Abstract
Retroviruses can cause tumors when they integrate near a protooncogene or tumor suppressor gene of the host. We infected >2,500 mice with the SL3-3 murine leukemia virus; in 22 resulting tumors, we found provirus integrations nearby or within the gene that contains the mir-17-92 microRNA (miRNA) cistron. Using quantitative real-time PCR, we showed that expression of miRNA was increased in these tumors, indicating that retroviral infection can induce expression of oncogenic miRNAs. Our results demonstrate that retroviral mutagenesis can be a potent tool for miRNA discovery.
Keywords: oncogene, retroviral mutagenesis
MicroRNAs (miRNAs) are short noncoding RNAs that regulate gene expression. They are initially transcribed by RNA polymerase II and contained within hairpins on a long primary transcript. The hairpins are then processed by two successive steps mediated by a double-stranded RNA-binding protein and RNase III (in mammals, DGCR8 and Drosha followed by TRBP and Dicer), to create the mature ≈21-nt miRNA. The miRNA is loaded into the RNA-induced silencing complex and in animals, the complex is directed to mRNAs by the complementarity of six or seven bases within the miRNA. This leads to either translational repression or mRNA cleavage (1). It has been predicted that one-third of human genes may be regulated in this way (2).
Several miRNA hairpins can be encoded as a cistron on a single primary transcript. Such is the case for the human gene, c13orf25, and its mouse homolog. Here, a primary transcript encodes (in order, 5′ to 3′) mir-17-5p, mir-17-3p, mir-18a, mir-19a, mir-20a, mir-19b-1, and mir-92-1. He et al. (3) used a miRNA microarray to profile human B cell tumor lines and found that miRNAs encoded by c13orf25 were overexpressed. They then showed that in mice, when B cells constitutively overexpressing c-Myc were transduced with part of the human cistron containing miRNAs 17-3p to 19b-1, lymphoma formed at an accelerated pace, suggesting that these miRNAs could be oncogenes (3). In addition, Hayashita et al. (4) found that the mir-17-92 cistron was overexpressed in human lung cancer. Also, O'Donnell et al. (5) showed that c-Myc expression leads to increased expression of miRNAs from the mir-17-92 cistron, and that mir-17-5p and mir-20 negatively regulate the cell proliferation factor E2F1, suggesting that these miRNAs could also have tumor suppressor properties.
Although microarray analysis of miRNA expression in tumors has proven quite useful in identifying candidates involved in cancer and has provided seminal insight, this method inherently cannot distinguish cause from correlation and so must be corroborated by additional data, expression of a transgene or identification of implicating deletions, translocations, and other mutations (3, –8). Alternatively, retroviral insertional mutagenesis might be used to identify causative cancer genes. In this method, slow-transforming retroviruses, which themselves carry no oncogene, insert provirus DNA into the host DNA. Because the provirus integrates into essentially random locations in the host genome, retroviruses can be used as a gene discovery tool to mutagenize on a genome-wide scale (9–12). When the provirus integrates in cis near an oncogene, the insertion of the viral promoter or enhancer can induce overexpression of the oncogene and drive tumor formation (9, –15). Or, when the provirus integrates within a tumor suppressor gene, this can truncate and disrupt the gene and thus also lead to tumorigenesis (16). Because barriers to superinfection keep the number of integrations per cell to a minimum, and because tumors are by nature clonal, the multiple yet few retroviral integrations found in a tumor are all likely to have activated or inactivated a cancer-causing gene. Thus, identification of the locations of integration leads to the discovery of oncogenes and tumor suppressor genes (10–12, 16–20). Here, we investigate the use of retroviral mutagenesis to modulate the expression of oncogenic miRNA. We show that murine leukemia virus can induce overexpression of miRNA from the mir-17-92 cistron and cause the formation of lymphoma in mice.
Results
Retroviral Integration Sites.
