mediated gene trapping has been demonstrated previously with native elements (42). To test the potential of using ORFeus as an in vivo mutagenesis tool, we modified the ORFeus construct by replacing the gfp indicator cassette with a gene-trap cassette (Fig. 10A, which is published as supporting information on the PNAS web site). This cassette consists of a bidirectional poly(A) signal sandwiched between two oppositely oriented splice acceptors (43, 44). The poly(A) signal in the sense orientation relative to the transcriptional direction of ORFeus is interrupted by an intron so that it remains nonfunctional until the intron is removed during retrotransposition. Thus, transcription termination of a target gene can be achieved independently of the orientation of an insertion after its integration into an intron of an endogenous gene. We transfected HeLa cells with this vector, harvested total RNA, and performed 5′ RACE by using adenoviral gene-trap sequence-specific primers. Gene-trapping events were readily recovered, and Fig. 10B shows a list of fusion transcripts between endogenous genes and gene-trap sequences. These results suggest that retransposition events by ORFeus can efficiently lead to gene disruption when it is equipped with appropriate gene-trap elements.
Discussion
It was of keen interest to determine whether unleashing a fully synthetic entity like ORFeus into the mouse germ line would lead to active retrotransposition. ORFeus essentially represents a new type of retrotransposon (indeed, it could even be considered a new species; ref. 45), which we have introduced into the mouse genome. Several lines of evidence suggest that the ORFeus element is in fact considerably more potent than previously described L1 elements in mice. First, our initial screen for founders discovered multiple pseudofounder animals. In most of these cases, insertions were able to transmit to F1 progeny, indicating that the insertions occurred early and were often incorporated into the germ line. Because these pseudofounders did not contain a donor element, retrotransposition likely occurred from an episomal donor in the short time between donor element transgene injection and its loss by cell division. Only a single definitive instance of such an event has been previously reported when a native human L1 isolate was used as the transgene (29). Second, from a single ORFeus mouse line, we were able to obtain a significant number of donorless, germ line insertion-containing mice and to calculate minimum germ line insertion frequencies unambiguously, which exceeds by >20 fold those in the literature (28, 29). Third, we detected somatic insertions in 100% of donor-containing animals, and an average of 17 different insertions were readily recovered from each donor-containing animal, which compares favorably with a recent report on transgenic mouse models using the most active native L1 isolate in which somatic insertions were detected in only ≈50% of L1LRE3-containing animals (30).
These results have immediate implications for the potential of L1s as a useful mutagenesis system in mammals, which has been limited by the low frequency of new insertions generated from native elements (28–30). A useful mutagenesis system in mice should easily generate clonable, highly abundant, and randomly inserted mutations. Engineered (retro)transposons are perfectly suited for this task because they mark mutations with a defined sequence that can serve as a molecular probe for easy mapping of the mutation (46). Two DNA transposons derived from heterologous hosts, Sleeping Beauty (SB) (47–51) and piggyBac (52), have been recently developed for use in mice. Data from SB studies have demonstrated its utility in germ line regional saturation mutagenesis (51) and in somatic mutagenesis for discovering cancer-susceptibility genes (53, 54). The high level of retrotransposition activities observed from the synthetic L1 element ORFeus represents a major step forward toward such directions (for optimization strategies, see Supporting Methods).
The difference in transposition mechanisms between retrotransposons and DNA transposons affords several unique properties to L1s as insertional mutagenesis tools (55, 56). First, L1s replicate by a copy-and-paste mode, and thus the number of mutations produced is not dictated by the initial copy number of transgenes as is the case for DNA transposons. Second, L1 insertions are not limited by “local hopping” observed with some DNA transposons, in which new insertions are closely linked to the donor site (47,48–50, 57). Our data directly confirm this finding in vivo because the distribution of L1 integration sites appears to be truly random, and there is no apparent preference into genes, AT-rich regions, or near-transcription start sites as is seen with certain retroviruses (58–60). Third, the L1 retrotransposition machinery operates on L1 RNA, and no rearrangement of donor L1 elements is expected. The ORFeus donor concatemer in line F210 has been stably transmitted to the third generation as evaluated by Southern blot analysis; iPCR profiling also indicates active retrotransposition in the N3 generation (data not shown). Conversely, DNA transposons transpose by first excising the donor element, and the excision of SB transposon is frequently accompanied by deletions of varied size and occasionally larger insertions at the excision sites (48, 57, 61, 62). Thus, DNA transposon-mediated mutagenesis may suffer from unintended mutations because of imprecise excision of the donor element, confounding the interpretation of the observed phenotype. Notably, such deletions at donor excision sites for DNA transposons differ from sequence deletions at the integration sites occasionally observed in L1 insertions in that the latter are molecularly tagged and thus can be easily detected. Fourth, L1 insertions, once integrated, are stable and not subject to excision. ORFeus insertions are expected to be even more stable than native L1 insertions because the ORFeus sequence is sufficiently divergent from endogenous mouse L1s to prevent homologous recombination between the two. Finally, unlike DNA transposons, L1 insertions are frequently 5′ truncated (2, 4–7), although the mechanism of truncation is unknown (30, 63). Our data indicate that ≈25% of ORFeus insertions are <502 bp long (Table 4), which places a restriction on the cargo size for L1 transgenes and demands the design of L1 vectors with compact reporter elements such as epitope tags. Nevertheless, the distinct transposition mechanisms used by retrotransposons make L1s particularly attractive and complementary mammalian mutagenesis tools.
