Abstract
The adverse effects of high salt on plants include Na+ toxicity and hyperosmotic and oxidative stresses. The plasma membrane-localized Na+/H+ antiporter SOS1 functions in the extrusion of toxic Na+ from cells and is essential for plant salt tolerance. We report here that, under salt or oxidative stress, SOS1 interacts through its predicted cytoplasmic tail with RCD1, a regulator of oxidative-stress responses. Without stress treatment, RCD1 is localized in the nucleus. Under high salt or oxidative stress, RCD1 is found not only in the nucleus but also in the cytoplasm. Like rcd1 mutants, sos1 mutant plants show an altered sensitivity to oxidative stresses. The rcd1mutation causes a decrease in salt tolerance and enhances the salt-stress sensitivity of sos1 mutant plants. Several genes related to oxidative-stress tolerance were found to be regulated by both RCD1 and SOS1. These results reveal a previously uncharacterized function of a plasma membrane Na+/H+ antiporter in oxidative-stress tolerance and shed light on the cross-talk between the ion-homeostasis and oxidative-stress detoxification pathways involved in plant salt tolerance.
Keywords: salt stress, reactive oxygen species, hydrogen peroxide stress
Soil salinity is a significant environmental factor that adversely affects plant growth and agricultural productivity. Excessive Na+ in saline soils is toxic to plants when it accumulates in the cytoplasm. The salt overly sensitive (SOS) pathway is responsible for Na+ homeostasis in plants (1). The Arabidopsis thaliana SOS1 protein resides at the plasma membrane, where it functions to extrude Na+ from the cytoplasm coupled to H+ influx (2). The Na+/H+ antiporter activity of SOS1 is regulated by SOS3 (a myristoylated calcium-binding protein) (3, 4) and SOS2 (a Ser/Thr protein kinase) (5, 6). Cytoplasmic calcium signals elicited by salt stress presumably are perceived by SOS3, which interacts physically with SOS2 (6), and this SOS2/SOS3 kinase complex activates the transport activity of SOS1 by phosphorylation (7). Overexpression of SOS1 or a constitutively activated form of SOS2 confers improved salt tolerance to transgenic plants (8, 9).
The SOS1 protein has 12 predicted transmembrane domains in the N-terminal region and a long cytoplasmic tail of ≈700 aa at the C-terminal side (10). The transmembrane portion has sequence similarities with plasma membrane Na+/H+ exchangers from animal, bacterial, and fungal cells (10). In animal cells, the ubiquitously expressed Na+/H+ antiporter NHE1 functions in pH homeostasis by exchanging intracellular H+ for extracellular Na+ (11). NHE1 has a C-terminal tail of ≈300 aa, which is important in regulating the Na+/H+ antiporter activity and other functions of the protein through phosphorylation and by binding to regulatory proteins and anchoring the actin cytoskeleton to the plasma membrane (12).
The predicted cytoplasmic tail of SOS1 does not contain any known sequence features. Still, the presence of such an unusually long hydrophilic tail in SOS1 suggests that the transporter may be regulated by diverse signals, and it may even have some other role besides the antiporter activity. It has been suggested that SOS1 is a possible Na+ sensor, like several other transporters with long cytosolic tails (1). In addition, the long tail of SOS1 may interact with various regulatory proteins involved in salt and other stress responses. However, there is no evidence as yet that would help to reveal the role of the cytoplasmic tail of SOS1.
In addition to ion toxicity, salt stress leads to the accumulation of high levels of reactive oxygen species (ROS) (13). The excessive production of ROS such as superoxide (·O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (·OH−) may disturb cellular redox homeostasis, leading to oxidative injuries. The Arabidopsis protein RCD1 (radical-induced cell death), also known as CEO1, when ectopically expressed in yeast, could protect the cells against oxidative damage imposed by H2O2, diamide, or tert-butyl peroxide (14). The Arabidopsis mutant rcd1–1 is sensitive to ozone and apoplastic superoxide, which induces cellular ·O2− accumulation and transient spreading of lesions (15). Interestingly, rcd1 mutants are resistant to methyl viologen (MV), which generates ROS in chloroplasts (16). Furthermore, the mutant exhibits reduced sensitivity to abscisic acid, ethylene, and methyl jasmonate and has altered expression of several hormonally regulated genes (17). RCD1 protein consists of a conserved globular domain called WWE, predicted to mediate specific protein–protein interactions, and a putative PARP (poly ADP-ribose polymerase) domain (17). However the exact function of these domains in RCD1 is still unknown. Belles-Boix et al. (14) carried out a yeast two-hybrid screening and found that RCD1 interacts with transcription factors such as the STO protein that can confer salt tolerance when ectopically expressed in yeast. Microarray analysis of the rcd1–1 mutant (17) identified several abiotic stress-responsive genes as having lower levels of expression in the mutant. All these studies indicate that RCD1 is an important transcriptional regulator of oxidative-stress responses and may be involved in regulating a wide range of abiotic stress responses in plants.
