Abstract
A major class of disease-resistance (R) genes in plants encode nucleotide-binding site/leucine-rich repeat (LRR) proteins. The LRR domains mediate recognition of pathogen-derived elicitors. Here we describe a random in vitro mutation analysis illustrating how mutations in an R protein (Rx) LRR domain generate disease-resistance specificity. The original Rx protein confers resistance only against a subset of potato virus X (PVX) strains, whereas selected mutants were effective against an additional strain of PVX and against the distantly related poplar mosaic virus. These effects of LRR mutations indicate that in vitro evolution of R genes could be exploited for enhancement of disease resistance in crop plants. Our results also illustrate how short-term evolution of disease resistance in wild populations might be toward broader spectrum resistance against multiple strains of the pathogen. The breadth of the disease-resistance phenotype from a natural R gene may be influenced by the tradeoff between the costs and benefits of broad-spectrum disease resistance.
Keywords: host parasite, transgenic plants, gene-for-gene model, potato virus X
Virus-resistant crop plants are conventionally based on genetically dominant disease-resistance (R) genes. These genes are introgressed into susceptible genotypes to generate new varieties that protect from losses associated with virus diseases in both the developed and the developing world (1). However, this breeding approach has the disadvantage that many backcross generations are required to eliminate unwanted characteristics of the virus-resistant parent in the breeding program. The selection process over several years is expensive and often too slow to be useful against newly emerging virus diseases, with trees or with some plants that are propagated vegetatively.
Alternative transgenic approaches have also been developed based on expression of antiviral proteins (2) or antibodies (3). Parasite-derived resistance has also been demonstrated in transgenic plants expressing viral proteins or RNA molecules (4), and, in some instances, the plants have been introduced as crops. One of the best examples, with papaya, resulted in revival of this crop in areas where it had been previously eliminated because of the effects of papaya ringspot virus (5). There are also other examples with viruses of potato, squash, tomato, and tobacco in which parasite-derived virus resistance has been tested in the field (6–9). However, concern about unwanted side effects of viral transgenes has inhibited the more general introduction of plants with virus-derived resistance. The contentious issues include transcapsidation of viral RNAs in coat proteins (CPs) encoded by the transgene, recombination between the transgene RNA and other viruses, enhancement of other viruses due to the effects of the transgene viral RNA or encoded protein, or toxicity of viral proteins encoded by the transgene (10).
A third approach to virus resistance involves transgenic transfer of R genes between species (11–13) and has the benefit of speed associated with the use of transgenes but without the concerns associated with parasite-derived resistance: the transgene is a plant gene moved into a related plant. It does have a limitation, however, in that R genes for many of the most-damaging virus diseases have not been characterized at the molecular level. In addition, there may be viral strains that overcome the R gene-mediated resistance. Consequently, as with R genes transferred by conventional means, there is the possibility that there would be selection of resistance-breaking strains, and the new disease-resistant cultivar would have only a short useful lifespan.
To overcome these limitations, we have explored the possibility of modifying an R gene so that it confers resistance against novel strains or types of virus. The target of our approach involved an R gene that encodes a nucleotide-binding (NB) site/leucine-rich repeat (LRR) protein. Similar proteins encoded by a major class of disease-resistance genes in plants confer resistance against bacteria, fungi, and invertebrate pests so that our findings could have more general relevance beyond viral disease resistance (14).
NB-LRR proteins mediate disease resistance through a multistage process initiated by molecular recognition of a pathogen-derived elicitor of disease resistance (14). This recognition mechanism may involve direct contact between the R protein and the elicitor or, in some instances, an indirect interaction involving other host proteins (15, 16). There is then an activation process involving conformational changes in the R protein (17) and ATP hydrolysis (18), leading to transduction of an uncharacterized signal and molecular changes in the cell so that the pathogen cannot thrive. In some instances, there may also be host cell death (19, 20).
Our approach to R gene modification was to target the molecular recognition step of this process. We predicted that a mutational change to the molecular structure of the R protein could alter the elicitor recognition specificity. Although most of the mutant proteins would lose elicitor recognition, we predicted that in some of them, a new structure would alter disease-resistance specificity. We targeted the LRR domain of an R protein for our mutation analysis, because several lines of evidence implicate this domain in the elicitor recognition function (21–24).
