Abstract
The molecular mechanism underlying the pathogenesis of the majority of cases of sporadic Alzheimer's disease (AD) is unknown. A history of stroke was found to be associated with development of some AD cases, especially in the presence of vascular risk factors. Reduced cerebral perfusion is a common vascular component among AD risk factors, and hypoxia is a direct consequence of hypoperfusion. Previously we showed that expression of the β-site β-amyloid precursor protein (APP) cleavage enzyme 1 (BACE1) gene BACE1 is tightly controlled at both the transcriptional and translational levels and that increased BACE1 maturation contributes to the AD pathogenesis in Down's syndrome. Here we have identified a functional hypoxia-responsive element in the BACE1 gene promoter. Hypoxia up-regulated β-secretase cleavage of APP and amyloid-β protein (Aβ) production by increasing BACE1 gene transcription and expression both in vitro and in vivo. Hypoxia treatment markedly increased Aβ deposition and neuritic plaque formation and potentiated the memory deficit in Swedish mutant APP transgenic mice. Taken together, our results clearly demonstrate that hypoxia can facilitate AD pathogenesis, and they provide a molecular mechanism linking vascular factors to AD. Our study suggests that interventions to improve cerebral perfusion may benefit AD patients.
Keywords: hypoxia–inducible factor 1α, amyloid–β protein, neuritic plaque, memory deficit, transcription
Deposition of amyloid-β protein (Aβ) in the brain is the hallmark of Alzheimer's disease (AD) pathology (1). Aβ, the major component of neuritic plaques, is derived from β-amyloid precursor protein (APP) after sequential cleavage by β- and γ-secretase. Early-onset familial AD caused by mutations in APP and in the presenilin 1 and 2 genes accounts for only ≈5% of total AD cases. The majority of AD cases are sporadic AD with late onset and have no defined cause. The major risk factors for AD include aging, atherosclerosis, diabetes mellitus, stroke, the ApoE ε4 polymorphism, and less education. Recent studies have shown that a history of stroke can increase AD prevalence by ≈2-fold among elderly patients (2–6). The risk is highest when stroke is concomitant with atherosclerotic vascular risk factors (7). Patients with stroke or cerebral infarction also show poorer cognitive performance and greater severity of clinical dementia (8). Hypoxia is a direct consequence of hypoperfusion, a common vascular component among the AD risk factors, and may play an important role in AD pathogenesis.
Oxygen homeostasis is essential for the development and physiology of an organism. Hypoxia-inducible factor 1 (HIF-1) is the principal molecule regulating oxygen homeostasis (9). HIF-1 is a member of the basic helix–loop–helix transcription factor family, and the basic region of the protein binds specifically to the 5′-RCGTG hypoxia-responsive element (HRE) in a gene promoter region. HIF-1 contains an oxygen-regulated expression subunit α (HIF-1α) and a constitutively expressed subunit β (HIF-1β) (Arnt). HIF-1α protein, mediated by its oxygen-dependent degradation domain, is rapidly degraded through the ubiquitin–proteasome pathway under normoxic conditions with a half-life of <5 min, but is quite stable under hypoxic conditions (10). The hypoxia signal transduction pathway plays a major role in vascular development and ischemia, as well as in neurodegeneration (11, 12). Although the brain accounts for only 2% of the body's mass, it utilizes ≈20% of total resting oxygen and is thus particularly susceptible to conditions of hypoxia. Neurons in the hippocampus and neocortex were shown to be selectively affected by cerebral ischemia (13, 14). When oxygen is in short supply, HIF-1 binds to HRE in promoters or enhancers, thereby activating a broad range of genes involved in angiogenesis, erythropoiesis, cell death, and energy metabolism (15). HIF-1 is up-regulated in the human frontal cortex with aging (16). Activation of the HIF-1 pathway by risk factors such as stroke, age, and cerebral vascular atherosclerosis may contribute to AD pathogenesis by facilitating Aβ deposition.
β-Site APP cleavage enzyme 1 (BACE1) is the β-secretase in vivo, and BACE2 is a homolog of BACE1. Even though BACE1 and BACE2 are highly homologous (17), they have distinct transcriptional regulation and function. BACE2 is not a β-secretase but rather functions as a novel θ-secretase to cleave APP within the Aβ domain (17, 18). In addition to processing APP at the β-secretase site, BACE1 was found to cleave other proteins (19–23). BACE1 interacts with reticulon family member proteins, and reticulon proteins block access of BACE1 to APP and reduce APP cleavage (24). BACE1 gene expression is tightly controlled at both the transcriptional and translational levels (17, 25–27). BACE1 protein and activity levels increase with aging and in some AD brains (28–32). Our recent finding suggests that abnormal BACE1 trafficking and maturation contribute to the AD pathogenesis in Down's syndrome (33). These studies indicate that up-regulated BACE1 gene expression at the level of transcription or translation could contribute to AD pathogenesis in some sporadic cases.
In the study described here, we found that hypoxia significantly increased BACE1 gene expression, resulting in increased β-secretase activity and Aβ production. Furthermore, hypoxia treatment markedly increased Aβ deposition and potentiated the memory deficit in AD transgenic mice. Our results demonstrate that hypoxia can facilitate AD pathogenesis, and they provide a molecular mechanism to link vascular factors to AD.
Results and Discussion
Hypoxia Up-Regulates BACE1 Promoter Activity.
