Abstract
Public responses where identical T cell receptors (TCRs) are clonally dominant and shared between different individuals are a common characteristic of CD8+ T cell-mediated immunity. Focusing on TCR sharing, we analyzed ≈3,400 TCR β chains (TCRβs) from mouse CD8+ T cells responding to the influenza A virus DbNP366 and DbPA224 epitopes. Both the “public” DbNP366-specific and “private” DbPA224-specific TCR repertoires contain a high proportion (≈36%) of shared TCRβs, although the numbers of mice sharing TCRβs in each repertoire varies greatly. Sharing of both the TCRβ amino acid and TCRβ nucleotide sequence was negatively correlated with the prevalence of random nucleotide additions in the sequence. However, the extent of TCRβ amino acid sequence sharing among mice was strongly correlated with the level of diversity in the encoding nucleotide sequences, suggesting that a key feature of public TCRs is that they can be made in a variety of ways. Using a computer simulation of random V(D)J recombination, we estimated the relative production frequencies and variety of production mechanisms for TCRβ sequences and found strong correlations with the sharing of both TCRβ amino acid sequences and TCRβ nucleotide sequences. The overall conclusion is that “convergent recombination,” rather than a bias in recombination or subsequent selection, provides the mechanistic basis for TCR sharing between individuals responding to identical peptide plus MHC class I glycoprotein complexes.
Keywords: diversity, repertoire, selection, public response
The immune T cell repertoire selected in response to any given peptide plus MHC class I glycoprotein (pMHCI) can be dominated by “public” T cell receptors (TCRs), defined on the basis of amino acid sequence identity in multiple individuals (1, 2). Such public TCRs have been observed in a variety of antigen-specific CD4+ and CD8+ T cell responses in different species (1–6). The recurrent contribution of identical TCRs to immune responses in different individuals is intriguing, given the possible extent of the TCR repertoire. For example, the potential size of the TCR repertoire in mice is >1015 (7), which greatly outnumbers both the total number of T cells (≈108) and the size of the actual (8) murine TCR α/β chain (TCRα/β) repertoire in a mouse (≈106).
Various explanations have been advanced to explain the prevalence of public TCRs in different immune responses. Early studies proposed that the need to maintain self tolerance to peptides with significant self homology restricts the capacity of TCRs to recognize some epitopes (1). More recently, peptide conformations in the MHCI groove that are flat (“vanilla;” refs. 9 and 10) or very prominent (“hot and spicy;” refs. 11 and 12) in the way they present to the TCR or unusual structural features of the public TCR and its interactions with pMHCI that somehow provide a high functional avidity (13) have been suggested as causes of public TCRs. Public TCRs may also be characterized by readily formulated near-germ-line recombination of the TCR V(D)J gene segments, involving no or minimal random nucleotide additions (2, 3, 14, 15). Other possibilities are that public TCRs represent primordial germ-line-encoded TCRs that are more degenerate in their peptide-binding specificity, have higher affinity for MHC, or are somehow different from “normal” TCRs (14, 16).
Independent of nucleotide addition, it is also known (17–19) that both codon usage and repetitive sequences in the germ-line Dβ segments may lead to preferential usage of particular amino acids in TCR complementarity-determining region (CDR)3, which interfaces directly with the pMHCI complex. This raises the possibility that underlying germ-line gene and codon biases may lead to some prevalent CDR3 amino acid motifs, a factor that may also influence the sharing of TCRs between individuals.
In this study, we investigated the sharing of TCRβ sequences in the H-2Db-restricted CD8+ T cell responses to the influenza virus nucleoprotein 366–374 peptide (DbNP366) and acid polymerase 224–233 peptide (DbPA224) in mice. The DbNP366 epitope selects public TCRβs that are clonally dominant (i.e., show dominance of a clonotype within an epitope-specific response) in the majority of mice (15, 20–22). In contrast, the response to the DbPA224 epitope has been characterized as private, with no public sequences found (23). Our analysis of >3,400 TCRβs revealed that both the public DbNP366- and private DbPA224-specific responses have a high degree of sharing, with a wide range in the number of different mice sharing both amino acid and nucleotide sequences. That is, TCR sharing does not fall neatly into categories of public or private, but rather there is a broad spectrum in the number of individuals sharing TCR, of which public and private are the extremes.
