Abstract
Transcription factor DREB2A interacts with a cis-acting dehydration-responsive element (DRE) sequence and activates expression of downstream genes involved in drought- and salt-stress response in Arabidopsis thaliana. Intact DREB2A expression does not activate downstream genes under normal growth conditions. A negative regulatory domain exists in the central region of DREB2A, and deletion of this region transforms DREB2A to a constitutive active form (DREB2A CA). We carried out microarray analysis of transgenic Arabidopsis-overexpressing DREB2A CA and found that the overexpression of DREB2A CA induces not only drought- and salt-responsive genes but also heat-shock (HS)-related genes. Moreover, we found that transient induction of the DREB2A occurs rapidly by HS stress, and that the sGFP-DREB2A protein accumulates in nuclei of HS-stressed cells. DREB2A up-regulated genes were classified into three groups based on their expression patterns: genes induced by HS, genes induced by drought stress, and genes induced by both HS and drought stress. DREB2A up-regulated genes were down-regulated in DREB2A knockout mutants under stress conditions. Thermotolerance was significantly increased in plants overexpressing DREB2A CA and decreased in DREB2A knockout plants. Collectively, these results indicate that DREB2A functions in both water and HS-stress responses.
Keywords: AP2-type transcription factor, drought-stress tolerance, heat-stress tolerance, knockout mutants, microarray analysis
Environmental stresses such as heat, cold, drought, and high salinity influence plant growth and productivity. Plants respond and adapt to these stresses to survive under stress conditions at physiological and biochemical levels. These stresses have been shown to induce the expression of genes with various functions in a variety of plants (1–4). The cis- and trans-acting elements that function in stress-responsive gene expression have been precisely analyzed to elucidate the molecular mechanisms of gene expression in response to drought stress (5). The dehydration-responsive element (DRE) with the core sequence A/GCCGAC was identified as a cis-acting promoter element that regulates gene expression in response to drought, salt, and cold stresses in Arabidopsis (6). A similar motif was identified as the C-repeat and low-temperature-responsive element in cold-inducible genes (7, 8).
Arabidopsis cDNAs encoding the ethylene-responsive element-binding factor/APETALA2 (ERF/AP2)-type DRE-binding proteins (DREB) CBF1, DREB1A, and DREB2A have been isolated with a yeast one-hybrid screening (9, 10). They specifically bind to the DRE/C-repeat sequence and activate the transcription of genes driven by the DRE sequence in Arabidopsis. Three DREB1/C-repeat-binding factor (CBF) proteins are encoded by genes that lie in tandem on chromosome 4 in the order of DREB1B/CBF1, DREB1A/CBF3, and DREB1C/CBF2 (10, 11). Arabidopsis also contains two DREB2 proteins, DREB2A and DREB2B (10). Although the expression of all three DREB1/CBF genes is induced by cold stress but not by drought and salt stress, both DREB2 genes are induced by drought and salt stress (10). Both DREB1/CBF and DREB2 proteins bind to DRE, but DREB1/CBFs are thought to function in cold-responsive gene expression, whereas DREB2s are involved in drought-responsive gene expression.
Overexpression of DREB1/CBF driven by the 35S CaMV promoter causes growth retardation under normal growth conditions and increases stress tolerance of drought, high-salinity, and freezing in transgenic Arabidopsis (10, 12, 13). More than 40 downstream targets of DREB1/CBF have been identified by using microarray analysis (14–17). Many of their protein products, such as RNA-binding proteins, sugar transport proteins, desaturases, late embryogenesis-abundant (LEA) proteins, and osmoprotectant biosynthesis-proteins, are known to function against stresses (4) and are probably responsible for the stress tolerance of the transgenic plants. Transcription factors were also downstream targets, which suggests the existence of further regulation of gene expression downstream of the DRE/DREB regulon (16–18). Conserved sequences in the promoter regions of the genes directly downstream of DREB1A were analyzed, and A/GCCGACNT was found in their promoter regions between −51 and −450 as a consensus DRE (16).