The murine leukemia virus strain SL3-3 causes T lymphoma almost exclusively (i.e., not B lymphoma; refs. 13 and 14) and has been used for retroviral insertional mutagenesis (16, 18, 19). We infected 2,545 newborn BALB/c mice with the retrovirus, and nearly all mice developed lymphoma. Tumors were excised from the spleen and thymus after an average of 84 ± 29 (average ± standard deviation) days. Genomic DNA flanking the retroviral integration sites was PCR-amplified and sequenced; integration sites were then mapped to locations in the genome. In 22 tumors (average latency, 92 ± 42 days), each from a different mouse, we found that the 8.9-kb SL3-3 provirus had integrated near the mir-17-92 miRNA cistron (Table 1 and Fig. 1). Because the exact integration location differed in 20 of 22 tumors, and because we were able to sequence the junction between the provirus LTR and the genome (Table 1), we believe it less likely that the integration locations near mir-17-92 were identified because of PCR artifacts. All of the LTR proviral sequences were two bases shorter than those contained in the retroviral RNA genome. The loss of these two bases occurs when a staggered cut is made at the ends of the provirus DNA before integration, and the lack of these bases at the junction sequences helps to rule out artifacts arising from any contamination from plasmid DNA reagents.
Table 1.
Location and junction sequence of proviruses near the mir-17-92 cistron in mouse T lymphomas
| No. | Chromosome 14 integration location | LTR genome junction sequence |
|---|---|---|
| 1 | 113914725 | CCTTATGAAGGGGTCTTTTAAGATGAGGAAATTGAGAAGT |
| 2 | 113915207 | CCTTATGAAGGGGTCTTTCAATTATTAAAAATTATAACAT |
| 3 | 113915504 | CCTTATGAAGGGGTCTTTCACTTTGCTAAAATGTAGCATT |
| 4 | 113915586 | CCTTATGAAGGGGTCTTTCAAAGGAGAGCTGATCAGGAGC |
| 5 | 113915928 | CCTTATGAAGGGGTCTTTCAATAGTACCTAGAGTGTCAAG |
| 6 | 113921419 | CCTTATGAAGGGGTCTTTCAGAACGAGACTAGGAAATAAG |
| 7 | 113921940 | CCTTATGAAGGGGTCTTTCAGCATGTGCTTTGCAAAGACT |
| 8 | 113921997 | CCTTATGAAGGGGTCTTTCACTTGTAGTAGATGTTATGAA |
| 9 | 113922010 | CCTTATGAAGGGGTCTTTCATTATGAATTCTCAATTCTTT |
| 10 | 113924219 | CCTTATGAAGGGGTCTTTCAGTTTTCTCCCCTCCGCGCCA |
| 11 | 113924350 | CCTTATGAAGGGGTCTTTCACCCCAATCAGGACCTCGTGG |
| 12 | 113924468 | CCTTATGAAGGGGTCTTTCACGTGCACGAATTAATGTGCC |
| 13 | 113924502 | CCTTATGAAGGGGTCTTTCAAAAATAAAGTTGAAAACCCA |
| 14 | 113915272 | CCTTATGAAGGGGTCTTTCATTAAATTACTGGCTTTTTCT |
| 15 | 113915299 | CCTTATGAAGGGGTCTTTCAAGTTATTTGTAACGCTTAGC |
| 16 | 113915504 | CCTTATGAAGGGGTCTTTCACTTTGCTAAAATGTAGCATT |
| 17 | 113917616 | CCTTATGAAGGGGTCTTCCAACACTGTTCTAGTGCGTGTG |
| 18 | 113921983 | CCTTATGAAGGGGTCTTTCAGTTTGAAATTGCTGCTTGTA |
| 19 | 113922010 | CCTTATGAAGGGGTCTTTCATTAGGAATTCTCAATCATTT |
| 20 | 113924395 | CCTTATGAAGGGGTCTTTCAAAGCTAGAGAATACTGGCTA |
| 21 | 113924434 | CCTTATGAAGGGGTCTTTCAAGATGAGTAGCAGCAAGCCT |
| 22 | 113924501 | CCTTATGAAGGGGTCTTTCAAAATAAAGTTGAAAACCCAC |
Junction sequence consists of 20 bases of the 5′LTR (reverse complement, normal type) and 20 bases of the adjacent genomic sequence (bold type). Base location is for the first base of the genomic sequence after the LTR as determined by the University of California, Santa Cruz, Mouse Genome Database (February 2006 assembly).
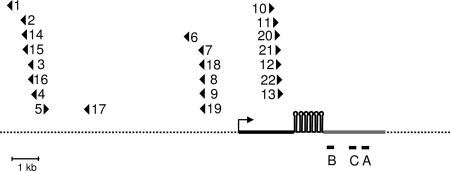
Fig. 1.