To explore the possibility that R gene modification could generate new disease-resistance specificity, we chose to investigate the NB-LRR protein Rx (w12). Rx confers resistance against strains of potato virus X [(PVX), Potexvirus group], with threonine and lysine at positions 121 and 127 of the elicitor CP (CP-TK) but not those with lysine and arginine (CP-KR; ref. 25). We mutated the LRR-encoding region of the Rx gene by error-prone PCR and used a transient expression assay to identify mutants with enhanced recognition of CP-KR. Transgenic expression of these mutant Rx proteins confirmed they confer resistance against a strain of PVX with CP-KR and also against a distantly related virus, poplar mosaic virus [Carlavirus group (PoMV)]. The novel property of these mutant proteins was a broader, rather than altered, spectrum of disease-resistance specificity, because they retained the ability to protect against the strain of PVX that is contained by the original Rxallele. Our findings indicate that R gene mutation may be a useful general approach to engineering of novel disease resistance in plants. The artificial evolution of Rxmay also be informative about R gene mutations that would be selected during natural evolution of R genes.
Results
Random Mutagenesis of the Rx LRR.
As an approach to analysis of Rx recognition specificity, we mutated the LRR by error-prone PCR (ref. ; Fig. 1A) under conditions in which there was an average of 1.8 mutations in each cloned mutant. The mutants were transiently coexpressed in Nicotiana tabacum leaves with a variant of the PVX CP with threonine and lysine residues at positions 121 and 127 (CP-TK). This is the most common variant of the CP, and it is a strong elicitor of Rx-mediated resistance (12). With wild-type Rx and the CP-TK, there was a strong cell death response that is indicative of disease resistance (12), whereas with the CP-KR of a resistance-breaking strain (12, 25, 27), the Rx-mediated cell death response was weak or absent (Fig. 1B). We predicted that, if the LRR plays a role in elicitor recognition, it would be possible to generate Rx LRR mutants exhibiting a cell death response to CP-KR.
Fig. 1.
Mutation of Rx and a cell death assay of Rx responses. (A) Procedure for generating an library of Rx LRR mutants in which error-prone PCR was used to introduce mutations into the Rx-HA LRR sequence between the ApaL1 and SacI sites of pB1:Rx-HA. The positions of these sites are shown relative to the coiled-coil (CC), NBS, and LRR domains in Rx. (B) Cell death assay for Rx responses. Agrobacterium cultures containing CP-TK or CP-KR were mixed with cultures containing mutant versions of Rx constructs and infiltrated into the leaves of 21-day-old N. tabacum plants. A necrotic response developed after 24 h with CP-TK coexpressed with wild-type Rx (◀) but not with CP-KR that was expressed either with wild-type Rx (○) or with many of the mutants tested in our screen (●). In some experiments, there was a weak response of wild-type Rx to CP-KR. The picture was taken after 6 d. (C) A diagrammatic representation of the Rx LRR between residues 473 and 949. The individual repeat units are numbered 1–15, and those with a mutation in M1–M4 are indicated with a red asterisk. The amino acid changes in M1–M4 are indicated above the diagram.
Of the 1,920 Rx mutants tested, there were 1,447 with a CP-TK response, 453 with a loss-of-function phenotype, and 20 exhibiting a gain of function. Seven of these gain-of-function mutants exhibited a cell death response in the absence of CP. It is likely that these autoactive mutants are modified in a recognition-independent function of the Rx LRR associated with activation of the disease-resistance mechanisms (17). The remaining 13 gain-of-function mutants exhibited a cell death response to CP-KR, and, as discussed below, they could be modified in either the elicitor recognition or the activation functions.