DREB2 also has a conserved ERF/AP2 DNA-binding domain and recognizes the DRE sequence. A genome search of Arabidopsis revealed at least six DREB2 homologues other than DREB2A and DREB2B. Because DREB2A and DREB2B are the only genes induced strongly by drought and high salinity among the eight genes of DREB2-type proteins, they are thought to be major transcription factors functioning under drought- and salt-stress conditions (19, 20). Domain analysis of DREB2A by using Arabidopsis protoplasts revealed that a negative regulatory domain exists in the central region of DREB2A, and deletion of this region transforms DREB2A to a constitutive active form (21). Overexpression of the constitutive active form of DREB2A (35S:DREB2A CA) significantly increased tolerance to drought stress but only slightly increased tolerance to freezing. Overexpression of this gene also resulted in growth retardation in transgenic Arabidopsis plants. Microarray analyses of the 35S:DREB2A CA plants revealed that DREB2A regulates the expression of many drought-inducible genes. However, some genes regulated by DREB2A are not regulated by DREB1A, which also recognizes DRE/C-repeat but functions in cold-stress-responsive gene expression (21). Promoter analysis of the DREB1A- and DREB2A-regulated genes and gel mobility-shift assay by using both recombinant proteins indicated that DREB1A and DREB2A exhibit different DNA-binding specificities. DREB1A has the highest affinity to A/GCCGACNT, whereas DREB2A preferentially binds ACCGAC (16, 21). This difference controls the induction of different downstream genes between DREB1A and DREB2A. Although the DREB2A-regulated genes play important roles in drought-stress tolerance, they are not sufficient to withstand freezing stress.
In contrast to drought- and cold-stress responses, the acquisition of tolerance to heat-shock (HS) stress is correlated to the induction of HS proteins (HSPs). The expression of some HSP genes is reported to be induced not only by HS but also by drought stress (22). HSPs act as molecular chaperones by maintaining homeostasis of protein folding and thus help to maintain the metabolic and structural integrity of cells (22–24). HSP expression is regulated at the transcriptional level by HS transcription factors (HSFs), which recognize the conserved HS element in the promoter regions of HSP genes (24–26). Arabidopsis contains 21 different HSF genes (27), and this high multiplicity suggests the importance of backup and diversification of HSFs. However, microarray and genetic data indicate that different regulatory systems contribute to heat tolerance at different stages of the plant life cycle, and that different gene sets may control the complex multigenic process for acquiring thermotolerance (28, 29).
In this study, we carried out a microarray analysis by using 35S:DREB2A CA plants and found that many HS-inducible genes were up-regulated. We showed that the expression of DREB2A was rapidly and transiently induced by HS and dehydration treatments, and that the DREB2A protein is accumulated by HS stress and localized in nuclei. Two T-DNA-tagged dreb2a mutants were used for further functional analysis. Many up-regulated genes in the 35S:DREB2A CA plants were down-regulated in the dreb2a mutants. Here we report the HS stress tolerance of the 35S:DREB2A CA transgenic plants and HS sensitivity of the dreb2a mutants and discuss the role of DREB2A in both HS and drought stress responses.
Results
Microarray Analysis of DREB2A Up-Regulated Genes in Transgenic Arabidopsis Plants Overexpressing the Constitutive Active Form of DREB2A.
We have shown that the DREB2A protein has a negative regulatory domain in the region between amino acid residues 136 and 165. Consequently, deletion of this region transforms DREB2A to a constitutive active form (DREB2A CA) (21). Furthermore, overexpression of DREB2A CA with the CaMV 35S promoter induces expression of many stress-responsive genes (21). To understand the entire transcriptional network of DREB2A, we analyzed up-regulated genes in the transgenic plants overexpressing DREB2A CA (35S:DREB2A CA) by using an Agilent Arabidopsis 2 Oligo Microarray (Agilent Technologies, Palo Alto, CA), covering >21,000 genes. We used 3-wk-old Arabidopsis plants of two independent lines overexpressing DREB2A CA, 35 S:DREB2A CA-a, and 35S:DREB2A CA-b (21). Two experiments were performed for each RNA sample by using different labels, cy3 or cy5. In comparison to vector control plants, 483 genes with the average of expression ratios greater than two times in the 35S:DREB2A CA plants are listed in Table 1, which is published as supporting information on the PNAS web site. Among the 483 up-regulated genes, 68 and 107 genes were induced by drought and salt stress, respectively, and 120 genes were induced by drought and/or salt more than two times (K.M. and K.Y.-S., unpublished data; Fig. 5, which is published as supporting information on the PNAS web site).