Considering the crucial role of the SOS pathway in Na+ homeostasis, it is of interest to test whether the pathway may also have a function in plant tolerance to oxidative stress, another component of salt stress. In the course of this study, we discovered that the C-terminal tail of SOS1 interacts with RCD1 under salt and oxidative stress. We obtained genetic evidence that SOS1 functions in oxidative-stress responses, and RCD1 also has a role in salt tolerance. Importantly, we found that the RCD1 protein resides in the nucleus under control conditions but is localized in both the nucleus and the cytoplasm near the cell periphery under salt or oxidative stress. Our study thus identified RCD1 as a previously uncharacterized component in salt-stress response pathways and revealed a surprising function of SOS1 in oxidative-stress tolerance.
Results
SOS1 Interacts with RCD1 in Vitro and in Vivo.
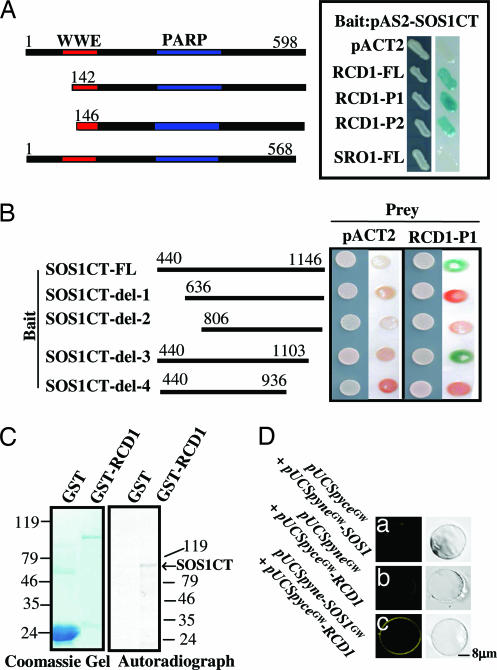
To try to understand the role of the SOS1 C-terminal tail, we carried out yeast two-hybrid screening to identify proteins that interact with the tail sequence. The C-terminal fragment (CT) of SOS1 cloned in the pAS2 vector was used as bait to screen a prey library. Only one positive clone was isolated, and subsequent DNA sequence analysis identified it as RCD1. Deletions of part of the WWE domain (RCD1-P1 and RCD1-P2) did not substantially affect the interaction with SOS1CT (Fig. 1A), suggesting that the WWE domain is not necessary for the interaction. SRO1 (similar to RCD one) is the most closely related member in the RCD1 protein family and has 76% amino acid similarity to RCD1. Unlike RCD1, SRO1 did not interact with SOS1CT (Fig. 1A). The result shows that the SOS1 C-terminal tail interacts specifically with RCD1.
Fig. 1.
Interaction between SOS1CT and RCD1 in vitro and in vivo. (A) Interactions between SOS1CT and different fragments of RCD1 in the yeast two-hybrid system. Yeast strains containing the pAS2-SOS1CT bait and pACT2-RCD1 and SRO1 prey were used for β-galactosidase filter assays. RCD1-FL (full-length RCD1), RCD1-P1, and RCD1-P2 are truncated RCD1, and SRO1-FL is full-length SRO1. The pAS2-SOS1CT and pACT2 combination was used as a negative control. The fragments used for interaction studies are shown on the far left, with the amino acid positions at the top. (B) The interaction between different portions of SOS1CT and RCD1. The combination of pACT2 and pAS2 containing different portions of SOS1CT was used as a negative control. The left side of each panel shows the growth of yeast colonies, and the right side shows the β-galactosidase filter assay. (C) In vitro binding assay. [35S]methionine-labeled SOS1CT was pulled down by RCD1-GST but not by GST alone. (D) In vivo interaction between SOS1 and RCD1 by the bimolecular fluorescence complementation assay (C). The protoplasts were treated with 25 mM NaCl for 2 h, followed by the addition of 1.5 mM H2O2 for 2 h. Negative controls are shown (a and b).
In an attempt to delineate sequences in the SOS1CT that may be responsible for interaction with RCD1, five deletions of the SOS1 C-terminal tail were generated and cloned into the pAS2 vector. The yeast strain Y190 was transformed with all of the combinations of the SOS1CT deletions and the RCD1-P1 prey. The β-galactosidase filter assay showed that both ends (between amino acids 440–806 and 936-1103) of the SOS1CT are important for the protein to interact with RCD1 (Fig. 1B).
To confirm the interaction of SOS1CT with RCD1, an in vitro pull-down assay was carried out. SOS1CT labeled with [35S]methionine was incubated with RCD1-GST protein (Fig. 1C). The proteins pulled down by RCD1-GST were loaded onto an SDS/PAGE gel. The autoradiograph showed a single strong band corresponding to the size of SOS1CT. In contrast, GST alone did not pull down the labeled SOS1CT. This experiment, together with yeast two-hybrid studies, strongly suggests that SOS1CT interacts with the RCD1 protein.