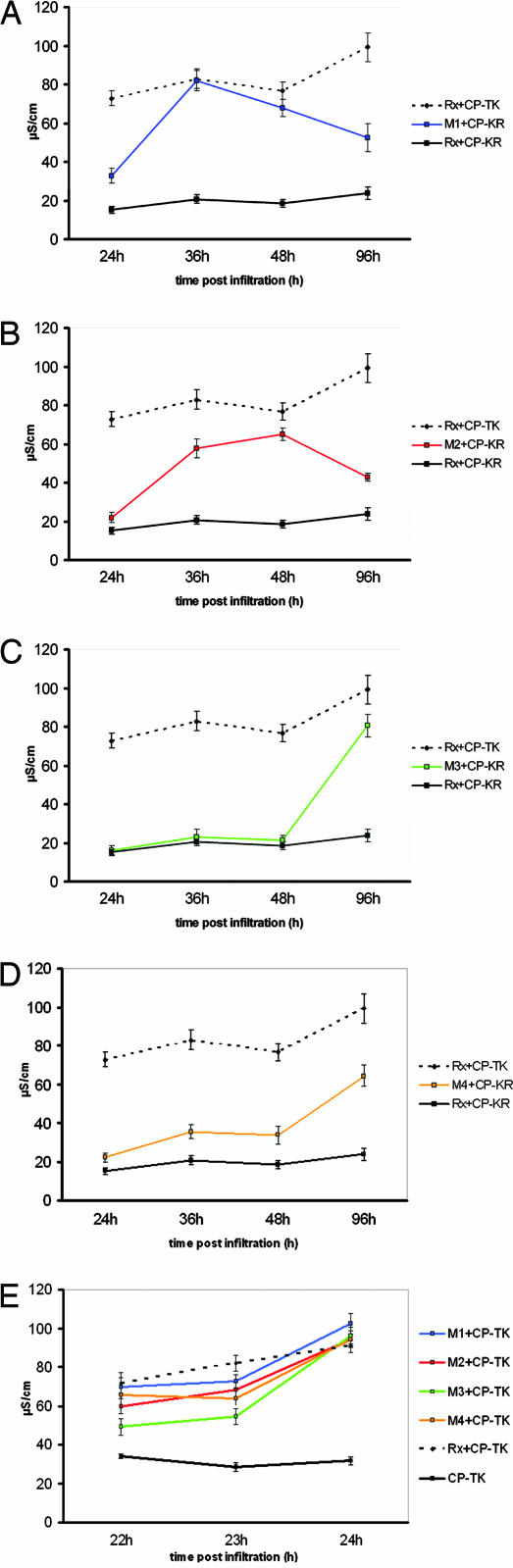
These 13 mutants exhibited varying degrees of enhanced response to CP-KR, and 4 of them (M1–M4) were selected for more detailed analysis, because their LRR domains contained only a single amino acid mutation (Fig. 1C). Fig. 2A–D illustrates the CP-KR response of M1–M4 using an electrolyte leakage assay (28) that provides a more quantitative response readout of cell death than the necrosis used in the initial screen. This assay confirmed that the mutants exhibited an enhanced CP-KR response, although with M2, M3, and M4, it was slower than with CP-TK, and with M1 and M2, it was transient. The four mutants accumulated at the same level as wild-type Rx (Fig. 5, which is published as supporting information on the PNAS web site) and retained full responsiveness to CP-TK (Fig. 2E).
Fig. 2.
Resistance responses of Rx mutants. Leaves of 21-day-old N. tabacum plants were infiltrated with Agrobacterium cultures containing constructs for expression of Rx and either CP-KR or CP-TK. Conductivity (μS·cm−1) measurements were derived from samples processed at the indicated time (h) postinfiltration. Higher conductivity levels are indicative of increased membrane leakage and imminent cell death. For each data point, a minimum of five independent replicates were sampled. Results are shown as the mean ± SEM. A–D show the response of the original Rx with CP-TK and CP-KR for reference. E does not show these reference data. The late decline in conductivity shown for the CP-KR response in A and B characterizes the post-cell-death response.
Gain of Rx Function Is Associated with Changes to the Recognition and Activation Steps of Disease Resistance.
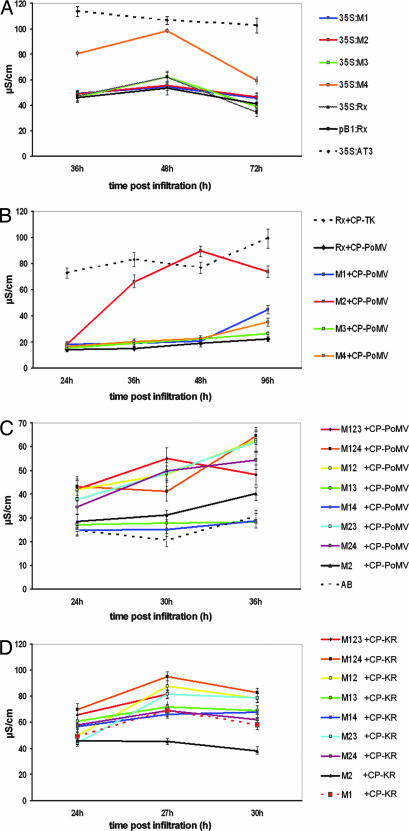
NB-LRR proteins mediate disease resistance through processes involving elicitor recognition and R protein activation. To investigate which of these stages is affected by the M1–M4 mutants, we exploited the weak HR response that is produced when Rx is overexpressed from the strong cauliflower mosaic virus (CaMV) 35S promoter in N. tabacum in the absence of elicitor (29). We reasoned that this response would depend on activation of Rx independent of elicitor recognition. If the mutations had affected the activation function of the LRR, they would enhance this response, but it would be unchanged if the mutations had affected elicitor recognition. Fig. 3A shows that, with M1, M2, and M3, the overexpression response in the electrolyte leakage assay was similar to that of the wild-type Rx(35S:Rx), but that it was enhanced with M4 to be similar to an autoactive mutant AT3. It is likely, therefore, that M1–M3 mutants are modified in elicitor recognition, and that M4 is altered for activation.