Location and orientation of proviruses in relation to the gene encoding the mir-17-92 cistron in mouse T lymphomas. Integration sites (triangles) from independent tumors mapped by genomic position relative to the gene (thick solid line) that encodes the mir-17-92 cistron (five hairpin loops). Triangles pointed to the right and left represent proviruses in the 5′ to 3′ and 3′ to 5′ orientations, respectively. The transcription start site (arrow) is positioned according to Cap Analysis Gene Expression tags (ID 4360008.1). Also shown: quantitative RT-PCR primers and probes targeted to three regions, A, B, and C (horizontal bars), of the primary RNA, and the region that comprises the EST AK053349 (gray region of the gene).
Tumors did contain copies of the provirus at other genomic locations and analysis located on average of 2 ± 1 additional sites per tumor. This result is in line with the known pathology of murine leukemia virus induced lymphoma; it is generally believed multiple cancer genes need to be activated or inactivated for tumorigenesis, and the multiple integrations accomplish this. The genes near other integration sites included Evi5, Notch1, and Jundm2.
The mir-17-92 integration sites were clustered together at three distinct regions; sites within these regions were separated by no more than 1.2 kb (Fig. 1). The proviruses in the first and second regions were primarily inserted in a transcriptional orientation opposite that of the mir-17–92 primary transcript. The sites were upstream of the putative promoter (Fig. 1) as indicated by numerous Cap Analysis Gene Expression tags (ID 4360008.1) sequenced by the Riken Institute. The proviruses of a third cluster were downstream of the promoter and transcribed in the same direction as the primary transcript. Unless there are retroviral biases or “hot spots” for integration, the clustering of sites suggests that there are certain positions and orientations optimal for tumorigenesis. We note that, to date, although mouse ESTs that include mir-17-92 sequences have not been deposited into the public EST database, they do exist for the human mir-17-92. For this study and in Fig. 1, the mouse EST AK053349, just downstream of the miRNA cistron, is presumed to be the 3′ end of the primary miRNA transcript; the 5′ boundary of the primary transcript is deduced from the location of the Cap Analysis Gene Expression tag sequences.
miRNA Primary Transcript Expression.
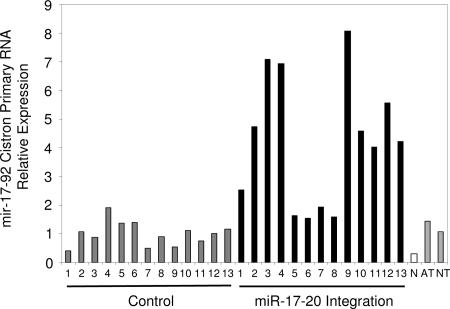
Using quantitative real-time PCR, we measured the steady-state levels of mir-17-92 primary RNA. Using primers and probes (Fig. 1) downstream of the miRNA cistron, we measured the expression level of the mir-17-92 primary RNA [Fig. 2; supporting information (SI) Fig. 4]. Expression of the primary miRNA transcript was on average 4.2 times greater in tumors where a provirus had integrated nearby the gene than in control tumors, i.e., tumors induced by retrovirus but without the mir-17-92 integration, and also 2.9 and 3.9 times greater than adult and neonate thymus tissue, respectively. NIH 3T3 fibroblasts had the lowest amount of the primary RNA, expressing a level less than one-third of those of the control tumors. Here comparison to the control tumors is the best for evaluating the oncogenicity of mir-17-92. The other tissues are informative but less ideal points of reference. NIH 3T3 cells, although immortalized, are of a different cell lineage, and healthy thymus tissue will contain different cell types, including dendritic cells and T cells of varying stages in development.
Fig. 2.
Expression of the primary RNA encoding the mir-17-92 cistron. Expression determined by quantitative RT-PCR (primer and probe set A) in tumors containing proviral integration near the mir-17-92 cistron (black), control tumors (gray), which were induced by retroviral infection but did not contain any integrations near the mir-17-92 cistron, NIH 3T3 fibroblasts (N, white), adult thymus (AT, gray), and neonate thymus (NT, gray). Expression was normalized so that the average expression of the control tumors equals one.
Mature miRNA Expression.