We subsequently focused on 36 DREB2A up-regulated genes that exhibited remarkable expression ratios greater than eight times, as summarized in Table 2, which is published as supporting information on the PNAS web site. Among the 36 genes, 14 and 19 are induced by drought and high salinity, respectively. The DRE sequence (A/GCCGAC), which is a recognition sequence of DREB2A, is found in the 1,000-bp promoter region of 29 genes among these 36 up-regulated genes. Expression for several of the identified up-regulated genes was confirmed by using quantitative RT-PCR and shown to increase in the 35S:DREB2A CA plants. With the exception of At1g52690 (LEA protein), their expression levels directly to DREB2A expression (see Fig. 6, which is published as supporting information on the PNAS web site). Among the 36 up-regulated genes in 35S:DREB2A CA, genes for LEA proteins, dehydrins, and COR15A, which function in acquisition of stress tolerance to drought and high salinity, were identified (5). Interestingly, a gene for a HSF, AtHsfA3, was included in the up-regulated genes, and its expression ratio was highest in the 35S:DREB2A CA plants (except DREB2A). The HSF gene family consists of genes that encode transcription factors involved in HS-inducible gene expression (27). In addition, genes for choloroplast-localized small HSP (CPsHSP, At1g52560), DnaJ-like protein (At5g37440), HSP70 (At3g12580), and HSP18.2 (At5g59720), which likely function in HS stress response, were also overexpressed in the transgenic plants. These results suggest that DREB2A functions not only in drought- and salt-stress responsive gene expression but also in HS stress-responsive gene expression.
To analyze the relationship between HS stress-inducible genes and the up-regulated genes in the 35S:DREB2A CA plants, a similar microarray analysis was performed. Total RNA samples were prepared from 3-wk-old plants treated at 37°C for 0.5 or 5 h and nontreated control plants (Tables 3 and 4, which are published as supporting information on the PNAS web site). Heat-inducible genes (268 and 778) with expression ratios greater than two times were identified at 0.5 and 5 h, respectively. Among the 36 up-regulated genes in the 35S:DREB2A CA plants listed in Table 2, nine genes were shown to be induced by heat. Expression of DREB2A is also induced by HS stress. Interestingly, induction for four of these nine genes, such as At1g52560 (CPsHSP), HSP18.2, At4G36010 (thaumatin-like protein), and AtCYP18-1, was heat-specific, whereas the other five genes and DREB2A were induced by drought, salt, and heat stress (Table 2).
Expression Analysis of DREB2A and Its Up-Regulated Genes Under High-Temperature Stress Condition.
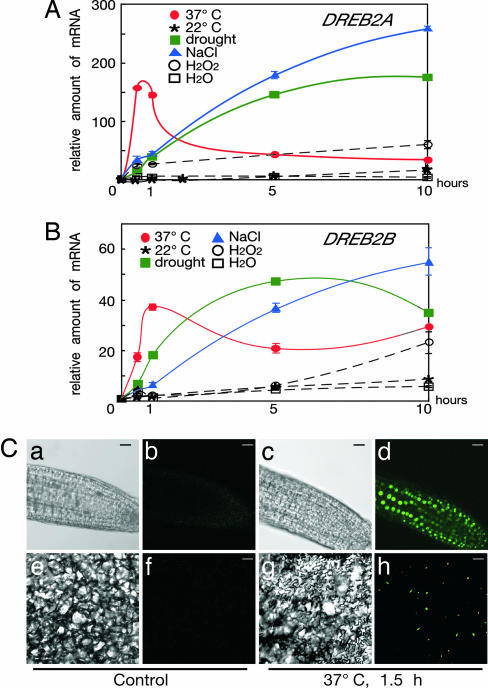
We performed quantitative RT-PCR to confirm the expression of DREB2A during heat stress (Fig. 1A). The expression of DREB2A was rapidly and transiently induced by HS treatment (37°C) and peaked ≈30 min. Then, the expression of DREB2A drastically decreased to a relatively low level that was maintained until 10 h after the treatment. In contrast, DREB2A was gradually induced by drought and salt stress during 10 h. We also analyzed the effect of H2O2-induced oxidative stress and found that, although DREB2A was also gradually induced by H2O2, the level of its induction was lower than those by drought and salt stress. We then analyzed the effects of various plant hormone treatments. DREB2A was not induced by any plant hormones such as abscisic acid, cytokinin, ethephon, auxin, methyl jasmonate, and salicylic acid (data not shown). We analyzed expression of a DREB2A homolog and found a similar rapid induction of DREB2B in response to HS stress (Fig. 1B). Similarly, the DREB2B gene was also gradually induced by drought, high salinity, and H2O2 (Fig. 1B).
Fig. 1.