The high degree of TCRβ sharing within the private DbPA224- and public DbNP366-specific responses suggests there is a spectrum of TCRβ sharing in all selected immune repertoires. Furthermore, these results are inconsistent with some explanations for public TCR selection that rely solely on the TCRβ amino acid sequence or clonal dominance within the response as a mechanism for TCR sharing. That is, clonal dominance cannot be central to TCR sharing, because we see sharing in the private DbPA224-specific response, which is not characterized by a strong clonal dominance hierarchy. Moreover, because a spectrum of sharing was also observed among TCRβ nucleotide sequences, TCRβ sharing cannot be explained solely by mechanisms such as TCR protein structure or overrepresentation of some amino acids in the CDR3 region because of a combination of codon usage and germ-line bias.
The present analysis focuses on two possible determinants of the sharing of both TCRβ amino acid and nucleotide sequences: (i) the near-germ-line nature of the TCRβ and (ii) the variety of ways in which the TCRβ can be generated by V(D)J recombination. The relative frequency of TCRβ production, accounting for both the near-germ-line nature and the variety of V(D)J recombination events, provided a good explanation of the spectrum of sharing for both TCRβ amino acid and nucleotide sequences.
Results
Extent of TCRβ Sharing in both Public DbNP366 and Private DbPA224 Repertoires.
The present analysis uses published and unpublished sequences from DbNP366-specific (22 mice) and DbPA224-specific (18 mice) CDR3β TCR repertoires (details are provided in Table 1). Those TCRβ with identical Vβ, Jβ, and CDR3β were considered to be shared when found in more than one mouse and highly shared when present in at least one-third of the mice.
Table 1.
The characteristics of the DbNP366- and DbPA224-specific CD8+ TCRβ repertoires
| Characteristic | DbNP366 | DbPA224 |
|---|---|---|
| No. of mice | 22 | 18 |
| No. of TCRβ sequences | 1839 | 1594 |
| Mean no. of TCRβ sequences per mouse | 83.6 | 88.6 |
| Range of no. of TCRβ sequences per mouse | 30–152 | 27–149 |
| Percent n.t. sequences encoding: | ||
| Shared a.a. sequences | 37.81 | 36.18 |
| Highly shared a.a. sequences | 25.37 | 8.31 |
| Amino acid sequences | ||
| No. of different a.a. sequences | 141 | 353 |
| No. of shared a.a. sequences | 16 | 70 |
| No. of highly shared a.a. sequences | 4 | 8 |
| Max. no. of mice sharing an a.a. sequence | 19 | 10 |
| Nucleotide sequences | ||
| No. of different n.t. sequences | 201 | 445 |
| No. of shared n.t. sequences | 30 | 48 |
| No. of highly shared n.t. sequences | 2 | 0 |
| Max. no. of mice sharing a n.t. sequence | 11 | 5 |
a.a., amino acid; n.t., nucleotide; No., number; Max., maximum; highly shared, present in at least one-third of mice; shared, present in at least two mice.
The public DbNP366-specific TCRβ repertoire was found to include four highly shared amino acid sequences, found in 19, 18, 16, and 11 of the 22 mice, and 12 other shared amino acid sequences. However, the DbPA224-specific repertoire (hitherto considered private) also included eight highly shared amino acid sequences, including one found in 10/18, three in 8/18, one in 7/18, and three in 6/18 mice. In addition, there were 62 other shared DbPA224 amino acid sequences (Table 1 and Fig. 3, which is published as supporting information on the PNAS web site). Thus, consistent with the public designation of the DbNP366-specific response, the most highly shared TCRβ amino acid sequence was found in a higher proportion of the mice than was the case for the DbPA224-specific response (19/22 vs. 10/18 mice, respectively). However, the proportion of unique nucleotide sequences encoding shared amino acid sequences was comparable for the DbNP366- (37.8%) and DbPA224-specific (36.2%) repertoires, suggesting there is no underlying difference in TCRβ sharing.