Fig. 3.
Activation and recognition phenotypes of Rx mutants. Leaves of 21-day-old N. tabacum plants were infiltrated with Agrobacterium cultures containing constructs for expression of 35S:Rx mutants (A), Rx mutants with CP-PoMV (B and C), and Rx mutants together with CP-KR (D). Conductivity (μS·cm−1) measurements were derived from samples processed at the indicated time (h) postinfiltration. In C and D, the mutants had one or more of the M1–M4 mutations. Thus, M123 carried the M1, M2, and M3 mutations. In A, the 35S:AT3 construct was a constitutive gain of function mutant (D543E) described (29) that is shown for reference. In C, AB is Agrobacterium infiltration buffer only. For each data point, a minimum of five independent replicates were sampled. Results are shown as the mean ± SEM.
Evidence that M1 and M2 are modified in elicitor recognition is reinforced by their response to the CP from PoMV (PoMV-ab strain; Fig. 3B). CP-PoMV and CP-TK are 60% identical in the region between 107 and 190 of CP-TK (Fig. 6, which is published as supporting information on the PNAS web site; ref. 30). Within this region of CP-TK, there are two residues, T residue at position 121 and K at position 127, that are both required for Rx recognition (27). CP-PoMV and CP-TK are identical at only one of these residues (T121; Fig. 6), and CP-PoMV did not elicit a response with wild-type Rx (Fig. 3B). However, CP-PoMV elicited a weak necrosis with M1 after 96 h (data not shown) and, when expressed with M2, it induced a strong electrolyte leakage response (Fig. 3B). Combinations of M2 with one or two of M1, M3, or M4 produced stronger CP-PoMV responses than M2 alone (Fig. 3C). Thus, the M2 mutation has modified Rx so that it recognizes CP-PoMV as well as the PVX CP elicitor. The double and triple mutants, including those without M2, also exhibited a stronger CP-KR response than the M1 or M2 single mutants (Fig. 3D).
Modified Disease Resistance by Rx Mutants.
Cell death in plant disease resistance is separate from the mechanisms causing suppression of the pathogen (12, 19, 20, 31). We therefore complemented the cell death assays described above (Figs. 1–3) with an assessment of the virus-resistance phenotype of single and multiple mutant forms of Rx.The mutant genes were transformed into Nicotiana benthamiana, and the plants were inoculated with either PoMV or the natural Rx resistance-breaking strain PVX-HB, in which the CP is similar to CP-KR (27). In the first generation (T1), there was variation in the resistance responses between lines with the same construct and, in some instances, between progeny of the same line because of segregation of the transgene (Figs. 7 and 8, which are published as supporting information on the PNAS web site). However, in homozygous T2 plants, the resistance phenotypes were uniform between different plants of a line (Fig. 4). In the most resistant lines, the absence of viral symptoms (Fig. 4 A and C) was associated with the corresponding absence of viral RNA accumulation (Fig. 4 B and D). These transgenic T2 lines also remained fully resistant against PVX-TK (data not shown). We can rule out that there was nonspecific virus resistance, because the M1–M4 lines, like the wild-type Rxgenotypes, were fully susceptible to cucumber mosaic cucumovirus (ref. 32; Fig. 9, which is published as supporting information on the PNAS web site). Strikingly, the PoMV symptoms were more severe on the M1 plants than on the wild-type Rx or the nontransgenic genotypes. We attribute these symptoms to the M1 CP-PoMV response (Fig. 3B) being either too weak or too slow to restrict spread of the virus from the site of initial infection.
Fig. 4.
Virus-resistance phenotypes of Rx mutants. N. benthamiana plants homozygous for wild-type Rx, M1, M2, or M123 transgenes were inoculated with PVX-HB (A and B) or PoMV (C and D). A and C show symptoms at 20 d postinoculation (dpi), and B and D show Northern analysis of the respective viral RNAs at 10 dpi. Viral RNA accumulation was assessed by RNA gel-blot analysis of 5 μg of total RNA extracted from the appropriate leaves by using probes derived from full-length CP-KR or CP-PoMV PCR products, respectively. The specific lines used were M1 (line 2B-10), M2 [line GR5-F4 in A–D, GR5-16 in D (right-hand lane)], or M123 (lines GR3F-11 in A and C and GR3F11 and GR3-14 in B and D, left to right, respectively).