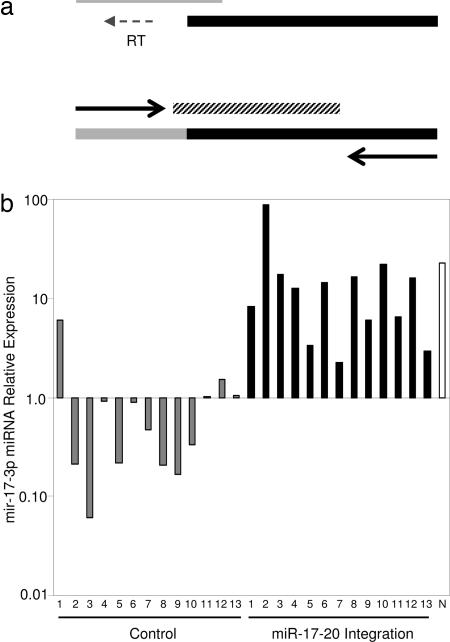
To ascertain whether the actual mature miRNA had increased because of provirus integration, we developed a method to amplify and measure miRNA by quantitative PCR. After first isolating total RNA from tumor tissue, silica gel columns were used to size-exclude long RNA and isolate shorter RNA. In doing this, we enriched our samples for miRNA [Qiagen (Valencia, CA) protocol]. A cDNA primer with a binding region (complementary seven bases) was used to create mir-17-3p cDNA from the miRNA enrichment. Because the cDNA primer also consisted of a long DNA overhang, the cDNA product was long enough to be amplified by PCR (Fig. 3a).
Fig. 3.
Quantitative RT-PCR measurement of miRNA 17-3p. (a) The 3′ end of a DNA primer (thick black line) with a long overhang pairs with target miRNA (thin gray line) and is extended by reverse transcriptase (RT) to create cDNA (lower thick line; black, primer sequence; gray, extended miRNA sequence). Quantitative PCR is then performed by using a fluorescent probe (diagonally shaded line) and DNA primers (arrows). (b) Relative expression of mir-17-3p in tumors containing proviral integration near the mir-17-92 cistron (black), control tumors (gray), which were induced by retroviral infection but did not contain any integrations near the mir-17-92 cistron, and NIH 3T3 fibroblasts (N, white). Expression was normalized so that the average expression of the control tumors equals one.
We took additional steps to establish this method's reliability. Using synthetic mir-17-3p we demonstrated that our protocol could effectively measure miRNA over concentrations varying by almost four orders of magnitude (SI Fig. 5). In addition, we could quantify mir-17-3p miRNA even in the presence of an RNA that differed only by a single base or of a randomly synthesized 21-nt oligomer (SI Fig. 6).
To analyze miRNA expression, absolute expression levels were first calculated per total RNA mass of the miRNA enrichment. Next we normalized the miRNA expression (Fig. 3b) so that the average control tumor expression equals one. The level of mir-17-3p in tumors with integrations near the mir-17-92 cistron was on average 16.6 times greater than the control tumors. Although the range of overexpression did vary, expression was increased in every one of these tumors. One control tumor (control 1) did express a relatively high amount of mir-17-3, and other mutations (spontaneous or proviruses at other locations) could have led to increased expression during the tumor evolution. The level of mir-17-3p was relatively high in NIH 3T3 fibroblasts. Considering that the primary miRNA transcript was low in the fibroblasts, the sizable expression of mir-17-3p intimates that the processing of miRNA from the primary transcript is more efficient in NIH 3T3 fibroblasts than in the T lymphomas. Gary Ruvkun (21) has recently pointed out that screens based on fibroblast cell transformation may have previously failed to find evidence of oncogenic miRNA, because, among other things, “the 3T3 fibroblasts may be deficient in components essential for miRNA-based regulation.” The high level of the mature miRNA that we detected in fibroblasts could possibly reflect such a difference in miRNA biology, perhaps a lower sensitivity to miRNA-regulated pathways. Or, it could be that fibroblasts in part become or are fibroblasts because they require an increased level of miRNA processing.
Other Factors Related to Provirus Integration and mir-17-92 Expression.