Results obtained from interaction studies in yeast and in vitro prompted us to investigate whether such interaction occurs in vivo in plants. We used bimolecular fluorescence complementation (BiFC) to directly visualize the interaction in living plant cells. Whereas protoplasts transformed with pUCSpyneGW-SOS1/pUCSpyceGW vector or pUCSpyceGW-RCD1/pUCSpyneGW vector produced no fluorescence (Fig. 1D a and b), a strong signal was observed when pUCSpyneGW-SOS1 was coexpressed with pUCSpyceGW-RCD1 (Fig. 1Dc). These data, together with the in vitro results, demonstrate that SOS1 can physically interact with RCD1 through its C-terminal cytosolic tail.
RCD1 Is Predominantly Localized in the Nucleus Under Unstressed Conditions.
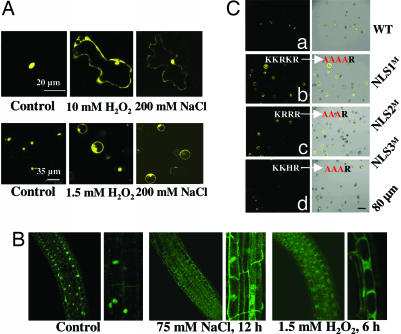
An analysis of the RCD1 amino acid sequence indicated the presence of three potential nuclear localization signals (NLS): KKRKR, KRRR, and KKHR (14). This finding suggests that RCD1 might be localized in the nucleus. However, SOS1 is localized at the plasma membrane (10). It would thus appear not physically possible for SOS1 to interact with RCD1 in vivo in plants. We therefore studied the localization of RCD1 in plants. We made yellow fluorescent protein (YFP)- and GFP-tagged RCD1 fusions that were expressed under the control of a constitutive cauliflower mosaic virus (CaMV) 35S promoter. Transient studies in Arabidopsis involving microprojectile bombardment of leaves or PEG-mediated transformation of mesophyll protoplasts with the RCD1-YFP plasmid construct showed that RCD1 is present predominantly in the nucleus under control conditions (Fig. 2A). Further studies in transgenic seedlings expressing RCD1-GFP also supported this observation (Fig. 2B). Root cells from stably transformed RCD1-GFP plants showed green fluorescence in the nuclei (Fig. 2B).
Fig. 2.
Localization of RCD1-GFP or RCD1-YFP fusion protein under control and stress conditions. (A) WT Arabidopsis leaves and mesophyll protoplasts were transfected with RCD1-YFP. Fourteen hours after the transfection, leaves and protoplasts were treated with H2O2 or NaCl and viewed under a confocal microscope. (B) RCD1-GFP transgenic seedlings were exposed to 75 mM NaCl or 1.5 mM H2O2, and the roots were examined for GFP localization. Enlarged pictures are shown on the right side of each panel. (C) Localization of RCD1-YFP containing WT (a) and mutated NLSs (b–d). Overlays of fluorescence and bright field are shown on the right side of each panel.
To identify the sequence motif involved in the nuclear import of RCD1 protein and to further confirm its nuclear localization, mutations were introduced in the three predicted NLSs of RCD1. We found that K-to-A and R-to-A mutations in NLS1 or NLS2 reduced RCD1 localization in the nucleus, because the fusion protein was present in the nucleus as well as cytoplasm (Fig. 2C b and c), unlike the protoplasts transformed with unmutated RCD1-YFP (WT), which exhibited localization in the nucleus only (Fig. 2Ca). However, K-to-A and H-to-A mutations in NLS3 did not alter the nuclear localization of the fusion protein (Fig. 2Cd). These results confirm the predominant nuclear localization of RCD1 under unstressed conditions and suggest that NLS1 and NLS2, but not NLS3, are responsible for the nuclear localization of RCD1.
RCD1 Is Localized in Both Nucleus and Cytoplasm Under Salt or Oxidative Stress.
We then investigated the localization of RCD1 protein under stress conditions. Leaves or protoplasts transiently transformed with RCD1-YFP and RCD1-GFP transgenic seedlings were exposed to either NaCl or H2O2 stress. Amazingly, we found that, when treated with salt (75 mM NaCl for 12 h for seedling roots, 200 mM NaCl for 6 h for leaves and for 2 h for protoplasts) or oxidative stress (1.5 mM H2O2 for 6 h for seedlings and for 2 h for protoplasts and 10 mM H2O2 for 6 h for leaves) the RCD1 fusion proteins accumulated in not only the nucleus but also the cytoplasm near the cell periphery (Fig. 2A and B).