The high degree of sharing in the DbPA224- vs. DbNP366-specific response was somewhat surprising, given that these have previously been characterized as private and public, respectively. However, the clonal dominance of a few clonotypes in the DbNP366-specific response (22) confounds the analysis of sharing. In previous studies, which focused on a smaller number of subjects and fewer TCRβs per individual, multiple identical copies of the clonally dominant DbNP366-specific TCRβ sequences were seen in the majority of mice, whereas the subdominant DbPA224-specific sequences were less likely to be sampled in multiple mice. Combining the TCRβ sequences from different studies and allowing for clonal dominance by counting individual sequences multiple times, the mean proportion of nucleotide sequences encoding a shared amino acid sequence in any given mouse is 78.6% for the DbNP366-specific response and 56.1% for the DbPA224-specific response. Thus, the major difference between these two responses is not the extent of TCRβ sharing but the fact that, in the public DbNP366-specific response, the shared sequences tend to be clonally dominant, whereas in the private DbPA224-specific response, they are clonally subdominant.
Sharing also Occurs at the Level of TCRβ Nucleotide Sequences.
Previous studies focused on shared TCRβ amino acid sequences and did not address the extent to which TCRβ nucleotide sequences are shared among mice. Within this combined cohort, we found a broad spectrum in the number of mice sharing nucleotide sequences in both the DbNP366- and DbPA224-specific repertoires. Two highly shared DbNP366 nucleotide sequences were each found in 11/22 individuals, and there were 28 others shared by two to six mice (Table 1). The DbPA224-specific repertoire contained 48 shared nucleotide sequences, with a maximum of five mice sharing a sequence. Thus, there was a high degree and broad spectrum of sharing of both TCRβ amino acid and nucleotide sequences in these two very different immune responses, suggesting the same may be true of other T cell repertoires that have not been analyzed in such detail.
Shared TCRβ Amino Acid Sequences Have Fewer Additions in Their Nucleotide Sequences.
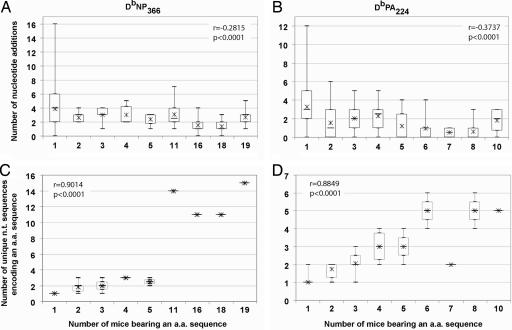
The DbNP366- and DbPA224-specific TCRβ nucleotide sequences were sequentially aligned with the Vβ, Jβ, and Dβ germ-line gene segments to calculate the germ-line contribution and the minimum number of nucleotide additions during the V(D)J recombination process. In support of the near-germ-line explanation for shared TCRs, the number of nucleotide additions was negatively correlated with the number of mice in which the amino acid sequence was present in both the DbNP366-specific (r = −0.28, P < 0.0001, Spearman) and DbPA224-specific (r = −0.37, P < 0.0001) repertoires (Fig. 1A and B). Despite this significant correlation, many of the shared amino acid sequences contained numerous nucleotide additions. For example, the most highly shared DbNP366 TCRβ amino acid sequence (found in 19/22 mice) could not be made without at least one nucleotide addition, and its median number of nucleotide additions was three, only one less than the median of four for the DbNP366 TCRβ sequences found in a single mouse. Similarly, for the DbPA224-specific response, the median number of nucleotide additions encoding the most highly shared amino acid sequence was two, only one less than the median of three for the unshared TCRβ amino acid sequences. Thus, correlating the number of nucleotide additions and TCRβ sharing does not explain why some TCRβs are shared so much more than others. Moreover, the most highly shared DbNP366 TCRβ amino acid sequence (considered public and present in 19/22 mice) had a higher median number of nucleotide additions than the most highly shared DbPA224 TCRβ amino acid sequence (present in 10/18 mice; median 3 vs. 2, respectively).
Fig. 1.
Sequence analysis of the DbNP366- and DbPA224-specific TCRβ repertoires. The relationship between the number of nucleotide additions in DbNP366- (A) and DbPA224-specific (B) TCRβ sequences and the number of mice in which an amino acid (a.a.) sequence was present. The relationship between the number of different nucleotide (n.t.) sequences encoding an amino acid sequence and the number of mice in which an amino acid sequence was found for the DbNP366- (C) and DbPA224-specific (D) responses. The box-and-whisker plots show the distributions of the number of nucleotide additions or the number of unique nucleotide sequences (vertical axis) for amino acid sequences present in a particular number of mice (horizontal axis). The median and mean are represented as a horizontal bar and an asterisk, respectively. The box represents the 25th and 75th centiles, and the lines represent the maximum and minimum values. The correlation and significance values are based on the Spearman test.