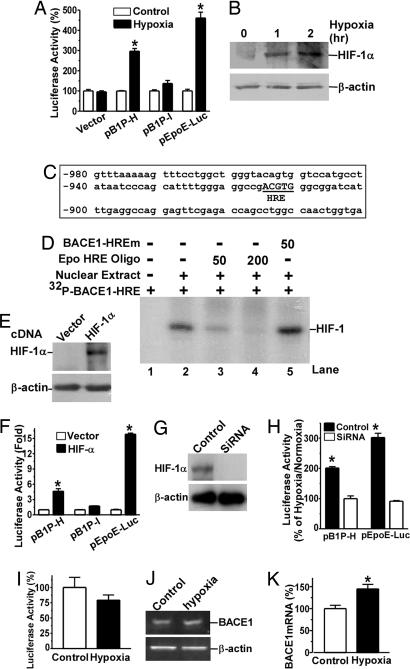
Previously, we cloned the human BACE1 gene promoter and found that BACE1 gene expression is tightly controlled at both the transcriptional and translational levels (26, 27). Here, a series of deletion luciferase reporter plasmids containing human BACE1 gene promoter regions were constructed to investigate whether BACE1 is one of the downstream target genes of the hypoxia signaling pathway. BACE1 promoter constructs containing base pairs −932 to +292 (pB1P-H) and base pairs −896 to +292 (pB1P-I) were transfected into SH-SY5Y cells. The transfected cells were treated with 2% O2 in a hypoxia incubator for 48 h. Luciferase activity was measured as an indication of promoter activity (Fig. 1A). Hypoxia treatment markedly increased the promoter activity of pB1P-H by (mean ± SEM) 296.80 ± 13.25% (P < 0.0001 relative to control) but had no significant effect on pB1P-I (P > 0.05). Consistent with previous studies (34), the promoter activity of positive control plasmid pEpoE-Luc was also significantly up-regulated under hypoxia treatment (P < 0.001), and hypoxia had no effect on the vector control (Fig. 1A). Western blot analysis showed that HIF-1α was significantly induced under hypoxic conditions (Fig. 1B). These results indicate that the region between base pairs −932 and −896 of the BACE1 promoter may contain a hypoxia-inducible enhancer element.
Fig. 1.
Up-regulation of BACE1 gene transcription by hypoxia. (A) Hypoxia increases human BACE1 promoter activity. BACE1 promoter constructs pB1P-H and pB1P-I were transfected into SH-SY5Y cells. Plasmid pGL3-Basic (vector) and pEpoE-Luc were used as negative and positive controls, respectively. Cells were exposed to 2% O2 (hypoxia) or 21% O2 (control) after transfection. Luciferase assay was performed 48 h after transfection to reflect promoter activity. ∗, P < 0.001 by ANOVA and Student's t test. (B) HIF-1α expression was rapidly induced by hypoxia in SH-SY5Y cells. Cells were treated with 2% O2 for 0, 1, and 2 h and then lysed in RIPA-Doc buffer (0.15 mM NaCl/0.05 mM Tris · HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). HIF-1α was detected by rabbit polyclonal anti-HIF-1α antibody (H206). (C) Diagram shows base pairs −980 to −900 of the human BACE1 promoter sequence (relative to the transcription start site). A HRE consensus site 5′-RCGTG was located at base pairs −915 to −911 (capitalized and underlined). (D) The human BACE1 promoter contains a HRE site. Gel shift assay was performed as described in Methods. A 32P-labeled double-stranded oligonucleotide probe, [32P]BACE1-HRE, corresponding to BACE1 promoter base pairs −924 to −907 was used as a probe. (E) Robust HIF-1α expression in HEK293 cells after pHIF-1α expression plasmid transfection. (F) BACE1 promoter activity was increased by HIF-1α overexpression. BACE1 promoter constructs pB1P-H, pB1P-I, and positive control pEpoE-Luc were cotransfected with HIF-1α expression plasmid into HEK293 cells. Luciferase assay was performed 48 h after transfection. HIF-1α overexpression can significantly increase promoter activity in cells transfected with pB1P-H but not pB1P-I. HIF-1α overexpression also significantly increased pEpoE-Luc promoter activity. ∗, P < 0.001 by ANOVA. (G) HIF-1α siRNA transfection reduced HIF-1α expression in HEK293 cells. (H) HIF-1α siRNA inhibited hypoxia's up-regulatory effect on the human BACE1 promoter activity. BACE1 promoter construct pB1P-H or positive control pEpoE-Luc plasmid were cotransfected with control siRNA or HIF-1α siRNAs. The transfected cells were then exposed to 2% O2 (hypoxia) or 21% O2 (control) 12 h after transfection. Luciferase assay was performed 24 h after hypoxia treatment to reflect promoter activity. pCMV-Rluc was cotransfected to normalize transfection efficiency. The numbers represent mean ± SEM; n = 4; ∗, P < 0.0001 by Student's t test. (I) Mutation in the HRE site abolishes the effect of hypoxia on BACE1 promoter activity. The HRE site in the BACE1 promoter of pB1P-H was mutated by site-directed mutagenesis. The mutant construct was transfected into SH-SY5Y cells and exposed to 2% O2 (hypoxia) or 21% O2 (normoxia) for 48 h. (J) Swedish APP stable SH-SY5Y cells were exposed to 2% or 21% O2 for 24 h before RNA extraction. (K) Hypoxia increased BACE1 mRNA by ≈1.5 times. ∗, P < 0.05 by Student's t test.