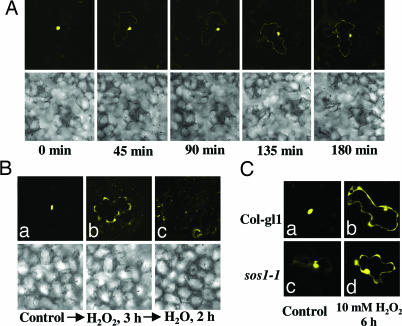
We carried out a time-course analysis of the stress-induced redistribution of RCD1 in transiently transformed Arabidopsis leaves. It was found that RCD1 localizes to the cell periphery as early as 45 min after exposure to H2O2 (Fig. 3A). We then determined what happens to the RCD1 protein localized to the cell periphery after removal of stress. Arabidopsis leaves bombarded with RCD1-YFP were exposed to 10 mM H2O2 for 3 h to allow protein accumulation near the cell periphery (Fig. 3B). Then stress was removed by placing the leaves in water for 2 h and localization of RCD1-YFP was observed. It is obvious that 2 h after stress removal, the protein was detectable only in the nucleus, and little fluorescence was observed elsewhere.
Fig. 3.
Kinetics of subcellular redistribution of RCD1-YFP. (A) Time course of stress-induced RCD1-YFP redistribution. Epidermal cells of mature Arabidopsis leaves were microprojectile-bombarded with RCD1-YFP construct, and, after 14 h of recovery, the leaves were treated with 10 mM H2O2. The epidermal cells were visualized under a confocal microscope every 45 min. (B) RCD1-YFP localization before (a) and after stress removal. After 3 h of H2O2 treatment (b), transiently transformed leaves were washed with water and floated on water for 2 h (c). Bright-field images are shown at the bottom of each panel. (C) Transient expression of RCD1-YFP in WT (a and b) and sos1–1 mutant (c and d) leaves.
We also investigated the localization of RCD1 in sos1–1 mutant cells. Transient expression of RCD1-YFP in leaves of sos1 mutants showed that RCD1 is localized in the nucleus under control conditions (Fig. 3C). When sos1 leaves bombarded with RCD1-YFP were exposed to H2O2 stress, distribution of the protein was similar to that in WT plants (Fig. 3C). These results indicate that SOS1 is not required for the subcellular redistribution of RCD1 under stress.
rcd1 and sos1 Mutants Are Hypersensitive to Salt and Hydrogen Peroxide but Tolerant to MV Stress.
We studied the extent of phenotypic similarity among sos1–1 (10), rcd1–1 (18), and rcd1–3 (isolated in this study). Previous studies in our laboratory have shown that sos1–1 mutants are hypersensitive to NaCl stress (10). Also, it is known that rcd1–1 exhibits resistance against MV but is sensitive to apoplastic ROS imposed by exposure to ozone (17). For the comparison, we tested the effects of salt and different oxidative stresses on seed germination and seedling growth of the mutants.
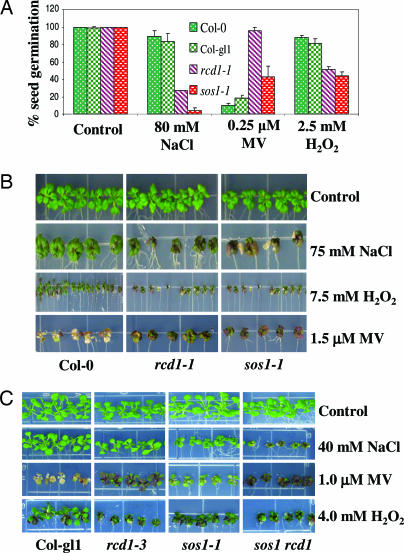
Seeds were plated on Murashige and Skoog (MS) medium supplemented with NaCl, H2O2, or MV, and seed germination was assessed as emergence of green cotyledons after 7 days of plating. As expected, sos1–1 seeds showed very strong germination inhibition (≈96%) by 80 mM NaCl (Fig. 4A). The rcd1–1 mutant seeds were also inhibited strongly (73%). In comparison, their WTs (Col-gl1 and Col-0) were inhibited only weakly (17% and 11% inhibition, respectively) by 80 mM NaCl. The response of the two mutants to oxidative stress was assessed by exposing them to MV, a herbicide known to function as a free-radical generator in chloroplasts. With 0.25 μM MV (Fig. 4A), rcd1–1 was inhibited by only 4% compared with 90% for its WT. sos1–1, on the other hand, was inhibited by 57%,compared with 82% inhibition of its WT. We tested the effect of another oxidative stress agent, H2O2. With 2.5 mM H2O2 (Fig. 4A), sos1–1 showed considerably more inhibition (56%) compared with its WT (19%). The rcd1–1 mutant (49% inhibition) was also more sensitive compared with its WT (12% inhibition). Thus, in the germination tests, the two mutants showed similar trends in their responses to salt and oxidative stresses.