Discussion
R Protein Mutation as an Approach to Disease Resistance in Crop Plants.
With these cell death and virus-resistance data, we have shown that the LRR mutations, as predicted, affected both the recognition and activation functions of Rx LRR. The M4 mutation enhanced the activation function of the LRR, whereas the M1–M3 mutations affected elicitor recognition (Figs. 2 and 3). A structural interpretation of these data assumes that the R protein LRR domains form a horseshoe-shaped structure similar to those of the human and porcine ribonuclease inhibitor LRR proteins (33–35). With just four mutants analyzed in detail, we cannot develop precise rules for directed R protein enhancement and modification. However, it is striking that none of the M1–M4 mutations affect the conserved nonpolar residues of the LxxLxLxx(N/C/T)xxL repeat that are predicted to project into the hydrophobic core of the horseshoe structure. It is likely, therefore, that the mutated residues are on the solvent-exposed part of the horseshoe structure and are available to interact with the elicitor or with host proteins that interact with the elicitor if they are involved in recognition. Alternatively, if these residues affect the activation function as in M4, they might influence interactions with other domains in Rx or with other host proteins.
NB-LRR R protein interactions with pathogen elicitors might be direct, as with the N gene and the tobacco mosaic virus p50 helicase (36). Alternatively, as with bacterial resistance genes RPM1 and RPS2, they could be indirect; there is a host protein RIN4 that interacts with both the elicitor and the R protein (37, 38). An indirect interaction could explain our results with Rx and CP-KR if the mutations had a simple quantitative effect on the Rx interaction with a cellular component that binds to the potexviral CP molecules. However, the strongest CP-PoMV response was with M2 (Figs. 3 and 4), whereas the strongest CP-KR response was with M1 (Fig. 2). From this difference, we infer there is a qualitative difference between M2 and the other mutants that is most easily explained by a direct LRR-CP interaction. However, we cannot rule out, for example, that the PVX-CP interactions are mediated by a host protein that binds strongly to M1 and that the CP-PoMV interaction involves a different protein that binds preferentially to M2.
In principle, the M1–M3 mutations affecting elicitor recognition could have generated LRR structures mediating direct or indirect interactions with CP-KR or CP-PoMV that are distinct from the CP-TK interactions of the original Rx. However, a more likely interpretation is that the CP-KR and CP-PoMV interactions involve features of the LRR that also mediate recognition of CP-TK. The original Rx, according to this idea, would not confer resistance against PVX with CP-KR or PoMV, because it has features on the solvent-exposed face of the LRR that destabilize or interfere with CP-KR and CP-PoMV interactions. In the absence of those features (in M1–M3), the mutant Rx would interact with common features of CP-TK, CP-KR, and CP-PoMV. We favor this second interpretation, because removal of a structural feature by mutation would be more frequent than the acquisition of a specific new structure. This second interpretation also allows for the observation that M1–M3 retain the interaction with CP-TK (Fig. 2E) and that there is a weak interaction of the original Rx with CP-KR (Fig. 1B).
Based on its effect in elicitor-independent activation of resistance, we propose that the M4 mutation affects the activation stage of Rx resistance (Fig. 2). Presumably the enhanced activation compensates for the weak recognition of CP-KR in the original Rx. M4, and other constitutive gain-of-function mutations affect the second and third LRR of Rx (29), and there is also evidence from other R proteins that the N-terminal region of the LRR has a signaling- or activation-related function (39, 40). Similarly, in Nod2, a mammalian NB-LRR protein, the N-terminal LRR repeats have a regulatory rather than recognition role (41). It is likely, therefore, that enhancement of the activation stage of disease resistance can be achieved by targeted modifications in the N-terminal region of the LRR.
The LRR mutation approach could be usefully extended to R genes other than Rx, including those that confer resistance against fungal or bacterial pathogens and invertebrate pests. If these genes, like Rx, mediate recognition of a polymorphic elicitor produced by most genotypes of the pathogen, it would be possible to screen LRR mutants for broader-spectrum disease resistance. The selected mutants would be expected to confer more durable resistance than with the original locus, provided the recognized elicitor is produced by most genotypes of the pathogen. It might also be possible to use LRR mutation to retrieve useful resistance from R genes that had been overcome in the field or to enhance the phenotype of R genes conferring only weak resistance against a pathogen.