The increased activity of synthetic L1s also provides a more sensitive in vivo model to follow retrotransposition, which is of great utility because our current understanding of the cellular conditions that regulate L1 mobilization is inadequate. Endogenous L1s are currently believed to be transcriptionally active in germ cells (64–66) and neurons (67), but the molecular details of how this activity occurs are not yet clear. The high frequency of retrotransposition reported here was achieved with an ORFeus transgene under the regulation of a constitutive heterologous promoter, which has allowed us to evaluate the integration preference of L1 in vivo on the basis of a large collection of recovered insertions. For example, we detected no integration bias toward AT-rich regions, in sharp contrast with observations made on the basis of preexisting endogenous L1 copies in mammalian genomes (1–3). This finding provides the most convincing evidence that the biased distribution of endogenous L1s in AT-rich regions of contemporary mammalian genomes is the result of selective accumulation rather than preferential integration at the first place, supporting a previous analysis on the distribution of a smaller set of recent L1 insertions in the human genome (68). Further, we found no unusually large deletions or rearrangements from 25 fully characterized insertions. The size distribution of target site deletion of host gDNA (1–238 bp from our data set) is contrary to findings from tissue culture experiments, which revealed a deletion size range from 1 to >71,000 bp (24, 25, 40), but it is consistent with recent reports on in vivo retrotransposon insertions in mammals (30, 41). However, our finding is largely based on the analysis of short insertions. More studies are needed to determine the frequency of large deletions associated with in vivo insertions.
Methods
Plasmids, Primers, and Transgenic Mice.
The ORFeus transgene is detailed in Fig. 1. The gene-trap version in Fig. 5 has a modified pCEP4 (Invitrogen, Carlsbad, CA) backbone that confers puromycin resistance in mammalian cells. Construction details are available on request. Primer sequences are summarized in Table 5, which is published as supporting information on the PNAS web site. The use of mice was approved by an Institutional Animal Care and Use Committee.
Mapping Transposition Events by Inverse PCR.
One microgram of mouse gDNA was digested with MspI, heat-inactivated, diluted, and ligated in a volume of 1 ml. The ligated material was concentrated to 50 μl by using a Microcon column (Millipore, Billerica, MA), and 1-μl samples were used as templates for a 50-μl PCR using primers JB8897 and JB8822. The PCR cycling parameters consist of an initial denaturation at 94°C for 2 min; 19 cycles of denaturing at 94°C for 15 s, annealing at 70°C (−0.5°C per cycle) for 30 s and 72°C for 60 s; 16 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 60 s; and a final extension step at 72°C for 7 min. The PCR product was purified either directly as a pool or as individual bands after agarose gel electrophoresis by a QIAquick column (Qiagen, Valencia, CA), and subcloned into a TA-cloning vector (Invitrogen). White colonies were selected for bidirectional sequencing analysis using universal vector primers.
Supplementary Material
Acknowledgments
We thank Y. Aizawa, R. Reeves, and K. O'Donnell for critical reading of the manuscript; C.-Y. Lee for sequence analysis; R. Yonescu for FISH analysis; and M. Strong for technical advice. This work was supported in part by an Affymetrix fellowship from the Life Sciences Research Foundation (to W.A.) and by National Institutes of Health Grants CA16519 and CA115604 (to J.D.B.).
Abbreviations
- gDNA
genomic DNA
- iPCR
inverse PCR
- L1
long interspersed element type 1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Smit AF. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Voliva CF, Jahn CL, Comer MB, Hutchison CA, 3rd, Edgell MH. Nucleic Acids Res. 1983;11:8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimaldi G, Skowronski J, Singer MF. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boissinot S, Chevret P, Furano AV. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 7.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning TG. Nucleic Acids Res. 1983;11:5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison CA., III Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanning T, Singer M. Nucleic Acids Res. 1987;15:2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer MF. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolosha VO, Martin SL. Proc Natl Acad Sci USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin SL, Bushman FD. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolosha VO, Martin SL. J Biol Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 16.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 18.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 19.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer MF, Thayer RE, Grimaldi G, Lerman MI, Fanning TG. Nucleic Acids Res. 1983;11:5739–5745. doi: 10.1093/nar/11.16.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostertag EM, Kazazian HH., Jr Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert N, Lutz-Prigge S, Moran JV. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 25.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 26.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 27.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 29.Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Proc Natl Acad Sci USA. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han JS, Szak ST, Boeke JD. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 32.Han JS, Boeke JD. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 33.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 34.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 36.Triglia T, Peterson MG, Kemp DJ. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochman H, Gerber AS, Hartl DL. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cost GJ, Boeke JD. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 39.Chen JM, Stenson PD, Cooper DN, Ferec C. Hum Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert N, Lutz S, Morrish TA, Moran JV. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 43.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 44.Nesic D, Cheng J, Maquat LE. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeke JD, Eickbush T, Sandmeyer SB, Voytas DF. In: Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. San Diego: Elsevier; 2005. pp. 397–407. [Google Scholar]
- 46.Carlson CM, Largaespada DA. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 47.Dupuy AJ, Fritz S, Largaespada DA. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 48.Fischer SE, Wienholds E, Plasterk RH. Proc Natl Acad Sci USA. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, Kokubu C, Kinoshita T, Akagi K, Jenkins NA, et al. Nat Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 52.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 54.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 55.Ivics Z, Izsvak Z. Trends Genet. 2005;21:8–11. doi: 10.1016/j.tig.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Bestor TH. Cell. 2005;122:322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Luo G, Ivics Z, Izsvak Z, Bradley A. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 59.Wu X, Li Y, Crise B, Burgess SM. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yant SR, Kay MA. Mol Cell Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Mol Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 63.Martin SL, Li WL, Furano AV, Boissinot S. Cytogenet Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 64.Branciforte D, Martin SL. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trelogan SA, Martin SL. Proc Natl Acad Sci USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 67.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 68.Ovchinnikov I, Troxel AB, Swergold GD. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.