Sharing of TCRβ Amino Acid Sequences Is Correlated with the Number of Encoding Nucleotide Sequences.
As reported in previous studies of public TCR repertoires, we also observed that highly shared TCRβ amino acid sequences were encoded by many different nucleotide sequences (4, 20, 24–26). The four most highly shared DbNP366 TCRβ amino acid sequences were derived from 11–15 nucleotide sequences, and the eight most highly shared DbPA224 TCRβ sequences were from two to six nucleotide sequences. The extent of sharing of a TCRβ amino acid sequence among different mice was highly correlated with the number of nucleotide sequences encoding that amino acid sequence for both the DbNP366-specific (r = 0.90, P < 0.0001, Spearman) and the DbPA224-specific (r = 0.88, P < 0.0001) responses (Fig. 1 C and D). Thus, the number of different nucleotide sequences encoding a TCRβ amino acid sequence appears to predict the extent of sharing of this sequence.
Shared TCRβ Nucleotide Sequences also Have Fewer Nucleotide Additions.
The spectrum in the number of mice sharing TCRβ nucleotide sequences was further analyzed by investigating the relationship between the number of nucleotide additions and the number of mice in which a nucleotide sequence was present, with significant correlations being found for both the DbNP366-specific (r = −0.35, P < 0.0001, Spearman) and the DbPA224-specific (r = −0.25, P < 0.0001) repertoires. However, as with the shared TCRβ amino acid sequences, many of the shared nucleotide sequences contained numerous nucleotide additions.
Shared TCRβ Nucleotide Sequences Can Be Made in a Variety of Ways.
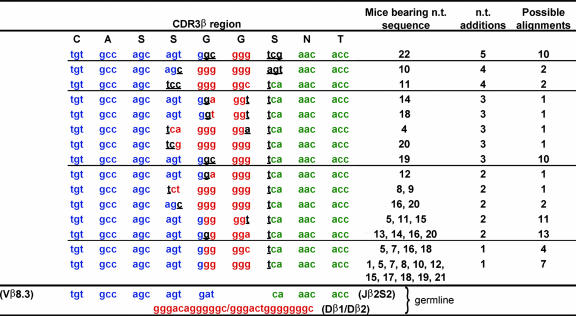
Because the sharing of TCRβ amino acid sequences is associated with the number of nucleotide sequences that encode them, it is possible that the sharing of nucleotide sequences is influenced by the number of ways they can be made by V(D)J recombination. However, we are unable to distinguish experimentally among different recombination events that may have produced identical nucleotide sequences and must rely instead on estimating the number of possible ways a sequence could have been generated. The 15 nucleotide sequences encoding the most highly shared DbNP366-specific amino acid sequence can be used to illustrate this point (Table 2). The two nucleotide sequences containing only one nucleotide addition were found in 4 and 11 mice. Similarly, sequences with two nucleotide additions were found in one to four mice. This suggests that some factor other than the number of nucleotide additions may contribute to TCRβ sharing. Examination of the number of ways these sequences could have been spliced from the TCRβ germ-line gene segments with only a minimal number of nucleotide additions provides insights into this hierarchy of TCRβ sharing. For example, of the two sequences with one nucleotide addition, the more highly shared could be spliced from the germ-line Dβ regions in multiple ways and in three different frames, because of homology between the 3′ end of the Vβ region and 5′ end of the Dβ regions (illustrated in Fig. 4, which is published as supporting information on the PNAS web site). By contrast, the less-shared sequence could be spliced fewer ways from the Dβ region. Thus, the number of ways that a TCRβ nucleotide sequence can be made, combined with the estimated minimal number of nucleotide additions, provides a good explanation for the hierarchy of sharing of the nucleotide sequences encoding the most highly shared DbNP366 amino acid sequence (Table 2).
Table 2.