An advantage of this approach over other strategies of transgenic disease resistance is that it mimics natural mechanisms of disease resistance, and there would be no requirement to transfer nonplant sequences among plants. In addition, in the absence of the inducing pathogen, there would be no effect of the disease-resistance gene on growth and development of the plant. A potential hazard, illustrated by the enhanced disease symptoms of the M1 plants infected with PoMV (Fig. 4), can be screened out by selection of the mutant R genes with a range of pathogens including those only distantly related to the target of the original resistance.
At present, the LRR mutation approach to R gene enhancement is restricted to systems in which the need is to extend the range of resistance targets within a group of related pathogens. Eventually, when more is understood about structure–function relationships in R proteins, it might be possible to use targeted mutation of LRR domains rather than the empirical approach described here. Such a targeted approach combined with a more high-throughput screen might also allow completely novel specificity to be engineered. Other technical developments might also allow transgenes to be avoided. For example, by TILLING point mutations at a defined genetic locus (42, 43) or by use of targeted gene modification (44), it might be possible to generate useful variation in the LRR coding sequence of endogenous R gene loci.
LRR Point Mutations in Natural R Protein Evolution.
The LRR point mutations like those described here would occur spontaneously in natural populations and, from their phenotype, we infer that a short-term trend in R gene evolution would be between narrow and broad spectrum resistance. In this context, narrow spectrum resistance would confer resistance against pathogens with one variant of the targeted elicitor. The broad-spectrum resistance would be targeted at the variant forms of an elicitor from different pathogen strains. If the pathogen isolates cause severe disease, the broad-spectrum resistance might confer a selective advantage to plants with the mutant R proteins. However, some of these mutant alleles might also have a cost to the plant. For example, those with a strong autoactive phenotype, like the seven mutants identified in our screen and others described previously (29), would be lethal due to the cell death phenotype and would be selected against. Other mutants with weaker autoactivity might have a more subtle penalty for the plant that would be detected only in extensive field experiments. The fitness cost associated with alleles of the bacterial disease-resistance locus RPM1(45) may be accounted for in this way. Other mutations like M1 might also have a cost. The M1 plants appeared normal in the absence of virus (Fig. 4C), but they developed severe symptoms to PoMV. These added symptoms are most likely because the resistance pathways and cell death response were activated too weakly or too slowly to restrict the virus to the site of inoculation. A similar exacerbation of disease symptoms was noted with weak mutants of the tobacco mosaic virus resistance gene N (46).
In wild populations, it is likely there is a tradeoff between these costs and benefits of LRR mutations affecting the breadth of the disease resistance. When pathogen infection pressure is high, the benefits of broad-spectrum resistance would compensate for the costs, because some plants in the population would remain disease-free. Conversely, in low-infection pressure, the costs would be greater than the benefits, and the mutants would be selected against. Support for this idea of a tradeoff between broad- and narrow-spectrum resistance is from the polymorphic alleles of a flax rust-resistance gene in flax (L) with either broad- or narrow-spectrum effectiveness. L5 is a broad-spectrum allele mediating recognition of two variants of a fungal avirulence (AVR) protein; L6 is an intermediate allele, because recognition is strong with only one of the AVR alleles, and L7 is a narrow-spectrum allele conferring weak recognition of this one AVR allele (47). Similarly, in Arabidopsis, there are alleles of the R gene RPP13 conferring broad- or narrow-spectrum resistance against the oomycete Hyaloperonospora parasitica (48, 49). Some alleles recognize three variants of the H. parasitica protein ATR13. Other RPP13 alleles are more specific and recognize subsets of either one or two ATR13 variants (48). Further understanding of natural R gene evolution will assist in the use of R genes in agriculture and in particular in the development of durable resistance with minimal cost to the crop plant.
Materials and Methods
General Methods and Biomaterials.