There is another gene, Gpc5, ≈50 kb downstream of the provirus integrations at mir-17-92. Because Gpc5 has been implicated as a potential oncogene (22), it was possible that the integrations near mir-17-92 were driving tumor formation through activation of Gpc5. Indeed, we did find elevated expression in several tumors, with tumors (tumors 1–4) of the first integration cluster (as in Fig. 1) expressing 5–35 times greater Gpc5 than the control tumors (SI Fig. 7). Yet tumors (tumors 6–9) from the second cluster (as in Fig. 1) produced little or no detectable Gpc5 mRNA (SI Fig. 7). Because mir-17-92 expression was elevated in all of the tumors with mir-17-92 integrations, and Gpc5 expression was not always elevated, this suggests that mir-17-92 was the causative oncogene activated by the retroviruses. Although Gpc5 may contribute in some way to tumorigenesis, it certainly is not a requirement.
We also measured c-Myc expression, because it has been suggested that it induces expression of the mir-17-92 cistron (5), and that it is an oncogenic commutation in B lymphoma (3). We found that c-Myc expression varied widely and did not correlate with provirus integration near the mir-17-92 gene (SI Fig. 8). In the past, retroviruses in tumors have been found to integrate near c-Myc (23), and this was the case for control tumor 4 and mir-17-92 integrant tumor 6. Although expression of c-Myc in these two tumors is not higher than in other tumors, this does not necessarily mean that the provirus integrations near c-Myc did not play a role in tumorigenesis. Instead, it may be that c-Myc expression is such a strong prerequisite for lymphoma that all tumors have been selected for events (e.g., provirus integration at c-Myc, provirus integration at genes that regulate c-Myc, or spontaneous mutations) that lead to adequately high c-Myc expression.
Discussion
Quantitative Real-Time PCR of miRNA.
The PCR method allowed us to quantify miRNA from small amounts (≈20 mg) of primary tissue. After extracting ≈50 μg of total RNA from tumor tissue and then enriching for miRNA, we were left with as little as 10 ng of RNA. PCR effectively amplified the cDNA generated from our extracts and allowed us to measure miRNA by quantitative real-time PCR. Although we could have conceivably performed Northern blot analysis, this method typically requires more RNA (≈50 μg per gel lane) and was not well suited for our limited quantity of tissue. As with the analysis of mRNA, real-time PCR of miRNA cannot distinguish the target RNA's size, yet better facilitates quantitative measurements. In our case, we enriched for short RNA (≈100 nt or less) and would not be able to distinguish a mature 21-nt miRNA from a 70-nt cleaved miRNA hairpin or even short degradation products of the full length transcript. However, assuming no differences in the proportions of these factors, our results should still reflect level of mature miRNA. Recently, Applied Biosystems (Foster City, CA) released a product for quantitation of human miRNA by real-time PCR (24). Although our design is similar, we note that we did not adopt their “clamped” primer approach. Our experience also suggests that the successful design of PCR primers and probes may vary depending upon the sequence of the miRNA, and that in the future, multiple primer and probe sets should be evaluated for new target miRNA.
Retroviral Insertional Mutagenesis as a miRNA Discovery Tool.
Our results indicate that the mir-17-92 cistron miRNAs behave as oncogenes in T lymphoma. We observed that tumors can arise when the SL3-3 provirus has integrated nearby the gene that encodes the mir-17-92 cistron. In the case of the first and second integration clusters (mir-17-92 tumors 1–9 and 14–19), it is probable that insertion of the retroviral enhancers caused an increase in expression of the primary transcript, consequently increasing the concentration of the oncogenic miRNA. With the third integration cluster (mir-17-92 tumors 10–13 and 20–22), the provirus integrated within the gene encoding the primary miRNA transcript and upstream of the miRNA cistron. Perhaps this is not disruptive, and the additional viral enhancer increases primary RNA levels, and/or the integration of the 3′ LTR viral promoter works to further drive transcription of the miRNAs.