Fig. 4.
Phenotypic evaluation of WT (Col-0 or Col-gl1), rcd1–1, sos1–1, rcd1–3, and sos1 rcd1 double mutant under salt and oxidative stresses. (A) Effects on seed germination in response to salt or oxidative stress. The average of three independent experiments is shown. (B and C) Effects on seedling response to stress. Photographs were taken after 15 days of stress treatments.
We also tested the growth of 5-day-old seedlings under different stress conditions. Seeds were germinated on MS agar medium for 5 days, and the seedlings were transferred to MS medium containing 75 mM NaCl, 1.5 μM MV, or 5 mM H2O2, and photographs were taken after 15 days (Fig. 4B). As expected, the sos1–1 mutant seedlings showed hypersensitivity to salt stress, because most of the seedlings died. The rcd1–1 seedlings, although not as sensitive as the sos1–1 mutant, also exhibited slower growth and increased damage compared with the WT (Fig. 4B, Col-0). H2O2 at 7.5 mM affected both mutants; the mutants were more sensitive than WT seedlings (Fig. 4B). When the seedlings were exposed to MV stress, WT seedlings showed complete bleaching, but both mutants were able to retain a trace of chlorophyll while accumulating considerable anthocyanin (Fig. 4B).
The above tests show that rcd1 and sos1 mutants exhibit similar responses to salt and oxidative stress. To test whether there is any genetic interaction between the RCD1 and SOS1 genes, we constructed a sos1 rcd1 double mutant by crossing sos1–1 with rcd1–3 and tested the sensitivity of the double mutant to salt and oxidative stresses. The rcd1–3 mutant is a T-DNA allele that exhibits responses to different stresses similarly to rcd1–1 (Fig. 4 B and C and data not shown). The single mutants were only slightly more sensitive to 40 mM NaCl than the WT (Fig. 4B, Col gl1), but the double mutant showed a strong sensitivity (Fig. 4C). Therefore, there was an additive effect of the two mutations on salt tolerance. In contrast, under oxidative stresses (H2O2 or MV), the double mutant behaved just like the rcd1–3 mutant, and no additive effect was seen (Fig. 4C).
SOS1 and RCD1 Regulate ROS Accumulation Under Salt Stress.
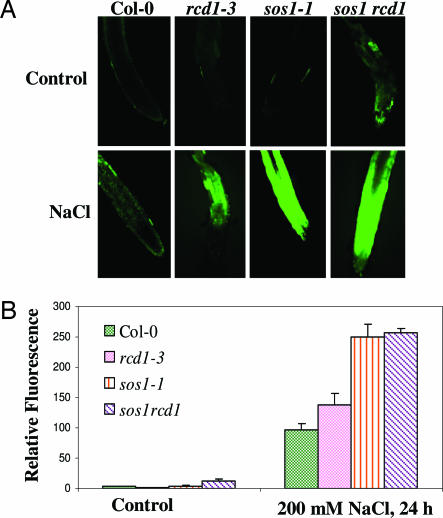
Salt stress is known to cause the accumulation of ROS. The ROS levels in sos1–1 and rcd1–1 mutants were examined after salt stress for 24 h (Fig. 5). Without NaCl treatment, there was no significant accumulation of ROS in either the WT or the single mutants. The sos1rcd1 double mutant exhibited slight accumulation of ROS under control conditions. In the roots of NaCl-treated plants, substantial levels of ROS were detected, particularly in the sos1–1 and sos1rcd1 mutants (Fig. 5A). The levels of ROS were quantified in terms of relative fluorescence, and the data show that the sos1–1 and rcd1–3 mutants accumulated higher levels of ROS compared with the WT (Fig. 5B). The double mutant appeared similar to sos1–1 in the level of ROS accumulation. The results suggest that SOS1 and RCD1 are important in controlling ROS accumulation under salt stress.
Fig. 5.
ROS accumulation in roots of rcd1–1 and sos1–1 mutant seedlings under salt stress. (A and B) Seedlings were exposed to 200 mM NaCl and then stained with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate for 15 min. Fluorescence images (A) and fluorescence intensity (B) are shown. Data plotted are the average of six roots.
The Expression Pattern of RCD1 Is Similar to That of SOS1.