Spectrum of sharing of nucleotide sequences encoding the most highly shared DbNP366-specific TCRβ amino acid sequence
The 15 unique nucleotide (n.t.) sequences that code for the amino acid sequence CASSGGSNTGQL are shown, along with one of the possible alignments with the germ-line gene segments, the mice in which the nucleotide sequences were found, the minimal number of nucleotide additions required to produce the sequence, and the number of possible different alignments to the germ-line gene segments involving minimal nucleotide additions (these alignments are detailed in Fig. 4). For the illustrated alignment, the germ-line Vβ8.3, Dβ1 or Dβ2, and Jβ2S2 gene segments are shown in blue, red, and green, respectively. Nucleotide additions are underlined and shown in black.
Analysis of Experimental Data Suggests Convergent Recombination Drives TCRβ Sharing.
The analysis of the experimental data suggests that the spectrum in the number of mice sharing TCRβ nucleotide sequences is driven by the frequency of production by V(D)J recombination, which is determined both by the number of nucleotide additions and the variety of ways a sequence can be made. Similarly, the sharing of TCRβ amino acid sequences is driven by the diversity of nucleotide sequences that can encode the same amino acid sequence and the V(D)J recombination mechanisms producing each of these nucleotide sequences. Thus, the level of sharing appears to be determined by the frequency of random V(D)J recombination events that converge to produce a given nucleotide or amino acid sequence. We term this phenomenon “convergent recombination.”
Testing the Convergent Recombination Hypothesis.
Further investigation of the convergent recombination hypothesis required knowledge of the specific V(D)J recombination event(s) that contributes to the TCRβ sequences, a definition that cannot be achieved by analyzing sequence data. This relationship between TCR sharing and convergent recombination was addressed by developing a computer simulation of unbiased V(D)J recombination to estimate the relative frequency with which different TCRβ amino acid or nucleotide sequences would be produced. To ensure that these estimates were not simply the number of times a few near-germ-line recombination events were repeated (i.e., the near-germ-line hypothesis of TCR sharing), we also monitored the variety of different V(D)J recombination events that produced each nucleotide and amino acid sequence.
The possibility of biased Vβ/Jβ pairing was avoided by restricting the analysis of each repertoire to a particular Vβ/Jβ combination (Vβ8.3/Jβ2S2 for DbNP366-specific and Vβ7.1/Jβ2S7 for the DbPA224-specific TCRs) that was commonly found among the known unshared and shared amino acid sequences. For each Vβ/Jβ combination, we simulated V(D)J recombination events to generate one million in-frame sequences. Analysis of the relationship between the in silico V(D)J recombination events of the simulation and the in vivo sharing of TCRβ sequences was restricted to those sequences that encoded the amino acid sequences found in the in vivo DbNP366- and DbPA224-specific repertoires.
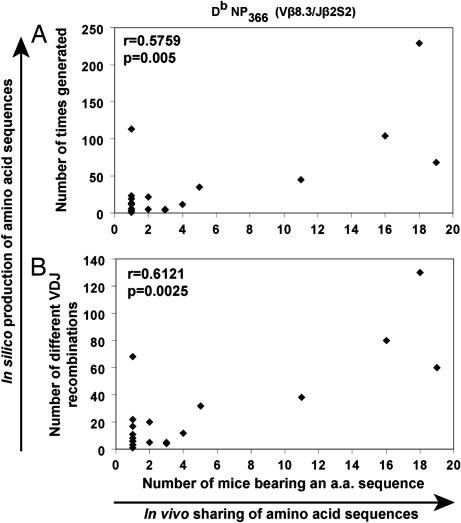
The number of mice in which an amino acid sequence was found in vivo was significantly correlated with the number of times the amino acid sequence was produced in silico by the simulations for both the DbNP366-specific (r = 0.58, P = 0.005, Spearman; Fig. 2A) and DbPA224-specific (r = 0.46, P < 0.0001) repertoires. Similarly, there was a significant correlation between the number of mice in which a nucleotide sequence was present in vivo and the number of times the nucleotide sequence was produced in the simulations (DbNP366, r = 0.47, P = 0.002; DbPA224, r = 0.39, P = 0.0005).
Fig. 2.
Analysis of an in silico TCRβ repertoire with respect to in vivo sharing. The relationship between the number of mice in which a DbNP366-specific (Vβ8.3/Jβ2S2) amino acid (a.a.) sequence was found in vivo and the number of times an amino acid sequence was generated in silico by the simulations (A) and the number of different V(D)J recombination mechanisms producing an amino acid sequence in silico (B). Each point on the graph represents an amino acid sequence that was found in vivo in the experiments, present in a particular number of mice (horizontal axis). On the vertical axis is the number of times (A) or the number of different ways (B) that each amino acid sequence was generated by the simulation. The correlation and significance values are based on the Spearman test.