A Functional HRE Site in the Promoter Regulates BACE1 Gene Expression.
Hypoxia up-regulates gene transcription by stabilizing transcription factor HIF-1, which binds to HRE in the promoter region of hypoxia-regulated downstream target genes. The HIF-1 heterodimer binds to HRE consensus sequence 5′-RCGTG. Sequence analysis reveals that the human BACE1 gene promoter contains a putative HRE site that is located at base pairs −915 to −911 (5′-ACGTG) (Fig. 1C). A gel shift assay was performed to determine whether transcription factor HIF-1 can bind to the putative HRE site in the human BACE1 gene promoter. BACE1-HRE, a 20-bp double-stranded oligonucleotide probe (5′-gggaggccgACGTGggcg) corresponding to BACE1 promoter region base pairs −924 to −907 was synthesized and end-labeled with 32P. A shifted DNA-protein complex band was detected after incubation of the BACE1-HRE probe with HeLa nuclear extract (Fig. 1D, lane 2). The binding intensity of this shifted band was significantly reduced by applying 50-fold molar excess of unlabeled Epo-HRE consensus competition oligonucleotides, and the shifted band was completely abolished by addition of 200-fold of unlabeled Epo-HRE consensus oligonucleotides (Fig. 1D, lanes 3 and 4). Preincubation of excessive mutant BACE1-HRE oligonucleotides containing the binding site mutations (5′-gggaggccgAAAAGggcg) with HeLa nuclear extract had no competitive effect on the BACE1-HRE shifted band (Fig. 1D, lane 5). These results clearly demonstrate that the human BACE1 promoter contains a HRE site.
To determine whether this binding element biologically responded to the transcription factor HIF-1 in the transcriptional regulation of the human BACE1 gene, SH-SY5Y cells were transfected with BACE1 promoter plasmids pB1P-H and pB1P-I and with HIF-1α mammalian expression plasmid. Transfection with pHIF-1α plasmid showed robust HIF-1α expression (Fig. 1E). Overexpression of HIF-1α had no effect on luciferase activity in cells transfected with pB1P-I (P > 0.05). In contrast, luciferase activity increased dramatically in cells cotransfected with pB1P-H and HIF-1α, showing a 4.63 ± 0.50-fold increase compared with the non-HIF-1α transfected cells (P < 0.001) (Fig. 1F). HIF-1α also significantly increased luciferase activity in cells transfected with positive control plasmid pEpoE-Luc (15.81 ± 0.23-fold, P < 0.001, relative to vector control) (Fig. 1F). To examine whether up-regulation of human BACE1 gene transcription by hypoxia was mediated by transcription factor HIF-1, an HIF-1α siRNA assay was performed to knock down HIF-1α expression. The HIF-1α siRNA significantly inhibited HIF-1α expression (Fig. 1G). In control siRNA transfected cells, hypoxia treatment markedly increased luciferase activity in the cells transfected with human BACE1 promoter plasmid pB1P-H or with positive control plasmid pEpoE-Luc. In contrast, hypoxia treatment had no effect on the promoter activity of human BACE1 promoter plasmids pB1P-H and pEpoE-Luc after transfection of HIF-1α siRNA (Fig. 1H). These data indicate that knock-down of HIF-1α expression blocked the effect of hypoxia on the human BACE1 gene transcription. To further confirm that HIF-1 can bind specifically to this HRE in the BACE1 promoter, we generated a mutant BACE1 plasmid, pB1P-HIFm. The putative HRE core binding sequence (5′aCGTg) in the BACE1 promoter plasmid pB1P-H was changed to 5′-aAAAg by site-directed mutagenesis, creating mutant plasmid pB1P-HIFm, which lacks the HIF-1 binding site. Hypoxia treatment had no effect on the promoter activity of pB1P-HIFm in SH-SY5Y cells (79.26 ± 8.52% under hypoxia vs. 100 ± 16.25% under normoxia; P > 0.05) (Fig. 1I). The mutation resulted in an inability of the BACE1 gene promoter to respond under hypoxic conditions. Taken together, these data demonstrate that the BACE1 gene promoter contains a hypoxia-inducible enhancer and that the HRE site is physiologically functional in transcriptional regulation of BACE1 gene expression.
To further investigate whether hypoxia plays an important role in the transcription of human BACE1, we analyzed endogenous BACE1 mRNA levels by using quantitative RT-PCR, with β-actin transcriptional levels serving as the internal control. Under hypoxic conditions, endogenous BACE1 mRNA levels in SH-SY5Y cells were increased to 144.8 ± 10.42% relative to normoxia (P < 0.05) (Fig. 1 J and K). The data are consistent with the BACE1 promoter assay results, indicating that hypoxia plays an important role in transcriptional regulation of the human BACE1 gene.
Hypoxia Increases APP Processing and Aβ Generation by Up-Regulating BACE1 Activity.