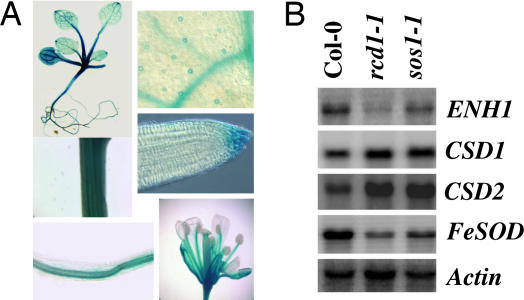
If RCD1 interacts with SOS1 and functions in salt tolerance, it should be expressed in the same tissues as SOS1, or, at least, the expression patterns of the two should overlap. It is known that SOS1 is strongly expressed in the vasculatures of roots, stems, and leaves and in the root tip, particularly at the epidermis (19). To examine the tissue-specific expression pattern of RCD1, we transformed Arabidopsis with an RCD1 promoter-GUS construct. Histochemical staining of transgenic Arabidopsis seedlings showed that RCD1 is ubiquitously expressed, and the expression is particularly strong in the vasculatures and in the root tip and epidermis (Fig. 6A). The results indicate a similar pattern of expression for the RCD1 and SOS1 genes.
Fig. 6.
RCD1 expression pattern and effect of sos1 and rcd1 mutations on gene expression. (A) Histochemical localization of GUS activity directed by RCD1 promoter in transgenic Arabidopsis. (B) Gene expression in WT and rcd1–1 and sos1–1 mutants. Actin was used as a loading control.
RCD1 and SOS1 Regulate the Expression of Genes Important for ROS Detoxification.
Superoxide dismutases (SODs) are enzymes critical for the detoxification of superoxide. The expression of genes for two Cu/Zn SODs (the cytoplasmic CSD1 and plastidic CSD2) and a FeSOD (plastidic) was examined. We found that both Cu/Zn SOD genes were expressed to higher levels in the sos1–1 and rcd1–1 mutants compared with the WT (Fig. 6B). However, the FeSOD gene showed lower levels of expression in the two mutants. We also analyzed the expression of ENH1, a gene encoding a rubredoxin-like protein. In anaerobic bacteria, rubredoxins function in ROS detoxification by serving as electron donors that divert electron flow from the electron-transport chain to superoxide-detoxification enzymes (20). Mutations in ENH1 cause ROS accumulation and enhance the salt-stress sensitivity of the sos3 mutant (J.Z., Y. D. Koo, F. E. Jenney, Jr., M. W. W. Adams, J.-K. Z., D. J. Yun, P. M. Hasegawa, and R. A. Bressan, unpublished work). Interestingly, ENH1 was expressed at lower levels in both the sos1–1 and rcd1–1 mutants (Fig. 6B). These results suggest that SOS1 and RCD1 control a common set of genes that are important for oxidative-stress responses.
Discussion
A previously uncharacterized role for the plasma membrane Na+/H+ antiporter, SOS1, is implied by this study. We found that, under stress, SOS1 interacts through its C-terminal predicted cytosolic tail with a protein known to be involved in oxidative stress tolerance, RCD1 (Fig. 1). Both SOS1 and RCD1 genes show preferential expression in vasculature and root tip. Genetic evidence confirmed the function of SOS1 in regulating oxidative-stress responses. sos1 mutant plants show excessive accumulation of ROS under salt stress. Like rcd1 mutants (17), sos1 mutant plants are more sensitive to apoplastic ROS imposed by H2O2 (Fig. 4). Interestingly, sos1 mutant plants are more tolerant to chloroplastic ROS imposed by MV (Fig. 4). This, too, is similar to rcd1 mutants (16). Like RCD1, SOS1 also controls the expression of genes important for ROS scavenging (Fig. 6). The lack of an additive effect between sos1 and rcd1 mutations on oxidative-stress tolerance indicate that SOS1 functions in the same pathway with RCD1 in oxidative-stress responses, and the role of SOS1 in oxidative stress may be mediated by RCD1.
Our protein interaction data also implied a role for RCD1 in salt-stress responses. Indeed, rcd1 mutants are more sensitive to salt stress (Fig. 4 A and B). The involvement of RCD1 in salt tolerance is possibly related to its role in oxidative stress, because oxidative stress is part of the complex effect of salinity on plants (21). Nevertheless, it is also conceivable that RCD1 may contribute to the ion-homeostasis pathway of salt tolerance, possibly by regulating the Na+/H+ antiporter activity of SOS1.
Regardless of the exact molecular mode of action, our genetic data suggest that RCD1 also functions in a salt-tolerance pathway unrelated to SOS1, because there is an additive effect between the rcd1 and sos1 mutations on salt tolerance (Fig. 4C). We suggest that RCD1 has two separate functions, both of which contribute to salt tolerance. One is a function in the nucleus. It has been shown that RCD1 interacts with transcription factors such as STO and DREB2A, which have been implicated in salt-stress tolerance (14). This nuclear function of RCD1 is presumably unrelated to SOS1. Another function of RCD1 is performed in the cytoplasm and near the cell periphery and is possibly related to its interaction with SOS1. The biochemical consequence of RCD1–SOS1 interaction is not known. Perhaps, this interaction regulates the transport of ROS and/or reductants across the cell membrane. The interaction likely also affects oxidative-stress signaling, because the expression of ROS-scavenging-related genes is controlled by RCD1 and SOS1. The altered expression of ENH1 and SOD genes in the rcd1 and sos1 mutants may explain, at least partly, the enhanced tolerance of the mutants to MV and the higher sensitivity of the mutants to H2O2.