To eliminate the possibility that these correlations arose because of a few repeated near-germ-line recombination events, we also analyzed the number of different V(D)J recombination events that produced each amino acid or nucleotide sequence. In support of the convergent recombination hypothesis of TCR sharing, we observed a strong correlation between the in vivo sharing of TCRβ amino acid sequences and the number of different V(D)J recombination mechanisms producing these sequences in the simulations for both the DbNP366-specific (r = 0.61, P = 0.003, Spearman; Fig. 2b) and DbPA224-specific (r = 0.48, P < 0.0001) repertoires. There was also a strong correlation between the number of different V(D)J recombinations in the simulation that produced a TCRβ nucleotide sequence and the number of mice in which it was found in vivo (DbNP366, r = 0.45, P = 0.004; DbPA224, r = 0.42, P = 0.0001, Spearman). Illustrations of the diversity of V(D)J recombination events in the simulations producing the most highly shared DbNP366 amino acid sequence and one of the most highly shared DbPA224 nucleotide sequences are provided in, which are published as supporting information on the PNAS web site.
The results of the simulations, which used an unbiased set of simulation parameters, provide a potent demonstration that the spectrum of sharing of TCRβ nucleotide and amino sequences can be explained by convergent recombination. That is, the relative frequencies with which sequences are produced are a good predictor of the spectrum of TCRβ sharing. Moreover, although some near-germ-line-encoded nucleotide sequences were produced repeatedly by the same V(D)J recombination mechanisms, many sequences were frequently produced with numerous nucleotide additions by multiple random recombination events because there were many independent ways to make them. Similarly, some amino acid sequences were frequently produced because they were encoded by highly recurrent nucleotide sequences, and/or rich in amino acids that are germ-line-encoded and/or have high codon degeneracy.
Discussion
The search for the cause of public T cell responses has been predicated on the assumption that sharing of TCRs is a rare event and must therefore reflect some special feature of the pMHCI complex or TCR conformation. However, our study of the DbNP366- and DbPA224-specific repertoires suggests that, when the repertoire of individual mice is sampled more intensively and larger groups of mice are considered, a high degree of sharing is observed (≈36% of all unique nucleotide sequences encoded a shared amino acid sequence in both the DbNP366- and DbPA224-specific repertoires). Moreover, rather than a TCRβ sequence simply being public or private, we characterized a spectrum in the number of mice sharing both TCRβ amino acid sequences and TCRβ nucleotide sequences that was not in accord with many of the proposed explanations of public TCR repertoires.
Although a negative correlation between the number of nucleotide additions and the number of mice sharing a TCRβ was observed in both the DbNP366- and DbPA224-specific repertoires (Fig. 1 A and B), the number of nucleotide additions could not fully explain why some TCRβ sequences were shared so much more than others or the hierarchy of sharing of nucleotide sequences encoding the same amino acid sequence (Table 2). Furthermore, a stronger correlation between the diversity of nucleotide sequences encoding a TCRβ amino acid sequence and the sharing of that TCRβ (Fig. 1 C and D) suggested the importance of the variety of different ways shared TCRβs can be made.
Using a computer-simulation approach to produce TCRβ sequences by random V(D)J recombination processes (involving random nucleotide addition), we found that the relative production frequencies and the variety of different ways a TCRβ sequence could be made was a much better predictor of TCRβ sharing than simply considering the number of random nucleotide additions. In the case of the DbNP366-specific response; for example, the number of different recombination events producing a TCRβ amino acid sequence explains ≈37% of TCRβ sharing, vs. only ≈8% of TCRβ sharing that is explained by the number of nucleotide additions. Thus, even with unbiased random recombination events, the probability of generating some nucleotide and amino acid sequences is higher than others because of convergent recombination.