Strains of PVX used in this study were described previously. PVX-TK is from an infectious cDNA of the CP4 isolate of PVX, and PVX-KR is a derivative of PVX-TK with the T121K and K127R mutations (12). PVX-HB-GFP is based on cDNA of a HB isolate that has the same amino acids at positions 121 and 127 of the CP as the PVX-KR mutant. It also has an inserted GFP reporter gene and the general structure of the originally described PVX-GFP (50). All infectious viral cDNAs used in this study were inserted into the expression cassette of an Agrobacterium binary Ti plasmid vector pBin61 (51), transformed into Agrobacterium, and inoculated to plants by leaf infiltration (52). The PoMV isolates were obtained from J. I. Cooper (Centre for Ecology and Hydrology, Oxford, U.K.) and were inoculated to plants as sap extracts. The CP sequences were cloned by RT-PCR of the viral RNA by using primers spanning the initiation and termination codons. The PCR-amplified cDNA was then cloned into pBin61 (51). Virus-infected material was carried out in containment glasshouses as authorized by Department for Environment, Food, and Rural Affairs (Defra) License PHL 24B/3654 (3/2001).
Clones of Rx cDNA, transient expression of Rx and CP, protein, and RNA detection methods were described previously (12, 17). The Rx template in all manipulations was derived from the binary vector pB1:Rx-HA (17). The Rx transgenic N. benthamiana lines were generated by Agrobacterium tumefaciens leaf disk transformation (53). Random mutagenesis of the Rx LRR used a two-stage error-prone PCR protocol (26). The PCR was carried out by using primers flanking the Rx LRR coding sequence and the SacI site in the 3′ UTR region. Sequencing 20 mutant Rx clones revealed a mutation frequency of 1 in 756 bases. Because the LRR region is 1,382 base pairs between the Apal1 site and the stop codon, there was an average of 1.82 mutations per clone. The gain-of-function clones all contained one or more mutations. Recombinant Rx clones containing multiple mutations were generated by chimeric PCR (54), and their identity was confirmed by sequencing.
Electrolyte Leakage Assays.
N. tabacum plants were infiltrated with A. tumefaciens cultures transformed with the appropriate Rx and CP constructs. Agrobacterium cultures were infiltrated at a final turbidity of 0.25 (OD600) in the CP analysis or 0.5 (OD600) in the overexpression analysis. Two 7-mm-diameter leaf disks from the same leaf were placed in a 1.5-ml Eppendorf (Hamburg, Germany) tube, and 1 ml of analytical grade H2O was added. Eppendorfs were placed in a Savant (Irvine, CA) Speedivac for 10 min at 35°C at a vacuum of 1 pascal. Leaf disks were then incubated at room temperature for 30 min and subsequently removed before storage of samples at −20°C. Conductivity was measured by using a Jenway (Essex, U.K.) 4010 conductivity meter with an A99 027-013 electrode calibrated with a 1 M solution of KCL according to the manufacturer's instructions. Electrolyte solutions were thawed at room temperature, and 0.8 ml of the solution was added to 3.2 ml of analytical grade H2O in a 15-ml Corning (Corning, NY) tube maintained at 22°C in a water bath. For each data point, a minimum of five independent replicates were sampled. Results are shown as the mean ± SEM.
Supplementary Material
Acknowledgments
We thank our colleagues, including Ian Crute, John Rathjen, Jonathan Jones, Becky Mosher, and Wladimir Tameling, for discussions and comments on the draft manuscript. This work was supported by the Gatsby Charitable Foundation.
Abbreviations
- R
resistance
- LRR
leucine-rich repeat
- CP
coat protein
- NB
nucleotide binding
- PVX
potato virus X
- PoMV
poplar mosaic virus.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Heath,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Hull R, Davies JW. Crit Rev Plant Sci. 1992;2:17–33. [Google Scholar]
- 2.Wang PG, Tumer NE. Adv Virus Res. 2000;55:325–355. doi: 10.1016/s0065-3527(00)55007-6. [DOI] [PubMed] [Google Scholar]
- 3.Nolke G, Fischer R, Schillberg S. Plant Pathol. 2004;86:5–17. [Google Scholar]
- 4.Baulcombe DC. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonsalves D. Annu Rev Phytopathol. 1998;36:415–437. doi: 10.1146/annurev.phyto.36.1.415. [DOI] [PubMed] [Google Scholar]
- 6.Accotto GP, Nervo G, Acciarri N, Tavella L, Vecchiati M, Schiavi M, Mason G, Vaira AM. Phytopathology. 2005;95:800–807. doi: 10.1094/PHYTO-95-0800. [DOI] [PubMed] [Google Scholar]
- 7.Gaba V, Zelcer A, Gal-On A. In Vitro Cell Dev Biol–Plant. 2004;40:346–358. [Google Scholar]
- 8.Kaniewski WK, Thomas PE. Mol Biotechnol. 1999;12:101–115. doi: 10.1385/MB:12:1:101. [DOI] [PubMed] [Google Scholar]