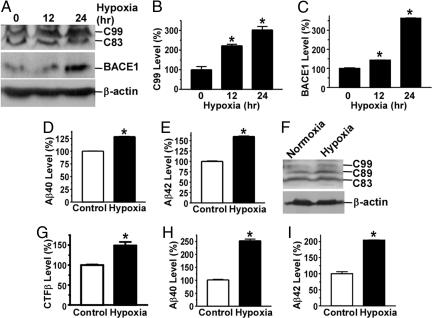
Aβ is derived from APP by β- and γ-secretase cleavages and is the major component of neuritic plaques. BACE1 is the major β-secretase in vivo. To determine whether up-regulation of BACE1 gene expression under hypoxic conditions affects APP processing at the β-secretase site to generate Aβ, we analyzed the protein levels of BACE1, APP C99, and Aβ in SH-SY5Y cells that stably overexpress Swedish mutant APP695. BACE1 protein and C99, the major β-secretase cleavage product of APP, were markedly increased under hypoxic conditions (Fig. 2A). Quantification showed that C99 levels were increased by 221.7 ± 7.32% and 302.1 ± 1.74% (P < 0.001) (Fig. 2B) and that BACE1 protein levels were increased to 143.4 ± 1.19% and 304.5 ± 1.69% after 12 h and 24 h of hypoxia treatment, respectively (P < 0.001) (Fig. 2C). The production of Aβ40 and Aβ42 was significantly increased to 128.6 ± 0.56% and 159.8 ± 1.17%, respectively, in hypoxia treated cells relative to control cells (P < 0.0001) (Fig. 2 D and E). To further confirm that hypoxia has a similar effect on wild-type (WT) APP processing, HAW1 cells that stably express a WT APP695 protein were treated with hypoxia. The β-secretase cleavage products, the C-terminal fragment β (CTFβ) including C99 and C89, were significantly increased under hypoxic conditions (Fig. 2F), and quantification showed that the CTFβ levels were increased by 148.97 ± 8.89% (P < 0.001 relative to control) (Fig. 2G). Hypoxia also markedly increased Aβ40 and Aβ42 levels in HAW1 cells to 252.6 ± 6.52% and 204.1 ± 0.96%, respectively (P < 0.0001 relative to controls) (Fig. 2 H and I). These data indicate that hypoxia can elevate BACE1 protein expression and in turn increase APP processing at the β-secretase site to potentiate Aβ generation.
Fig. 2.
Hypoxia increases Aβ production by up-regulating BACE1 activity. (A) SH-SY5Y cells stably overexpressing Swedish mutant APP were exposed to 2% O2 for 0, 12, or 24 h. C20 antibody was used to detect APP CTFs; 208 antibody was used to detect BACE1. (B and C) Quantification of C99 (B) and BACE1 protein (C) levels. ∗, P < 0.001 by ANOVA. (D and E) ELISA was performed to measure Aβ40 (D) and Aβ42 (E) in culture media from SH-SY5Y cells stably overexpressing Swedish mutant APP exposed to 2% O2 (hypoxia) or 21% O2 (normoxia) for 24 h. ∗, P < 0.0001 by Student's t test. (F) WT APP695 cell HAW1 was cultured under 2% O2 for 12 h, and APP CTFs were detected with C20 antibody. (G) The levels of CTFβ including C99 and C89 were quantitated. ∗, P < 0.001 by Student's t test. (H and I) The conditioned media from the same hypoxia-treated cells were analyzed for Aβ40 (H) and Aβ42 (I) levels. The numbers represent mean ± SEM; n = 3; ∗, P < 0.0001 by Student's t test.
Hypoxia Facilitates β-Secretase Cleavage of APP and Aβ Deposition in APP23 Transgenic Mice.
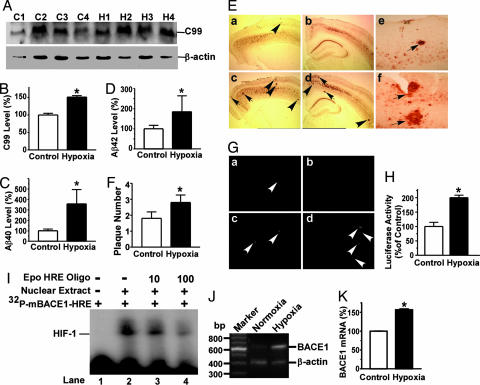
Our data indicate that hypoxia facilitates BACE1 gene transcription and increases APP processing at the β-secretase site to generate Aβ. To investigate whether hypoxia would facilitate AD pathogenesis in vivo, APP23 transgenic mice, a valued AD mouse model, were subjected to hypoxia treatment. APP23 mice carry the human Swedish mutant APP751 transgene driven by the Thy1.2 promoter element, which drives the transgene to be specifically expressed in neurons (35). APP23 mice were subjected to 8% O2 for 16 h/day for 1 month; age-matched control APP23 mice were exposed to normal air (21% O2). To determine whether hypoxia affects APP processing in vivo, the levels of APP CTFs in APP23 brain tissue lysates were assayed using Western blot analysis (Fig. 3A). C99, the major β-secretase product, was significantly increased in the hypoxic mice by 151.1 ± 4.15% relative to controls (P < 0.01) (Fig. 3B). To examine the effect of hypoxia on Aβ generation in the transgenic mice, Aβ ELISA analysis was performed. Hypoxia treatment increased the levels of Aβ40 generation by 357.5 ± 137.01% relative to controls (P < 0.001) and increased the levels of Aβ42 by 184.6 ± 79.08% (P < 0.001) (Fig. 3 C and D). These data demonstrate that hypoxia up-regulates β-secretase cleavage of APP and thus Aβ production. APP23 mice develop amyloid plaques in neocortex and hippocampus as early as 6 months of age and progressively with age (35). To investigate whether the altered APP processing caused by hypoxia resulted in Aβ deposition and an increase in neuritic plaque formation in vivo, the hypoxia-treated and control APP23 mice were killed and 4G8 immunostaining (Fig. 3E) and thioflavin S staining (Fig. 3G) were used to detect Aβ-containing neuritic plaques in brain. Neuritic plaque formation was significantly increased in APP23 mice under hypoxic conditions relative to normoxic controls (Fig. 3 E and G). More and larger plaques were seen in hypoxic mice than in normoxic mice (Fig. 3E). Quantification showed that hypoxia-treated APP23 mice had ≈1.5-fold more plaques than normoxic mice (2.8 ± 0.5 vs. 1.8 ± 0.4 per slice; P < 0.001) (Fig. 3F).