Stress-inducible expression of the DREB2A gene and accumulation of the DREB2A protein. (A and B) Expression of DREB2A and DREB2B in response to HS (37°C), drought, NaCl, and H2O2. Total RNA was prepared from 3-wk-old Arabidopsis plants that had been heated at 37°C (37°C), incubated at 22°C (22°C), dehydrated (drought), transferred to hydroponic growth in 250 mM NaCl, transferred to hydroponic growth in 20 mM H2O2, and transferred to water (H2O). Treatments were carried out for 30 min, 1 h, 5 h, and 10 h. Accumulation of the DREB2A and DREB2B mRNAs was analyzed by quantitative RT-PCR. Data represent means and standard errors of three replications. The expression level under control conditions was defined as 1.0 (C) Nomarski microscope images (a, c, e, and g) and confocal microscope images of sGFP fluorescence (b, d, f, and h) of the promoter:sGFP-DREB2A plants. The root (a–d) and leaf (e–h) tissues were observed under microscope before or immediately after incubation at 37°C for 1.5 h. [Scale bars, 20 μm (a–d) and 50 μm (e–h).]
To ascertain that HS stress induces accumulation of the DREB2A protein, we generated transgenic plants harboring a sGFP-DREB2A construct driven by the DREB2A promoter (promoter:sGFP-DREB2A). The same DREB2A promoter sequence was used as reported to be inducible under dehydration and salt stresses (19). Three-week-old seedlings grown on agar plates were incubated at 37°C for 1.5 h. Roots and leaves of these plants were immediately observed with confocal microscopy. As shown in Fig. 1C, GFP fluorescence was detected in the nuclei of both root and leaf tissues, especially in root tips and cotyledons. DAPI staining was used to confirm the nuclear localization of the protein (data not shown). In contrast, under control conditions, almost no GFP fluorescence could be detected. These data indicate that the DREB2A protein is accumulated in nuclei by HS stress. Interestingly, these protein accumulation and localization patterns that were identified in leaves were not observed in guard cells, suggesting that DREB2A does not function in guard cells (data not shown).
Quantitative RT-PCR was used to confirm the drought- and HS-induced expression of some DREB2A up-regulated genes identified by the microarray (Fig. 2). Induction of AtHsfA3, AtGolS1, At4g36010 (thaumatin-like protein), and RD17 was detected under both HS- and drought-stress conditions (Fig. 2A). HSP70, AtCYP18-1, HSP18.2, and A1g52560 (CPsHSP) showed HS-inducible expression but did not clearly respond to drought stress (Fig. 2B). In contrast, expression of At1g52690 (LEA protein) and RD29A was induced by drought stress but not by HS stress (Fig. 2C). These results suggest that DREB2A is involved not only in drought- and salt-stress response but also in HS response.
Fig. 2.
Quantitative RT-PCR analysis of DREB2A up-regulated genes in response to HS or drought stress. Data represent means and standard errors of three replications. (A) Genes induced by both HS and drought. (B) Genes induced by HS. (C) Genes induced by drought (the highest expression level was set to 100).
Expression of the DREB2A Up-Regulated Genes in DREB2A Knockout Mutants.
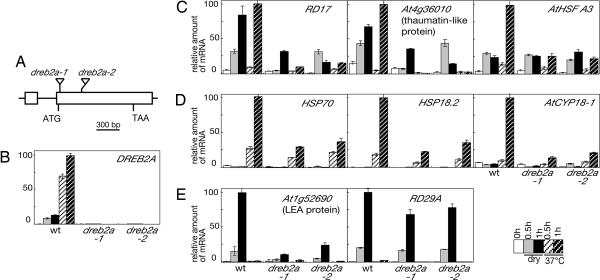
To further analyze the function of DREB2A in HS- and drought-stress responses, we analyzed two knockout mutants, dreb2a-1 and dreb2a-2. T-DNAs were inserted 468 and 835 bp downstream of the translational start site of the DREB2A gene in dreb2a-1 and dreb2a-2, respectively (Fig. 3A). These insertions were confirmed by the sequences of their respective PCR products. DREB2A expression was repressed in both dreb2a-1 and dreb2a-2 homozygous lines, as shown in the quantitative RT-PCR analysis (Fig. 3B). Quantitative expression of DREB2A up-regulated genes was also analyzed both in the DREB2A mutants and wild-type plants (Columbia ecotype) with and without drought or HS-stress treatments for 0.5 and 1 h. Fig. 3C shows the expression of RD17, At4g36010 (thaumatin-like protein), and AtHsfA3 that are induced by both HS and drought stress. In contrast to wild-type plants, HS-inducible expression of these genes was decreased remarkably in both dreb2a-1 and dreb2a-2. Drought-inducible expression of the genes was also decreased, but their low-level expression was still detected even under stress conditions. Expression of HSP70, HSP18.2, and AtCYP18-1 that are induced by HS stress but not drought stress was also decreased in dreb2a-1 and dreb2a-2 (Fig. 3D). Although a significant decrease of drought-stress-inducible expression of At1g52690 (LEA protein) was observed in knockout mutants, the response of RD29A was unclear (Fig. 3E).