Although retroviral induction of conventional oncogenes has been known for quite some time, here we demonstrate retrovirus-mediated induction of oncogenic miRNA. Our finding that expression of the mir-17-92 cistron miRNAs can be increased by retroviral infection demonstrates, in principle, that retroviral insertional mutagenesis can be used to identify miRNAs, protooncogenes and tumor suppressors, that play a role in cancer. In our screen, we have also identified or predicted (computationally) >30 other miRNAs as potential oncogenes. One candidate among these other miRNAs is another miRNA cistron, closely related to mir-17-92 (25), that encodes mir-106a, -19b, -92-2 and -363. Here three clusters of integrations 1.5–22 kb away from the miRNA cistron were found in 75 independent tumors. Although in the past provirus insertion near or within noncoding genes may have received inadequate attention, now, armed with a quickly expanding miRNA database (26) and prediction algorithms (27), retroviral mutagenesis screens [including those already performed (28)] are poised to be a powerful tool for miRNA discovery.
Materials and Methods
Retroviral Induction of Tumors of Mice.
BOSC23 retroviral packaging cells were transfected with plasmids encoding the complete SL3-3 provirus. Viral particles from culture supernatant were injected interperitoneally into newborn (<3 days) BALB/c mice. The fathers of the injected mice (except for the mouse that yielded mir-17-92 integrant tumor 3) were also mutagenized by ethyl-nitroso-urea as part of another study (16). Mice were monitored every day for general sickness as well as tumor development. When sickness or tumors of defined size were discovered, mice were killed, and tumors of the spleen and thymus were removed and frozen at −80°C.
Identification of Provirus Integration Sites.
The genomic locations of the proviral integrations were determined by using the splinkerette-based PCR method described previously (12). This method recovers genomic DNA directly flanking the 5′ LTR of the integrated provirus. Briefly, genomic DNA was isolated from tumors by using the DNeasy Tissue kit (Qiagen) and digested by using restriction enzymes BstYI or NspI. A double-stranded splinkerette adapter molecule (29) containing the appropriate restriction site was ligated to the digested genomic DNA by using the Quick Ligation kit (New England Biolabs, Ipswich, MA). These ligation products were then digested with EcoRV to prevent subsequent amplification of internal viral fragments. The resulting mixture was purified by using QIAquick PCR purification kits (Qiagen) and subject to three rounds of PCR by using nested PCR primers that had homology to the adapter DNA and to the 5′ LTR sequence of the SL3-3 virus. After resolving the PCR products by gel electrophoresis, the desired bands were purified by using QIAquick Gel Extraction kits (Qiagen) and subject to standard DNA sequencing. All procedures were performed according to the manufacturer's protocols.
Quantitative Real-Time PCR of mRNA and miRNA Primary RNA.
Total RNA was extracted from frozen tumor tissue by using the Qiagen RNeasy Mini kit. Using random DNA hexamer primers and reverse transcriptase, we created cDNA by using the Invitrogen (Carlsbad, CA) SuperScript III First-Strand Synthesis System for RT-PCR. PCR analysis was performed by using the ABI PRISM 7700 (Applied Biosystems) machine. PCR primers and probes were as follows: mir-17-92 primary transcript region A: 5′-AATTAAGCTGCTTTCTGCACTAAGG-3′, 5′-GCCCCCTGTGCGGAA-3′, 5′-[6-FAM]-CTTCACGCAGCAACGCACTTGTTCA-[TAMRA]-3′; region B: 5′- TGTGACCAGAAGATGTGAAAATGA-3′, 5′-CACTCCATCAGCTCGTGAACA-3′, 5′-[6-FAM]-AATATTGCTGAAGATGCCGATTTCCACTGTAA-[TAMRA]-3′; region C: 5′-AAACGTGATAAAGCTCTAATGTCAGC-3′, 5′-TGGTGACAGACCCCTCCCT-3′, 5′-[6-FAM]- CGATCCACATGATCCATGTGCCTCC-[TAMRA]-3′; beta-actin: 5′-AGGTCATCACTATTGGCAACGA-3′, 5′-CACTTCATGATGGAATTGAATGTAGTT-3′, 5′-[6-FAM]-TGCCACAGGATTCCATACCCAAGAAGG-[TAMRA]-3′; c-Myc: 5′- CCTAGTGCTGCATGAGGAGACA-3′, 5′-CCTCATCTTCTTGCTCTTCTTCAGA-3′, 5′-[6-FAM]-CGCCCACCACCAGCAGCG-[TAMRA]-3′.
Quantitative Real-Time PCR of miRNA.