Fig. 3.
Hypoxia increases β-secretase cleavage of APP and Aβ deposition in APP23 transgenic mice. Eight-month-old APP23 mice were treated with 8% O2 (hypoxia) for 16 h/day for 1 month. Ten mice per group were used for hypoxia and normoxia treatment. (A) Half brains from hypoxic and age-matched normoxic control mice were lysed in RIPA-Doc lysis buffer and separated with 16% Tris-Tricine SDS/PAGE gel. C99 was detected by C20 polyclonal antibody. β-actin was detected by anti-β-actin antibody AC-15 as the internal control. (B) Quantification showed that C99 was significantly increased in hypoxia-treated mice. ∗, P < 0.01 by Student's t test. (C and D) ELISA was performed to measure Aβ40 (C) and Aβ42 (D) levels in mouse brain tissue lysates. ∗, P < 0.001 by Student's t test. (E) 4G8 immunostaining. The other half brains were dissected from hypoxic and age-matched normoxic control mice, fixed, and sectioned. Neuritic plaques were detected by Aβ-specific mAb 4G8 (Signet Laboratories) and the DAB method. The plaques were visualized under a microscope at ×40 magnification. More neuritic plaques were stained in hypoxic mice (c and d) compared with age-matched control mice (a and b). Arrows point to plaques. Magnification at ×200 reveals more and larger plaques in hypoxic mice (f) than in normoxic mice (e). (F) Quantification of neuritic plaques. ∗, P < 0.001 by ANOVA. (G) Thioflavin S staining. Neuritic plaques were further confirmed by using thioflavin S fluorescent staining and were visualized under a microscope at ×100 magnification. a and c are sections of frontal cortex; b and d are sections of hippocampus. More neuritic plaques were seen in hypoxia-treated mice (c and d) than in age-matched controls (a and b). Arrows point to green fluorescent neuritic plaques. (H) A mouse BACE1 promoter was cloned into the pGL3-Basic vector and transfected into cells. Hypoxia increased luciferase activity in pB1Pm-A transfected cells that contained two putative HRE consensus sites. ∗, P < 0.01 by Student's t test. (I) Gel shift assay demonstrated that the mouse BACE1 promoter also contains an HIF-1 transcription factor binding site. Gel shift assay was performed using a 32P-labeled [32P]mBACE1-HRE probe in which the two putative HRE consensus sites were juxtaposed. Lane 1, probe alone without nuclear extract; lane 2, incubation of HeLa nuclear extract with probe retarded the migration of free probes to form a new HIF-1-DNA complex; lanes 3 and 4, competition assays adding 10- and 100-fold molar excess of Epo-HRE consensus oligonucleotides. (J) Endogenous BACE1 mRNA level in the WT control mice. The control mice were subjected to the hypoxia treatment, and total RNA samples were extracted from the mouse brains for RT-PCR assay. β-actin was amplified as the internal control. (K) Hypoxia significantly increased endogenous BACE1 mRNA in WT mice by ≈1.5 times. ∗, P < 0.0001 relative to normoxia WT mice controls by Student's t test.
To confirm that hypoxia facilitates APP processing and Aβ generation in the transgenic mice by up-regulating mouse BACE1 gene transcription, we cloned the mouse BACE1 promoter. Sequence analysis reveals that there are two putative HRE consensus sites in the mouse BACE1 5′UTR. Hypoxia markedly increased luciferase activity by 199.81 ± 8.97% in cells transfected with mouse promoter plasmid pB1Pm-A, which contains the HRE sites (P < 0.01) (Fig. 3H). Gel shift assay showed that the mouse BACE1 promoter region contains HRE (Fig. 3I). To determine whether hypoxia up-regulates mouse BACE1 gene transcription in vivo, WT control mice were subjected to hypoxia treatment and the mouse endogenous BACE1 mRNA levels were measured using quantitative RT-PCR. Under hypoxic conditions, endogenous BACE1 mRNA levels in the WT mice were significantly increased to 156.7 ± 2.45% relative to normoxia (P < 0.0001) (Fig. 3 J and K). These results suggest that mouse BACE1 gene expression, like human BACE1 gene expression, can be up-regulated by hypoxia at the transcriptional level.
Hypoxia Potentiates the Memory Deficit in APP23 Mice.