Fig. 3.
Analysis of DREB2A knockout mutants. (A) A scheme of the Arabidopsis DREB2A gene. Exons (open boxes) and an intron (line) are indicated. The positions of the T-DNA insertions are shown (not to scale). (B–E) Expression of DREB2A (B), DREB2A downstream genes induced by both HS and drought (C), DREB2A downstream genes induced by HS (D), and DREB2A downstream genes induced by drought (E) was analyzed in wild-type and dreb2a plants by quantitative RT-PCR (the highest expression level was set to 100). Data represent means and standard errors of three replications.
Then, we compared the expression profiles in the two DREB2A mutants of 3-wk-old plants under drought- or HS-stress conditions with that in the wild-type plants by using the Agilent Arabidopsis 2 Oligo Microarray. Genes with expression ratios <0.5 on the average of two mutant lines are listed in Tables 5 and 6, which are published as supporting information on the PNAS web site. Eleven and 50 genes showed decreased expression in the DREB2A mutants under drought- and HS-stress conditions, respectively. Among these down-regulated genes, 31 were overexpressed more than two times in the 35S:DREB2A CA plants (Table 7, which is published as supporting information on the PNAS web site). We searched for conserved sequences in the 31 promoter regions of the 36 DREB2A up-regulated genes by using the MEME program (30). The 1,000-bp promoter regions upstream of putative transcriptional initiation sites that are the 5′ end of the full-length cDNA clones were used for this search (31), and we found a highly conserved sequence (A/GCCGAC) that included the DRE-core motif. We previously reported that DREB2A has a higher affinity to ACCGAC than GCCGAC (21). Therefore, the frequency of ACCGAC and GCCGAC in the promoter region of the 35 genes was verified. Twenty-eight genes had DRE, A/GCCGAC, sequence(s) within 1,000-bp promoter regions. Among the 28 genes, 24 (86%) have at least one ACCGAC sequence in their promoter regions.
HS-Stress Tolerance of the 35S:DREB2A Plants and the DREB2A Knockout Mutants.
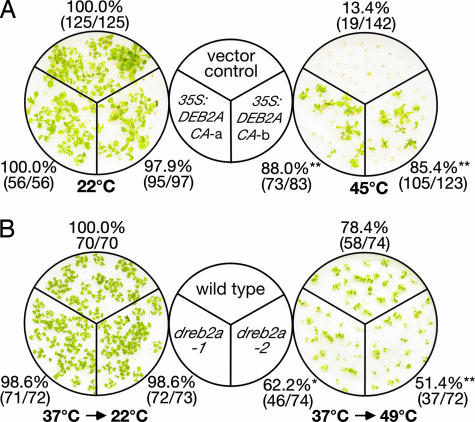
Expression of DREB2A was induced by HS transiently and significantly. Moreover, microarray analysis of the 35S:DREB2A CA plants showed that DREB2A up-regulates the expression of many HS-inducible genes in the plants. We were interested to determine whether the 35S:DREB2A plants had an improved tolerance to heat stress. Seven-day-old vector control and transgenic plants, germinating on a filter paper premoistened by liquid germination medium, were grown at 22°C and then subjected to HS treatment at 45°C. As shown in Fig. 4A, two transgenic lines of the 35S:DREB2A CA-a, 35S:DREB2A CA-b, and vector control plants grew well under 22°C. However, when the plants were exposed at 45°C for 1 h, only 13.4% of the vector control plants survived, whereas 88.0% of 35S:DREB2A CA-a and 85.4% of 35S:DREB2A CA-b survived during the subsequent 7-day recovery at 22°C. These results clearly indicate augmented thermotolerance of the 35S:DREB2A CA plants.
Fig. 4.
HS-stress tolerance of the 35S:DREB2A CA and dreb2a plants. (A) One-week-old seedlings of the vector control or the 35S:DREB2A CA plants were treated at 22°C or 45°C for 1 h. (B) One-week-old seedlings of wild-type or dreb2a plants were preincubated at 37°C for 1 h and then treated at 22°C or 49°C for 1 h. After treatment, plants were grown under normal conditions for 1 wk. Percentages of surviving plants and numbers of surviving plants per total numbers of tested plants are indicated around the photographs. More than 15 plants were used per test, and each test was repeated three times. ∗, These plants had significantly lower survival rates than the wild-type plants (χ2 test, P < 0.05). ∗∗, These plants had significantly higher or lower survival rates than the vector control or the wild-type plants (χ2 test, P < 0.01).