Although convergent recombination provides a mechanistic explanation for TCR sharing, it does not explain the clonal dominance of public TCRs. Convergent recombination may play a role in TCR precursor frequency, but there are other factors, such as TCR affinity for the pMHCI complex and stochastic events, which may also influence clonal dominance. If neither the peptide shape nor the germ-line-like character of the TCR provides a consistent explanation for the occurrence of public TCRs, what then is the mechanism underlying this phenomenon? The present analysis suggests that the underlying degree and spectrum of TCR sharing is similar across different responses, and the apparent “public-ness” of the response is determined by the clonal dominance of T cells expressing shared TCRs. If the antigen-specific repertoire were randomly drawn from the naïve repertoire, in which there is also a high degree of sharing, (18–27% of TCR sequences shared between two mice (27)), we should expect to see shared TCRs emerge frequently in the response to different antigens. However, the experimental detection of these shared TCRs depends on both the sampling effort and the clonal dominance of these shared TCRs. In some responses, shared TCRs will be clonally dominant, more likely to be detected in multiple individuals, and thus characterized as public. In other responses, the shared TCRs will be clonally subdominant, and the extent of their sharing will be detected only by analyzing (as here) large numbers of TCRs from many individuals.
In summary, the mechanistic basis underlying public T cell responses has been an important question in immunology for over a decade. Although a variety of explanations have been advanced from individual limited data sets, there has been no consistent explanation of TCR sharing in different responses. Our analysis illuminates the mechanistic basis for this phenomenon by demonstrating that convergent recombination is a good predictor of the extent of TCR sharing in both public and private responses. Recent experiments suggest that the extent of TCR diversity in virus-specific CD8+ T cell responses to persistent viruses correlates directly with the limitation of immune escape (24). Moreover, public TCRs tend to be prominent in persistent viral infections (3, 4). Thus, understanding the basis of public T cell responses not only is important for our understanding of immune repertoire and diversity and hierarchy, but it also has implications for immune control of pathogens and vaccine design.
Methods
TCRβ Repertoires.
The TCRβ sequences for CD8+ T cell responses to influenza A in C57BL/6J mice (summarized in Table 1) were obtained in previous studies by single-cell sorting of CD8+Vβ 8.3+DbNP366-tetramer+ and CD8+Vβ 7.1+DbPA224-tetramer+ cells and subsequent amplification using Vβ-specific primers. The experimental procedures are described in detail in refs. 20, 22, and 28.
Estimating the Number of Nucleotide Additions.
The Vβ, Dβ, and Jβ germ-line gene segments used in the sequence alignments were obtained from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov). We adopted a basic process to align each sequence to the germ-line gene segments and estimate the minimum number of nucleotide additions. This involved initially aligning the 5′ and 3′ ends of the sequence with the Vβ and Jβ gene segments, respectively, and then matching the remaining nucleotide sequence to the Dβ gene segments. A match to a string of two or more nucleotides was considered as originating from a Dβ gene segment. Any nucleotides that were not identified with the germ-line gene segments were counted as nucleotide additions.
Simulation of TCRβ Recombination.
The simulations involved a specific Vβ/Jβ germ-line gene segment pair and one of the two Dβs randomly chosen for each recombination event. Nucleotides were randomly removed from the 3′ end of the Vβ, the 5′ end of the Jβ, and both ends of the Dβ, followed by random nucleotide addition between the truncated Vβ and Dβ, and Dβ and Jβ, gene segments (Fig. 7, which is published as supporting information on the PNAS web site). We analyzed the in vivo frequency of the addition or deletion of different numbers of nucleotides of a portion of the naïve TCRβ repertoire (Fig. 8, which is published as supporting information on the PNAS web site). These distributions of nucleotide removal/addition are biased by the alignment process toward being near-germ-line and may also reflect the effects of thymic selection and peripheral survival. To avoid these biases, we allowed the simulation to randomly remove between 0 and 10 nucleotides from the Vβ and Jβ with equal probability, randomly remove between 0 and 12/14 nucleotides from Dβ1/Dβ2, and randomly add between 0 and 10 nucleotides (effectively biasing the simulation toward producing a greater proportion of sequences with a high number of nucleotide additions than demonstrated by the distributions). The simulations were performed using Matlab 7.0.1 (The Mathworks, Natick, MA).
Statistical Analysis.