- 9.Xu DM, Collins GB, Hunt AG, Nielsen MT. Crop Sci. 1999;39:1195–1202. [Google Scholar]
- 10.Tepfer M. Annu Rev Phytopathol. 2002;40:467–491. doi: 10.1146/annurev.phyto.40.120301.093728. [DOI] [PubMed] [Google Scholar]
- 11.Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, Prins M. Mol Breed. 2001;7:151–161. [Google Scholar]
- 12.Bendahmane A, Kanyuka K, Baulcombe DC. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitham S, McCormick S, Baker B. Proc Natl Acad Sci USA. 1996;93:8776–8781. doi: 10.1073/pnas.93.16.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dangl JL, Jones JDG. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 15.vanderBiezen EA, Jones JDG. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 16.Belkhadir Y, Subramaniam R, Dangl JL. Curr Opin Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Moffett P, Farnham G, Peart JR, Baulcombe DC. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tameling WIL, Elzinga SDJ, Darmin PS, Vossen JH, Takken FLW, Harling MA, Cornelissen B. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu A-J, Rathjen JP, Bendahmane A, Day L, Baulcombe D. EMBO J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole AB, Kiraly L, Ross H, Schoelz JE. Mol Plant–Microbe Interact. 2001;14:31–41. doi: 10.1094/MPMI.2001.14.1.31. [DOI] [PubMed] [Google Scholar]
- 21.Innes RW. Plant Physiol. 2004;135:695–701. doi: 10.1104/pp.104.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodds PN, Lawrence GJ, Ellis JG. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen QH, Zhou FS, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendahmane A, Köhm BA, Dedi C, Baulcombe DC. Plant J. 1995;8:933–941. doi: 10.1046/j.1365-313x.1995.8060933.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Petersen EI, Petersen SB, El-Gewely MR. BioTechniques. 1999;27:1102–1108. doi: 10.2144/99276bm02. [DOI] [PubMed] [Google Scholar]
- 27.Goulden MG, Köhm BA, Santa Cruz S, Kavanagh TA, Baulcombe DC. Virology. 1993;197:293–302. doi: 10.1006/viro.1993.1590. [DOI] [PubMed] [Google Scholar]
- 28.Aviv DH, Rusterucci C, Holt BF, Dietrich RA, Parker JE, Dangl JL. Plant J. 2002;29:381–391. doi: 10.1046/j.0960-7412.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- 29.Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 30.Henderson J, Gibbs MJ, Edwards ML, Clarke VA, Gardner KA, Cooper JI. J Gen Virol. 1992;73:1887–1890. doi: 10.1099/0022-1317-73-7-1887. [DOI] [PubMed] [Google Scholar]
- 31.Jakobek JL, Lindgren PB. Plant Cell. 1993;5:49–56. doi: 10.1105/tpc.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohm BA, Goulden MG, Gilbert JE, Kavanagh TA, Baulcombe DC. Plant Cell. 1993;5:913–920. doi: 10.1105/tpc.5.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kajava AV. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 34.Kajava AV, Kobe B. Protein Sci. 2002;11:1082–1090. doi: 10.1110/ps.4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DA, Jones JDG. Adv Bot Res. 1997;24:89–167. [Google Scholar]
- 36.Ueda H, Yamaguchi Y, Sano H. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- 37.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl J. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 39.Warren RF, Henk A, Mowery P, Holub E, Innes RW. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang C-F, Williamson VM. Plant J. 2003;34:585–593. doi: 10.1046/j.1365-313x.2003.01749.x. [DOI] [PubMed] [Google Scholar]
- 41.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stemple DL. Nat Rev Genet. 2004;5:145–150. doi: 10.1038/nrg1273. [DOI] [PubMed] [Google Scholar]
- 43.Henikoff S, Till BJ, Comai L. Plant Physiol. 2004;135:630–636. doi: 10.1104/pp.104.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 45.Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- 46.Dinesh-Kumar SP, Tham WH, Baker BJ. Proc Natl Acad Sci USA. 2000;97:14789–14794. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose LE, Bittner-Eddy PD, Langley CH, Holub EB, Michelmore RW, Beynon JL. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 50.Baulcombe DC, Chapman SN, Santa Cruz S. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 51.Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. Plant J. 2000;21:73–81. doi: 10.1046/j.1365-313x.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- 52.Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. Plant J. 2002;29:569–579. doi: 10.1046/j.1365-313x.2002.029005569.x. [DOI] [PubMed] [Google Scholar]
- 53.Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. Science. 1985;227:1229–1231. [Google Scholar]
- 54.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.