We then compared the thermotolerance of the DREB2A knockout mutants to that of wild-type plants. Seven-day-old plants grown on a filter paper were subjected to 37°C for 1 h, then followed by 49°C for 1 h. Wild-type plants (78.4%) survived during the subsequent 7-day recovery at 22°C, whereas 62.2% and 51.4% of dreb2a-1 and dreb2a-2 survived, respectively (Fig. 4B). Thus, the phenotype of these mutants in survival under HS stress contrasted with those of the transgenic plants overexpressing DREB2A CA, indicating that DREB2A functions in improving HS-stress tolerance.
Discussion
The DRE contains the core sequence A/GCCGAC and has been identified as a cis-acting promoter element in regulating gene expression in response to drought, salt, and cold stresses in Arabidopsis (6). DREB2A was subsequently isolated as a cDNA encoding a DREB protein by using a yeast one-hybrid screening (10). Therefore, the function of DREB2A under these stresses has been well studied (10, 21), but the effect of HS stress has not been previously studied. However, in our current study, we clearly identified the up-regulation of many HS-related genes such as AtHsfA3 and HSPs, as a result of overexpressing DREB2A CA (Table 2 and Fig. 6). We subsequently confirmed that these genes were induced by HS stress (Table 2 and Fig. 2). Moreover, we found that the DREB2A gene is also strongly induced by HS stress (Fig. 1A), and that the sGFP-DREB2A protein is stabilized in the nuclei in plant cells by HS stress (Fig. 1C). We hypothesized that DREB2A is important for the regulation of gene expression in response to heat stress as well as drought and salt stress. We later confirmed this hypothesis by using the dreb2a mutants and the 35S:DREB2A CA plants. Elimination of the DREB2A gene decreased HS- and drought-stress-inducible expression of DREB2A up-regulated genes (Fig. 3). Finally, we found that the 35S:DREB2A CA plants showed enhanced thermotolerance, and that the dreb2a mutants displayed reduced survival under HS condition (Fig. 4).
Four of five HS-related genes, AtHsfA3 and three HSPs, that are up-regulated in the 35S:DREB2A CA plants with remarkable expression ratios greater than eight times, showed strong inducible expression by HS stress (Table 2). AtHsfA3 is a member of the HSF gene family, which consists of 21 genes belonging to three major classes that are based on structural differences, HsfA, HsfB, and HsfC. HsfAs, have been shown to be the major transcription factors that function in heat-induced expression of HSP genes (24). AtHsfA3 might act as a transcription factor in HS-stress-responsive gene expression, but detailed function of AtHsfA3 has not yet been identified. HSP70 (At3g12580) encodes one of the HSP70 chaperones that have an essential function in preventing aggregation and in assisting refolding of nonnative proteins under stress conditions. They are also involved in the degradation of unstable proteins by targeting the proteins to lysosomes or proteasomes (22). Overexpression of HSP70 genes in transgenic plants has been shown to correlate positively with the acquisition of thermotolerance and results in enhanced tolerance to drought, salt, and heat stress (22). Two genes, At1g52560 (CPsHSP) and HSP18.2, encode small HS proteins (sHSPs). Although the sHSPs alone are not able to refold nonnative proteins, they probably bind to unfolded proteins and facilitate further refolding by HSP complexes (22). These HSPs probably contribute to the acquisition of stress tolerance not only to heat stress but also to drought and salt stress in the 35S:DREB2A CA plants. Five more genes such as RD17 encoding a group two LEA protein (dehydrin), AtGolS1 encoding a galactinol synthase (32), MT2A encoding a putative detoxification-related gene (33), AT4g36010 (thaumatin-like protein) encoding a PR-5 like protein related to biotic stress, and AtCYP18-1 encoding an unknown protein, were shown to be HS-inducible. These genes may have important roles to increase HS stress tolerance in the transgenic plants. On the other hand, thermotolerance of the dreb2a mutants was not dramatically reduced (Fig. 4). Our microarray analysis revealed that 956 genes showed HS-responsive expression, and 110 of them are up-regulated by DREB2A (Tables 3 and 4). Functionally redundant proteins are encoded by DREB2A-regulated and -unregulated HS-responsive genes. It appears that the latter genes are capable of complementing the DREB2A mutation.