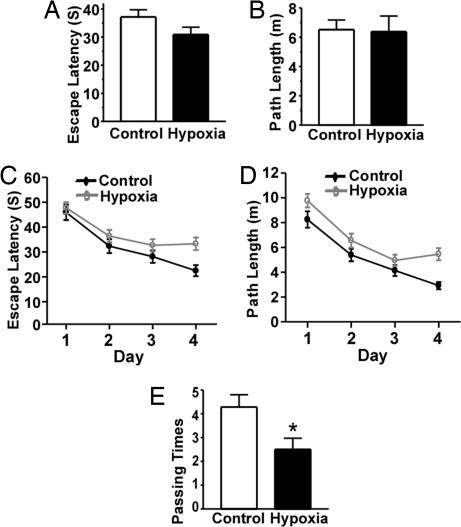
To further investigate whether hypoxia affects learning and memory in AD pathogenesis, behavioral tests were performed after APP23 mice underwent 1 month of hypoxia treatment. The Morris water maze was used to determine the effect of hypoxia on spatial memory. In visible platform tests, the hypoxic and normoxic APP23 mice had a similar escape latency (37.15 ± 2.58 s and 30.90 ± 2.60 s, respectively; P > 0.05) (Fig. 4A) and path length (6.52 ± 0.66 m and 6.38 ± 1.07 m, respectively; P > 0.05) (Fig. 4B), indicating that hypoxia did not affect mouse motility or vision. In the hidden-platform swimming test, hypoxia-treated APP23 mice showed a longer latency (P < 0.005; Fig. 4C) and swam a significantly longer distance (P < 0.001; Fig. 4D) to reach the platform compared with normoxic mice. Furthermore, hypoxic APP23 mice had significantly fewer platform-passing times in the probe trial (2.5 ± 0.48 s vs. 4.29 ± 0.52 s; P < 0.05) (Fig. 2E). These data demonstrate that hypoxia significantly potentiates the memory deficit in APP23 mice.
Fig. 4.
Hypoxia potentiates the memory deficit in Swedish mutant APP transgenic mice. A Morris water maze protocol with 1 day of visible platform tests and 4 days of hidden platform tests, plus a probe trial on day 6. Animal movement was tracked and recorded by an HVS 2020 Plus image analyzer (HVS Image, Hampton, UK). Ten mice per group were used for hypoxia and normoxia treatment. (A) During the first day of visible platform tests, mice were trained in five contiguous trials. The hypoxic and normoxic APP23 mice exhibited a similar latency to escape onto the visible platform. P > 0.05 by Student's t test. (B) Hypoxic and normoxic APP23 mice swam a similar distance before escaping onto the visible platform in the visible platform test. P > 0.05 by Student's t test. (C) In hidden platform tests, mice were trained with six trials per day for 4 days. Hypoxic APP23 mice showed a longer latency to escape onto the hidden platform. P < 0.005 by two-way ANOVA. (D) Hypoxic APP23 mice swam farther before escaping onto the hidden platform. P < 0.001 by two-way ANOVA. (E) In the probe trial on day 6, hypoxic APP23 mice crossed the target platform significantly fewer times than controls. ∗, P < 0.05 by Student's t test.
Hypoxia is one of the major common components of vascular risk factors for AD pathogenesis such as stroke, hypertension, atherosclerosis, and diabetes. Hypoxia can also arise from cerebral amyloid angiopathy. Previous studies (32, 36) showed that BACE1 expression and enzymatic activity are elevated in the brains of sporadic AD patients, and the degree of elevation is correlated with Aβ production. The mechanism by which vascular factors facilitate AD neurodegeneration in AD pathogenesis has not yet been defined. It has been reported that HIF-1 is increased in the brains of elderly people (16). By subjecting AD transgenic model mice to low-oxygen conditions, we clearly demonstrated that hypoxia facilitates plaque formation and enhances memory deficits. Our study shows that BACE1 is one of the downstream target genes of the hypoxia signaling pathway and that hypoxia potentiates APP processing to generate Aβ by activating BACE1 transcription through an HRE in its promoter region, leading to increased BACE1 expression. A slight increase in BACE1 expression could lead to a dramatic increase in Aβ production (37). HIF-1α can also be up-regulated by Aβ (38). Because our data showed that hypoxia increases Aβ production, a positive feedback circuit may dramatically increase Aβ production and deposition in AD pathogenesis. It has been reported that chronic hypoxic insults could alter APP processing at the α-secretase site, resulting in reduction of C83 and sAPPα (39–41). Although we have not observed a significant decrease in the α-secretase pathway, our data also show that chronic hypoxia can alter APP processing by its effects on the β-secretase pathway in vivo. Furthermore, hypoxia can trigger the transcription of many other genes besides BACE1. Prolonged or severe hypoxia can also trigger apoptosis in the brain, which may contribute to the neuronal loss and memory impairment seen in AD patients. Transient hypoxic injury to cortical neurons can cause the mitochondrial dysfunction, impaired membrane integrity, and altered APP processing that occur in AD (42). Ergoloid mesylates (e.g., Hydergine) have been the longest used putative cognitive enhancement drugs and are one of the classes of drugs used in AD treatment. As a mild cerebral vasodilator, Hydergine can decrease hypoxia in the early stages of AD and improve symptoms in AD patients (43). Our study suggests that enhancing oxygen supply by improving cerebral perfusion or by using vasodilators may have pharmaceutical potential for treating cognitive impairments, thereby benefiting AD patients.
Methods
Transgenic Mice and Hypoxia Treatment.