Our proposed dual functions of RCD1 are consistent with the change in its subcellular localization pattern in response to salt or oxidative stress. RCD1 resides in the nucleus under control conditions, but a portion of it is found outside the nucleus when plants are exposed to salt or H2O2 stress (Fig. 2). This finding further strengthens the possibility that SOS1 interacts with RCD1 in planta. How this change in the cellular localization of RCD1 is brought about in response to stresses is an important question to be addressed in the future. Posttranslational disulfide-bond formation plays important roles in modulating the structure and function of proteins when cells are subject to oxidative-stress conditions (22, 23). It is possible that, during normal conditions, the RCD1 protein is in a reduced form that allows it to localize to the nucleus. However, oxidative conditions within the cell may lead to intra- and/or intermolecular disulfide-bond formation. Disulfide-bond formation may permit the RCD1 protein to be exported to the cytoplasm or prevent new RCD1 protein from entering the nucleus. It is also possible that, during stress, the phosphorylation status of RCD1 is changed, which results in the unmasking of nuclear export signal and its export to the cytoplasm. Future experiments will determine whether the nuclear import and/or export of RCD1 are affected by stress and whether this involves disulfide-bond formation.
In summary, we have discovered a previously uncharacterized function of a plasma membrane Na+/H+ antiporter in oxidative-stress responses. Our results suggest that this function is mediated by an interaction between the predicted cytoplasmic tail of the transporter and RCD1, a regulator of oxidative-stress responses. Our work also reveals a function for RCD1 in salt-stress tolerance. Furthermore, we found that salt stress causes a change in the subcellular localization of a protein. These findings open up many new questions regarding the cross-talk between ion-homeostasis and oxidative-stress pathways, and the biochemical mechanism of function of SOS1 and RCD1 in oxidative-stress tolerance.
Experimental Procedures
Plant Growth and Stress Treatments.
For seed germination assays, seeds of WT and mutant Arabidopsis were sown on MS plates containing 0.6% agar and 3% sucrose supplemented with NaCl, H2O2, or MV. The seeds were stratified at 4°C for 2 days and then transferred to 22°C under continuous light for germination and growth. Seed germination was determined as emergence of green cotyledons. To study the effect of different stresses on seedling growth and development, 5-day-old seedlings of WT and mutants were transferred onto either MS agar plates or MS agar plates containing different concentrations of NaCl, H2O2, or MV. Ten-day-old seedlings grown on MS agar medium were used for Northern blot analysis as described (10).
ROS-Detection Assay.
Seven-day-old Arabidopsis seedlings were treated with salt stress (200 mM NaCl solution prepared in half-strength MS medium containing 1% sucrose) for 24 h. Seedlings were washed and stained with 25 μM 5-(and -6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Molecular Probes, Carlsbad, CA) for 15 min, following the protocol of Shin and Schachtman (24). Roots were then cut and washed thoroughly and viewed under a Leica (Leica Microsystems, Wetzlar, Germany) SP2 laser scanning confocal microscope. All images were scanned under same conditions, such as laser power, gain, offset, pinhole, and zoom. For quantification of ROS in roots, fluorescence was measured at three different positions along the root length by using a Leica LCSlite and its mean was calculated. For each set of experiments, at least six seedlings were analyzed.
Yeast Two-Hybrid Screen and Interaction Assays.
The C-terminal cytosolic tail of SOS1 (corresponding to amino acids 440-1146) was amplified by PCR and cloned in-frame in pAS2 to obtain the bait plasmid, pAS2-SOS1CT. The screening of pACT plasmid library CD4–10 was performed as described (6). For interaction assays, bait and prey constructs were transformed into the Saccharomyces cerevisiae strain Y190. The transformants were grown overnight at 30°C in synthetic complete (SC) media lacking tryptophan, uracil, and leucine. Twenty microliters of the cell suspension containing ≈4 × 104 cells was dropped onto SC agar plates lacking tryptophan, uracil, and leucine, and the cells were grown for 2 days at 30°C. After colonies were transferred onto a nylon membrane (Hybond-N; Amersham, Buckinghamshire, U.K.), β-galactosidase (β-gal) filter assays were carried out as described (6).
in Vitro Pull-Down Assay.
In vitro pull-down assays were carried out to confirm the physical interaction of SOS1CT and RCD1. SOS1CT was subcloned in pCITE4a for in vitro transcription and translation. In vitro transcription and translation were carried out in the presence of [35S]methionine by using the Quickcouple transcription and translation kit (Promega, Madison, WI) according to the manufacturer's instructions. Full-length RCD1 was cloned in pGex4T-1. RCD1-GST protein was purified from Escherichia coli by using affinity chromatography. Pull-down assay was carried out as described (6).