Our microarray analysis of 35S:DREB2A CA plants confirmed that many drought-stress- or HS-inducible genes are potentially downstream of DREB2A (Table 2 and Fig. 2). However, an analysis with a gain-of-function method with constitutive and ectopic gene expression sometimes creates false responses. Therefore, we confirmed the expression of these DREB2A up-regulated genes in the dreb2a mutants by using quantitative RT-PCR and microarray analyses. Six DREB2A up-regulated genes showed clear down-regulation of stress-inducible expression in both dreb2a-1 and -2 plants (Fig. 3 C and D). Similar to DREB2A, most of the genes such as RD17, HSP70, AtCYP18-1, and HSP18.2 showed rapid and transient expression in response to HS stress, but their expression was slightly delayed. Although their expression peaked 1 h after stress treatment (Fig. 2 A and B), HS-inducible DREB2A expression peaked between 30 min and 1 h (Fig. 1A). This time lag supports the hypothesis that these genes are downstream targets of DREB2A. Drought-stress-inducible expression of At1g52690 (LEA protein) also decreased in the dreb2a mutants (Fig. 3E). In addition, stress-inducible expression of At1g01470 (LEA14), COR15A, At3g17520, XERO2, and RD29A was reduced in the microarray analysis of the dreb2a mutants (Table 7). All of these genes have DRE in their promoter regions, suggesting they are direct downstream genes of DREB2A.
Among 483 DREB2A up-regulated genes with the expression ratios greater than two times, 68 and 107 genes showed drought- and salt-responsive gene expression, respectively (Fig. 5). Fifty-five (80%) of the 68 drought-stress-inducible genes are also induced by salt stress. This extensive overlap suggests that a common regulatory mechanism exists between drought- and salt-induced gene expression of the DREB2A up-regulated genes. One hundred ten genes showed HS-responsive expression, and only 38 of them showed drought- and/or salt stress-responsive expression (Fig. 5). HS- and drought-specific expression of the DREB2A up-regulated genes was also detected by quantitative RT-PCR. Genes shown in Fig. 2B responded to HS but not to drought stress. In contrast, genes shown in Fig. 2C responded to drought but not to HS stress. These results suggest that distinct regulatory mechanisms exist between HS- and drought-/salt-induced gene expression of the DREB2A up-regulated genes. Recently, Diaz-Martin et al. (34) reported that a sunflower DREB2-type protein, HaDREB2, interacts with a heat-stress factor A9 (HaHSFA9), and these proteins synergistically induce expression of HaHSP17.6. Interaction of HS or drought-/salt-stress specific cofactors such as HaAHSFA9 may be necessary to activate DREB2A. Otherwise, HS- or drought-/salt stress-specific repressors may inhibit the gene expression activated by DREB2A.
When sGFP was fused to full-length (FL) DREB2A (sGFP-DREB2A) and expressed in Arabidopsis, only weak green fluorescence was observed in the nucleus under unstressed control conditions. In contrast, strong sGFP signals were observed in the nucleus of the transgenic plants expressing chimeric genes with sGFP fused to DREB2A CA. DREB2A CA lacks the negative regulatory domain that contains a PEST sequence (21). The FL DREB2A protein containing the PEST sequence may be degraded rapidly by the ubiquitin–proteasome system (21). However, strong GFP fluorescence was detected in the nuclei of transgenic plants expressing sGFP-DREB2A under HS-stress conditions (Fig. 1C). These data indicate that the DREB2A protein is stabilized by HS stress and nuclear-localized during stress conditions. The PEST sequence of DREB2A (RSDASEVTSTSSQSEVCTVETPGCV) contains many phosphorylation target sites of protein kinases such as PKC and CKII. Because the phosphorylation of the PEST sequence has been reported to be important for protein degradation (35), it is plausible that the modification of the DREB2A protein by these kinases might be important for its stabilization under stress conditions.
In summary, DREB2A is shown to play an important role in crosstalk between drought- and heat-stress responses. DREB2A is HS-stress-inducible and contains an HS element in its promoter. Moreover, DREB2A controls HSF as one of the downstream genes. This complex cascade of transcription may enable plants to properly respond to both heat and drought stress.
Materials and Methods
Plant Materials.
Arabidopsis plants were grown, transformed, and treated as described (21). The dreb2a-1 (line 379F02) and dreb2a-2 (line 179C04) mutants were obtained from the GABI-Kat (36). For HS-stress treatment, Arabidopsis seedlings were grown on agar plates at 22°C for 3 wk and then transferred to 37°C.
Quantitative RT-PCR and Microarray Analysis.