APP23 transgenic mice carry the human APP751 cDNA with the Swedish double mutation at positions 670/671 (KM→NL) under control of the murine Thy-1.2 expression cassette (35). Littermates of APP23 mice carrying no human Swedish mutant APP751 cDNA were used as WT controls. APP23 mice (8 months of age, n = 20, 50% female) were assigned randomly to hypoxia and control groups. The hypoxia group was treated in a hypoxia chamber at 8% O2 for 16 h/day for 1 month. The oxygen level was regulated by infusing nitrogen into a semisealable chamber controlled by a PROOX model 110 controller (BioSpherix, Redfield, NY).
Morris Water Maze.
The water maze test was performed in a pool 1.5 m in diameter. In the hidden platform trials, a 10-cm-diameter platform was placed in the southeastern quadrant of the pool. The procedure consists of 1 day of visible platform tests and 4 days of hidden platform tests, plus a probe trial 24 h after the last hidden platform test. In the visible platform test, mice were tested for five contiguous trials, with an intertrial interval of 30 min. In the hidden platform tests, mice were trained for six trials, with an intertrial interval of 1 h. Tracking of animal movement was achieved with an HVS 2020 Plus image analyzer (HVS Image). Data were analyzed by two-way ANOVA.
Neuritic Plaque Staining.
Mice were killed after the behavioral tests, and the half brains were fixed and sectioned with a Leica (Deerfield, IL) cryostat to 40 μm thickness. Every sixth slice with the same reference position of the mice was mounted onto the slides for staining. The slices were immunostained with biotinylated monoclonal 4G8 antibody (Signet Laboratories, Dedham, MA) at 1:500 dilution. Approximately 24 slices were stained for each mouse. Plaques were visualized by the avidin-biotin-peroxidase complex (ABC) and diaminobenzidine (DAB) method and counted under microscopy with ×40 magnification. Plaques were quantitated by average plaque count per slice for each mouse. The data were analyzed by two-way ANOVA. Thioflavin S staining of plaques was performed with 1% thioflavin S, and the green fluorescence-stained plaques were visualized with fluorescent microscopy.
Cell Culture, Transfection, Hypoxia Treatment, and Luciferase Assay.
SH-SY5Y cells, HEK293 cells, and N2A cells were grown in complete DMEM. All cells were maintained at 37°C in an incubator containing 5% CO2. Cells were transfected with plasmid DNA by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). A Swedish mutant APP stable cell line was generated by transfecting pZ-APPsw into SH-SY5Y cells and selected by 1,000 μg/ml Zeocine (Invitrogen). HAW1 cells are the WT APP695 stably expressing HEK293 cells. Hypoxia treatments were performed by incubating cells in a 37°C incubator containing 2% O2 and 5% CO2. The Renilla (sea pansy) luciferase vector pCMV-Rluc was cotransfected to normalize transfection efficiency. Luciferase assay was performed 48 h after transfection by using a dual-luciferase reporter assay system (Promega, Madison, WI) as described previously (17).
Plasmids, Gel Shift Assay, Immunoblotting, siRNA Assay, Quantitative RT-PCR, and Aβ40/42 Sandwich ELISA.
Details of these procedures are provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Brain MacLean, Xiaojian Sun, and Jane Wang for technical assistance, and Anthony G. Phillips and Kelley Bromley for helpful discussion. This work was supported by the Canadian Institutes of Health Research (CIHR), The Jack Brown and Family Alzheimer's Research Foundation, and The Michael Smith Foundation for Health Research (W.S.). W.S. is the holder of The Canada Research Chair in Alzheimer's Disease. W.Z. was the recipient of The Arthur and June Willms Fellowships. G.H. was supported by The Chinese Scholarship Council Award. F.D. was the recipient of a Natural Sciences and Engineering Research Council (NSERC) graduate fellowship.
Abbreviations
- Aβ
amyloid-β protein
- AD
Alzheimer's disease
- APP
β-amyloid precursor protein
- BACE1
β-site APP cleavage enzyme 1
- HIF
hypoxia-inducible factor
- HRE
hypoxia-responsive element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Mattson MP. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White L, Petrovitch H, Hardman J, Nelson J, Davis DG, Ross GW, Masaki K, Launer L, Markesbery WR. Ann NY Acad Sci. 2002;977:9–23. doi: 10.1111/j.1749-6632.2002.tb04794.x. [DOI] [PubMed] [Google Scholar]
- 3.Snowdon DA, Greiner LH, Mortimer JA, Riley KP, Greiner PA, Markesbery WR. J Am Med Assoc. 1997;277:813–817. [PubMed] [Google Scholar]
- 4.Altieri M, Di Piero V, Pasquini M, Gasparini M, Vanacore N, Vicenzini E, Lenzi GL. Neurology. 2004;62:2193–2197. doi: 10.1212/01.wnl.0000130501.79012.1a. [DOI] [PubMed] [Google Scholar]
- 5.Schneider JA, Wilson RS, Cochran EJ, Bienias JL, Arnold SE, Evans DA, Bennett DA. Neurology. 2003;60:1082–1088. doi: 10.1212/01.wnl.0000055863.87435.b2. [DOI] [PubMed] [Google Scholar]