In Vivo Interaction Studies.
For in vivo interaction studies, the bimolecular fluorescence complementation method (25) was used. pUCSpyce and pUCSpyne (25) were converted into Gateway-compatible vectors (named as pUCSpyceGW and pUCSpyneGW) by using the gateway vector conversion kit (Invitrogen, Carlsbad, CA). RCD1 and SOS1 were cloned in pUCSpyceGW and pUCSpyneGW, respectively, by using LR recombinase as per the manufacturer's instructions. The plasmid DNA constructs were delivered into protoplasts isolated from 12-day-old seedlings (27). Protoplasts were also transformed with pUCSpyneGW+pUCSpyceGW-RCD1 and pUCSPpyneGW-SOS1+ pUCSpyceGW as controls. Transformed protoplasts were incubated at 23°C for 14 h, subjected to stress treatments (25 mM NaCl for 2 h, followed by the addition of 1.5 mM H2O2 for 2 h), and visualized for YFP signal.
Transgenic Plants and RCD1-Localization Studies.
RCD1 was cloned into the binary vector pEGAD downstream of the cauliflower mosaic virus 35S promoter and was transformed into WT Arabidopsis plants. Roots from 5-day-old T2 transgenic seedlings were visualized under a confocal laser scanning microscope.
For transient experiments, full-length RCD1 ORF was amplified by using the following primers: forward 5′-GGAAGATCTTCCATGGAAGCCAAGATCGTCAA-3′ and reverse 5′-CTAGTCTAGACTAGTGATATTCGTCATCATCATC-3′. After digestion with BglII and XbaI, the amplified product was cloned in a transient expression vector, pBS35SYFP, under the control of cauliflower mosaic virus 35S promoter and fused in-frame to the N terminus of YFP. RCD1-YFP construct was delivered into either 20- to 25-day-old Arabidopsis leaves by microprojectile bombardment (26) or protoplasts isolated from 3-week-old Arabidopsis plants by the PEG method, essentially as described (27). To study the effect of mutation in NLSs of RCD1, site-directed mutagenesis was carried out by using the QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA).
To obtain RCD1 promoter-GUS transgenic plants, a 2.0-kb promoter fragment was amplified from Arabidopsis thaliana ecotype Col-0 genomic DNA by using the following primers: forward 5′-CCGGAATTCAGTAAACCCAATCACCAACACAG-3′ and reverse 5′-CCCAAGCTTCGTAGTCACGGCCGGTCCATC-3′. The PCR product was digested with EcoRI and HindIII and cloned into the pCAMBIA1391Z vector. T1 plants were stained in X-gluc buffer to visualize GUS expression.
Acknowledgments
This work was supported by National Institutes of Health Grant R01GM059138 (to J.-K.Z.).
Abbreviations
- CT
C-terminal fragment
- MS
Murashige and Skoog
- MV
methyl viologen
- NLS
nuclear localization signal
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- SOS
salt overly sensitive
- YFP
yellow fluorescent protein.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
Conflict of interest statement: No conflicts declared.
This article is a PNAS direct submission.
References
- 1.Zhu J-K. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu Q, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 4.Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM. Proc Natl Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Lee BH, Wu SJ, Zhu J-K. Nat Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y, Qiu Q, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK. Plant Cell. 2004;16:435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Ishitani M, Kim C-S, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Counillon L, Pouyssegur J. J Biol Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Putney LK, Denkerm SP, Barber DL. Annu Rev Pharmacol Toxicol. 2002;42:527–552. doi: 10.1146/annurev.pharmtox.42.092001.143801. [DOI] [PubMed] [Google Scholar]
- 13.Borsani O, Valpuesta V, Botella MA. Plant Physiol. 2001;126:1024–1030. doi: 10.1104/pp.126.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belles-Boix E, Babiychuk E, Van Montagu M, Inze D, Kushnir S. FEBS Lett. 2000;482:19–24. doi: 10.1016/s0014-5793(00)02016-0. [DOI] [PubMed] [Google Scholar]
- 15.Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjarvi J. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT. Plant Physiol. 2004;134:275–285. doi: 10.1104/pp.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overmyer K, Brosche M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinanen M, Saarma M, Scheel D, Kangasjarvi J. Plant Physiol. 2005;137:1092–1104. doi: 10.1104/pp.104.055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, Quintero FJ, Pardo JM, Zhu J-K. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenney FE, Jr, Verhagen MFJM, Cui X, Adams MWW. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J-K. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 22.Mou Z, Fan W, Dong X. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 23.Foyer CH, Noctor G. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin R, Schachtman DP. Proc Natl Acad Sci USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 26.Ohta M, Guo Y, Halfter U, Zhu J-K. Proc Natl Acad Sci USA. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovtun Y, Chiu WL, Tena G, Sheen J. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]