Total RNA was extracted with TriZol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and quantitative RT-PCR was performed as described (21). The primer pairs used in the real-time PCR are shown in Table 8, which is published as supporting information on the PNAS web site. Microarray analysis and data mining were carried out as described (37).
Observation of Subcellular Localization of Green Fluorescent Signals in Transgenic Plants.
The DNA sequence of sGFP fused with the DREB2A coding sequence and Nos terminator (NosT) of 35S:GFP-DREB2A plasmid (21) was amplified by PCR and cloned into the pGreenII0029 vector (38) by PstI and SalI sites. The 1.8 kbp of the DREB2A promoter sequence (19) was subsequently inserted in front of sGFP-DREB2A-NosT by the PstI site. Plant transformation was carried out as described (10). GFP fluorescence was analyzed by a confocal laser-scanning microscope LSM5 PASCAL (Zeiss, Oberkochen, Germany).
HS Tolerance Test.
Arabidopsis seeds were germinated and grown on two layers of filter paper containing 4 ml of germination medium (6) in 90-mm plastic Petri dishes. Petri dishes were sealed with Micropore surgical tape (3M Health Care, St. Paul, MN) and incubated at 22°C. Twenty-five to 30 seeds were sown for each experiment, and seedlings that germinated within 7 days after sowing were subjected to the HS-tolerance test. Throughout the incubation period, 2 ml of sterilized water was added to Petri dishes every 3 days. Seven-day-old seedlings were subjected to HS treatment on a paper box (width 13 mm × depth 130 mm × height 50 mm) that was placed in a hybridization oven (HB-80, TAITEC, Saitama, Japan). After HS treatment, plants were grown under normal conditions for 1 wk, and the number of surviving plants was counted.
Supplementary Material
Acknowledgments
We thank Dr. Y. Niwa (Shizuoka Prefectural University, Shizuoka, Japan) for providing the sGFP gene. We are grateful for the excellent technical support provided by Kyoko Yoshiwara, Hiroko Sado, Ekuko Ohgawara, and Kaori Amano, and for skillful editorial assistance by Masami Toyoshima of the Japan International Research Center for Agricultural Sciences. We also thank the National Institute of Agrobiological Sciences for supporting the 22k microarray analysis. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences, Core Research for Evolutional Science and Technology, and by a project grant from the Ministry of Agriculture, Forestry, and Fisheries, Japan.
Abbreviations
- DRE
dehydration-responsive element
- HS
heat shock
- HSP
HS protein
- HSF
HS transcription factor
- CBF
C-repeat-binding factor.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The complete set of microarray data has been deposited in the European Bioinformatics Institute ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession nos. E-MEXP-601, MEXP-602, and MEXP-603).
References
- 1.Thomashow MF. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinozaki K, Yamaguchi-Shinozaki K. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 4.Bartels D, Sunkar R. Crit Rev Plant Sci. 2005;24:23–58. [Google Scholar]
- 5.Yamaguchi-Shinozaki K, Shinozaki K. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker SS, Wilhelm KS, Thomashow MF. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 8.Jiang C, Iu B, Singh J. Plant Mol Biol. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- 9.Stockinger EJ, Gilmour SJ, Thomashow MF. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 12.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 13.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 14.Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowler S, Thomashow MF. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 17.Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 18.Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/a:1006321900483. [DOI] [PubMed] [Google Scholar]
- 20.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 21.Sakuma Y, Maruyama K, Osakabe K, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Vinocur B, Shoseyov O, Altman A. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Vierling E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- 24.Sung DY, Guy CL. Plant Physiol. 2003;132:979–987. doi: 10.1104/pp.102.019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 26.Miller G, Mittler R. Ann Bot. 2006;98:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nover N, Bharti N, Doring P, Mishra SK, Ganguli A, Scharf KD. Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkindale J, Hall JD, Knight MR, Vierling E. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busch W, Wunderlich M, Schoffl F. Plant J. 2006;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- 30.Bailey TL, Elkan C. Proc Int Conf Intell Syst Mol Biol; 1995. pp. 21–29. [PubMed] [Google Scholar]
- 31.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 32.Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Goldsbrough P. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz-Martin J, Alguera C, Prieto-Dapena P, Espinosa JM, Jordano J. Plant Physiol. 2005;139:1483–1494. doi: 10.1104/pp.105.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmeron J, Janzen J, Soneji Y, Bump N, Kamens J, Allen H, Ley SC. J Biol Chem. 2001;276:22215–22222. doi: 10.1074/jbc.M101754200. [DOI] [PubMed] [Google Scholar]
- 36.Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 37.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.