A miRNA enrichment was created by using a protocol supplied by Qiagen. In this method, total RNA is extracted from tissue and then loaded on to an RNeasy Mini silica gel column. Low-molecular-weight RNA, including miRNA, is eluted from this column by using 100% ethanol. The eluate is then loaded onto the RNeasy MinElute silica gel column. The bound RNA is then washed and eluted with water. RNA concentrations were measured by a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Using the Invitrogen SuperScript III First-Strand Synthesis System for RT-PCR, mir-17-3p cDNA was created from this RNA by using the cDNA primer, 5′- CGTCGGTGGTAGGTCGAGCGACGTACAAGT-3′, where TACAAGT is complementary to mir-17-3p. Here we used reaction conditions identical to that prescribed for random hexamer priming. A mixture containing the cDNA primer, dNTP, and RNA was first incubated at 65°C for 5 min and then placed on ice for at least 1 min. Next, a premixed solution containing reverse transcriptase buffer, MgCl2, DTT, RNaseOUT, and SuperScriptIII reverse transcriptase was added. The mixture was then incubated at 10 min for 25°C, followed by 50 min at 50°C (elongation) and 85°C for 5 min (inactivation).
Quantitative real-time PCR was performed by using the ABI PRISM 7700 (Applied Biosystems) machine by using an annealing temperature of 50°C and an elongation temperature of 60°C. The primers and probes for mir-17-3p were 5′-GACTGCAGTGAGG-3′, 5′-CGTCGGTGGTAGG-3′, 5′-[6-FAM]-CACTTGTACGTCGCTCG-[TAMRA]-3′. Synthetic RNA used for the calibration curve and doping experiments were: 17-3p, 5′-ACUGCAGUGAGGGCACUUGUA-3′, 17-3p with one nucleotide mismatch (underlined), 5′-ACUGCAGUGAGCGCACUUGUA-3′, and a randomly synthesized 21-nt RNA 5′-NNNNNNNNNNNNNNNNNNNNN-3, where N is an A, G, C, or U.
Supplementary Material
Acknowledgments
We thank Jeff Bluestone and the University of California, San Francisco, Diabetes Center for use of their quantitative real-time PCR machine; Michael McManus for discussion; and Finn Pedersen for critical guidance. This work was supported by National Institutes of Health Grants AG20684, CA100266, and AI39506; the Natural Sciences and Engineering Research Council of Canada; and Synergenics, LLC.
Abbreviation
- miRNA
microRNA.
Footnotes
Conflict of interest statement: Except for M.K. and N.K., all authors have a financial interest in Picobella.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609030103.DC1.
References
- 1.Du T, Zamore PD. Development (Cambridge, UK) 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 7.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uren AG, Kool J, Berns A, van Lohuizen M. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 12.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 13.Ethelberg S, Lovmand J, Schmidt J, Luz A, Pedersen FS. J Virol. 1997;71:7273–7280. doi: 10.1128/jvi.71.10.7273-7280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallberg B, Schmidt J, Luz A, Pedersen FS, Grundstrom T. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorensen KD, Quintanilla-Martinez L, Kunder S, Schmidt J, Pedersen FS. J Virol. 2004;78:13216–13231. doi: 10.1128/JVI.78.23.13216-13231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glud SZ, Sorensen AB, Andrulis M, Wang B, Kondo E, Jessen R, Krenacs L, Stelkovics E, Wabl M, Serfling E, et al. Blood. 2005;106:3546–3552. doi: 10.1182/blood-2005-02-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen AB, Lund AH, Ethelberg S, Copeland NG, Jenkins NA, Pedersen FS. J Virol. 2000;74:2161–2168. doi: 10.1128/jvi.74.5.2161-2168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen MH, Sorensen AB, Morris DW, Dutra JC, Engelhard EK, Wang CL, Schmidt J, Pedersen FS. Virology. 2005;337:353–364. doi: 10.1016/j.virol.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruvkun G. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 22.Yu W, Inoue J, Imoto I, Matsuo Y, Karpas A, Inazawa J. J Hum Genet. 2003;48:331–335. doi: 10.1007/s10038-003-0026-2. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen AA, Sorensen AB, Schmidt J, Pedersen FS. J Virol. 2005;79:67–78. doi: 10.1128/JVI.79.1.67-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanzer A, Stadler PF. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. Nucleic Acids Res. 2004;32:D523–7. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devon RS, Porteous DJ, Brookes AJ. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.