All correlations were performed by using the Spearman rank correlation and GraphPad Prism software (GraphPad, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Tadeusz Davenport for assistance with sequence analysis. This work was supported by the James S. McDonnell Foundation 21st Century Research Award/Studying Complex Systems; the National Institutes of Health; the Australian National Health and Medical Research Council (NHMRC); and a Burnet Award of the NHMRC and Science, Technology, and Innovation funds from the Government of Victoria, Australia (to P.C.D.). M.P.D. is a Sylvia and Charles Viertel Senior Medical Research Fellow. D.A.P. is a Medical Research Council (U.K.) Clinician Scientist, K.K. is an NHMRC Peter Doherty Postdoctoral Fellow, and S.J.T. is an NHMRC R. D. Wright Fellow.
Abbreviations
- TCR
T cell receptor
- TCRα/β
TCR α/β chain
- CDR
complementarity-determining region.
Footnotes
The authors declare no conflict of interest.
References
- 1.Casanova JL, Cerottini JC, Matthes M, Necker A, Gournier H, Barra C, Widmann C, MacDonald HR, Lemonnier F, Malissen B, et al. J Exp Med. 1992;176:439–447. doi: 10.1084/jem.176.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibotti R, Cabaniols JP, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos JM, Kourilsky P. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaet VP, Schmidt CW, Burrows SR, Silins SL, Kurilla MG, Doolan DL, Suhrbier A, Moss DJ, Kieff E, Sculley TB, Misko IS. J Exp Med. 1994;180:2335–2340. doi: 10.1084/jem.180.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, et al. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levraud JP, Pannetier C, Langlade-Demoyen P, Brichard V, Kourilsky P. J Exp Med. 1996;183:439–449. doi: 10.1084/jem.183.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis MM, Bjorkman PJ. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 8.Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. J Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 9.Davis MM. Nat Immunol. 2003;4:649–650. doi: 10.1038/ni0703-649. [DOI] [PubMed] [Google Scholar]
- 10.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 11.Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, Fontes MR, McCluskey J, Rossjohn J. J Immunol. 2002;169:5153–5160. doi: 10.4049/jimmunol.169.9.5153. [DOI] [PubMed] [Google Scholar]
- 12.Miles JJ, Elhassen D, Borg NA, Silins SL, Tynan FE, Burrows JM, Purcell AW, Kjer-Nielsen L, Rossjohn J, Burrows SR, McCluskey J. J Immunol. 2005;175:3826–3834. doi: 10.4049/jimmunol.175.6.3826. [DOI] [PubMed] [Google Scholar]
- 13.Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 14.Gavin MA, Bevan MJ. Immunity. 1995;3:793–800. doi: 10.1016/1074-7613(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 15.Fazilleau N, Cabaniols JP, Lemaitre F, Motta I, Kourilsky P, Kanellopoulos JM. J Immunol. 2005;174:345–355. doi: 10.4049/jimmunol.174.1.345. [DOI] [PubMed] [Google Scholar]
- 16.Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Siu G, Kronenberg M, Strauss E, Haars R, Mak TW, Hood L. Nature. 1984;311:344–350. doi: 10.1038/311344a0. [DOI] [PubMed] [Google Scholar]
- 18.Moss PA, Bell JI. Hum Immunol. 1996;48:32–38. doi: 10.1016/0198-8859(96)00084-5. [DOI] [PubMed] [Google Scholar]
- 19.Wallace ME, Bryden M, Cose SC, Coles RM, Schumacher TN, Brooks A, Carbone FR. Immunity. 2000;12:547–556. doi: 10.1016/s1074-7613(00)80206-x. [DOI] [PubMed] [Google Scholar]
- 20.Kedzierska K, Turner SJ, Doherty PC. Proc Natl Acad Sci USA. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner SJ, Kedzierska K, La Gruta NL, Webby R, Doherty PC. Semin Immunol. 2004;16:179–184. doi: 10.1016/j.smim.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Kedzierska K, La Gruta NL, Davenport MP, Turner SJ, Doherty PC. Proc Natl Acad Sci USA. 2005;102:11432–11437. doi: 10.1073/pnas.0504851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner SJ, Diaz G, Cross R, Doherty PC. Immunity. 2003;18:549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 24.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, et al. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. J Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 26.Lim A, Trautmann L, Peyrat MA, Couedel C, Davodeau F, Romagne F, Kourilsky P, Bonneville M. J Immunol. 2000;165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- 27.Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 28.Kedzierska K, Venturi V, Field K, Davenport MP, Turner SJ, Doherty PC. Proc Natl Acad Sci USA. 2006;103:9184–9189. doi: 10.1073/pnas.0603289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.