- 6.Vermeer SE, Prins ND, den Heijer T, Hofman A, Koudstaal PJ, Breteler MM. N Engl J Med. 2003;348:1215–1222. doi: 10.1056/NEJMoa022066. [DOI] [PubMed] [Google Scholar]
- 7.Honig LS, Kukull W, Mayeux R. Neurology. 2005;64:494–500. doi: 10.1212/01.WNL.0000150886.50187.30. [DOI] [PubMed] [Google Scholar]
- 8.Heyman A, Fillenbaum GG, Welsh-Bohmer KA, Gearing M, Mirra SS, Mohs RC, Peterson BL, Pieper CF. Neurology. 1998;51:159–162. doi: 10.1212/wnl.51.1.159. [DOI] [PubMed] [Google Scholar]
- 9.Huang LE, Willmore WG, Gu J, Goldberg MA, Bunn HF. J Biol Chem. 1999;274:9038–9044. doi: 10.1074/jbc.274.13.9038. [DOI] [PubMed] [Google Scholar]
- 10.Huang LE, Bunn HF. J Biol Chem. 2003;278:19575–19578. doi: 10.1074/jbc.R200030200. [DOI] [PubMed] [Google Scholar]
- 11.Pugh CW, Ratcliffe PJ. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 12.Halterman MW, Miller CC, Federoff HJ. J Neurosci. 1999;19:6818–6824. doi: 10.1523/JNEUROSCI.19-16-06818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulsinelli WA, Brierley JB, Plum F. Ann Neurol. 1982;11:491–498. doi: 10.1002/ana.410110509. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Nagayama T, Jin K, Stetler RA, Zhu RL, Graham SH, Simon RP. J Neurosci. 1998;18:4914–4928. doi: 10.1523/JNEUROSCI.18-13-04914.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp FR, Bernaudin M. Nat Rev Neurosci. 2004;5:437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 16.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Wang Y, Qing H, Christensen MA, Liu Y, Zhou W, Tong Y, Xiao C, Huang Y, Zhang S, et al. FASEB J. 2005;19:739–749. doi: 10.1096/fj.04-3426com. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, He G, Song W. FASEB J. 2006;20:1369–1376. doi: 10.1096/fj.05-5632com. [DOI] [PubMed] [Google Scholar]
- 19.von Arnim CAF, Kinoshita A, Peltan ID, Tangredi MM, Herl L, Lee BM, Spoelgen R, Hshieh TT, Ranganathan S, Battey FD, et al. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Sudhof TC. J Biol Chem. 2004;279:10542–10550. doi: 10.1074/jbc.M310001200. [DOI] [PubMed] [Google Scholar]
- 21.Pastorino L, Ikin AF, Lamprianou S, Vacaresse N, Revelli JP, Platt K, Paganetti P, Mathews PM, Harroch S, Buxbaum JD. Mol Cell Neurosci. 2004;25:642–649. doi: 10.1016/j.mcn.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y. Proc Natl Acad Sci USA. 2001;98:13554–13559. doi: 10.1073/pnas.241509198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenthaler SF, Dominguez DI, Westmeyer GG, Reiss K, Haass C, Saftig P, De Strooper B, Seed B. J Biol Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- 24.He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Nat Med. 2004;10:959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- 25.Ge YW, Maloney B, Sambamurti K, Lahiri DK. FASEB J. 2004;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W, Song W. Mol Cell Biol. 2006;26:3353–3364. doi: 10.1128/MCB.26.9.3353-3364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W. Mol Cell Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 29.Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo C, Schettini G, Saido TC, Hulette C, Lippa C, Lannfelt L, Ghetti B, Gambetti P, Tabaton M, Teller JK. Nature. 2000;405:531–532. doi: 10.1038/35014735. [DOI] [PubMed] [Google Scholar]
- 31.Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Ann Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 32.Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, Beach T, Sue L, Wong P, Price D, et al. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- 33.Sun X, Tong Y, Qing H, Chen CH, Song W. FASEB J. 2006;20:1361–1368. doi: 10.1096/fj.05-5628com. [DOI] [PubMed] [Google Scholar]
- 34.Madan A, Curtin PT. Proc Natl Acad Sci USA. 1993;90:3928–3932. doi: 10.1073/pnas.90.9.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, et al. Proc Natl Acad Sci USA. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, et al. Proc Natl Acad Sci USA. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Zhou W, Tong Y, He G, Song W. FASEB J. 2006;20:285–292. doi: 10.1096/fj.05-4986com. [DOI] [PubMed] [Google Scholar]
- 38.Soucek T, Cumming R, Dargusch R, Maher P, Schubert D. Neuron. 2003;39:43–56. doi: 10.1016/s0896-6273(03)00367-2. [DOI] [PubMed] [Google Scholar]
- 39.Auerbach ID, Vinters HV. J Neuropathol Exp Neurol. 2006;65:610–620. doi: 10.1097/00005072-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 40.Webster NJ, Green KN, Peers C, Vaughan PF. J Neurochem. 2002;83:1262–1271. doi: 10.1046/j.1471-4159.2002.01236.x. [DOI] [PubMed] [Google Scholar]
- 41.Webster NJ, Green KN, Settle VJ, Peers C, Vaughan PF. Brain Res Mol Brain Res. 2004;130:161–169. doi: 10.1016/j.molbrainres.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 42.Chen GJ, Xu J, Lahousse SA, Caggiano NL, de la Monte SM. J Alzheimers Dis. 2003;5:209–228. doi: 10.3233/jad-2003-5305. [DOI] [PubMed] [Google Scholar]
- 43.Olin J, Schneider L, Novit A, Luczak S. Cochrane Database Syst Rev. 2001 doi: 10.1002/14651858.CD000359. CD000359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.