Abstract
Vitamin E is an essential nutrient for humans and is obtained primarily from food, especially oil, derived from the seed of plants. Genes encoding the committed steps in vitamin E synthesis in plants (VTE, loci 1–5) have been isolated and used for tocopherol pathway engineering with various degrees of success. As a complement to such approaches we have used quantitative trait loci analysis with two sets of Arabidopsis thaliana recombinant inbred lines and have identified 14 QVE (quantitative vitamin E) loci affecting tocopherol content and composition in seeds. Five QVE intervals contain VTE loci that are likely QVE gene candidates. Nine QVE intervals do not contain VTE loci and therefore identify novel loci affecting seed tocopherol content and composition. Several near-isogenic lines containing introgressions of the accession with increased vitamin E levels were shown to confer significantly elevated tocopherol levels compared with the recurrent parent. Fine-mapping has narrowed QVE7 (a γ-tocopherol quantitative trait loci) to an 8.5-kb interval encompassing two genes. Understanding the basis of the QVE loci in Arabidopsis promises to provide insight into the regulation and/or metabolism of vitamin E in plants and has clear ramifications for improving the nutritional content of crops through marker-assisted selection and metabolic engineering.
Keywords: biofortification, quantitative trait loci, seed metabolism, tocopherol
Humans and most other mammals require a select number of essential vitamins and minerals in their diets to maintain proper health. Plant-derived foods have the biochemical potential to produce and/or accumulate all of these essential micronutrients, with the exception of vitamins D and B12, and can theoretically provide balanced nutrition as dietary staples. However, most crop plants contain several micronutrients at levels too low to fulfill minimal nutritional needs, even when the crop represents the major proportion of daily caloric intake. As a result, there has been growing interest in delivering a more balanced micronutrient content directly in food crops, especially those that are staples for the poor in developing countries.
During the past 5 years, several large international efforts have been initiated to understand the basis of differing mineral and vitamin levels in plants, with the long-term goal of applying this information to elevate the levels of targeted micronutrients in world crops (the HarvestPlus research program and the Grand Challenges in Global Health initiative). A key component of these global efforts is using the full power of genomic, genetic, and molecular tools within model systems to provide the fundamental knowledge necessary to achieve the desired micronutrient contents in a staple crop. Partly as a result of these efforts, significant progress has been made in our understanding of the synthesis of provitamin A carotenoids, vitamin E, folate, biotin, and most other vitamins and in the transport and accumulation of selected minerals in model plants (1–3).
The engineering of various organic micronutrient levels (vitamins) using selected biosynthetic pathway genes has yielded many notable successes (4–7). However, these efforts also have highlighted that, even when the genes for the core biosynthetic pathways are known and the encoded proteins are well characterized, much still remains to be understood about the genetic and biochemical control of micronutrient accumulation, turnover, and bioavailability in plant tissues and different foodstuffs of the human diet (3). In an effort to fill such gaps in our knowledge, we have undertaken a quantitative genetic approach to determine the genetic basis of tocopherol (vitamin E) levels during seed development in the model plant Arabidopsis thaliana.
Understanding the control of micronutrient levels, such as tocopherols, in seed is of particular importance because it is this tissue that provides the bulk of the micronutrients in the diet. We report here that even with a well studied biosynthetic pathway like that for tocopherols, this quantitative genetic approach can identify additional loci and alleles that impact the micronutrient content in seed. Isolating the genes identified by these quantitative trait loci (QTL) and understanding the molecular and biochemical basis of their impact on micronutrient levels has significant potential to enhance the micronutrient levels of crops. The combined genetic, genomic, and biochemical resources in Arabidopsis make it an ideal system to pursue this approach.
Tocopherols are a group of four lipid-soluble amphipathic molecules that are synthesized exclusively by photosynthetic organisms and collectively are an essential component of the human diet, vitamin E. The α-, β-, δ-, and γ-tocopherols produced by plants differ only in the position and number of methyl groups on their chromal ring (Fig. 1). Although all four tocopherols have vitamin E activity, the activity of α-tocopherol is 2–50 times higher than that of the other three tocopherols because of the affinity of the mammalian hepatic α-tocopherol transfer protein for α-tocopherol. The different tocopherol species vary greatly in amounts and proportions in different plants and tissues, with seed most often containing the highest levels of total tocopherols, although α-tocopherol is often only a minor component. Nonetheless, seeds are by far the major source of vitamin E in the human diet because of the high per capita intake of seed oils (8).
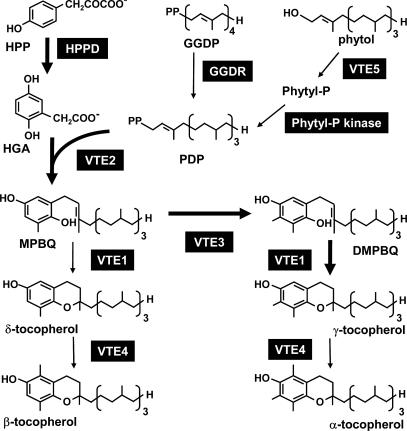
Fig. 1.
The tocopherol biosynthetic pathway in A. thaliana. Enzymes are in black boxes. Bold arrows show the biosynthetic route leading to the most abundant tocopherol in seed, γ-tocopherol. HPP, hydroxyphenylpyruvate; GGDP, geranylgeranyl-diphosphate; Phytyl-P, phytyl-monophosphate; PDP, phytyl-diphosphate; HGA, homogentisic acid; MPBQ, 2-methyl-6-phytyl-1,4-benzoquinol; DMPBQ, 2,3-dimethyl-6-phytyl-1,4-benzoquinol; HPPD, hydroxyphenylpyruvate dioxygenase; GGDR, geranylgeranyl-diphosphate reductase; VTE1, tocopherol cyclase; VTE2, homogentisic acid phytyltransferase; VTE3, 2-methyl-6-phytyl-1,4-benzoquinol methyltransferase; VTE4, γ-tocopherol methyltransferase; VTE5, phytol kinase.
All tocopherols consist of a polar chromal head group that remains at the membrane surface and a hydrophobic prenyl tail that associates with the membrane lipids. The tocopherol biosynthetic pathway has been elucidated in plants (3) and is summarized in Fig. 1. The genes encoding the enzymes VTE1 through VTE5 have been identified in Arabidopsis and are encoded as single copies whose mutation changes the content and composition of tocopherols in seed and/or leaves (9–13). With the cloning of the five VTE loci, much effort has been focused on using these genes to engineer plant tissues to increase tocopherol production and vitamin E content (8, 14). Although some engineering experiments have drastically increased the levels of α-tocopherol, with corresponding increases in vitamin E activity (9, 15, 16), others have led to unanticipated or less desirable results, such as marginal effects on tocopherol levels, large increases in tocotrienols instead of tocopherols, or a decrease in ascorbate and glutathione levels (reviewed in refs. 3 and 17). Thus, there is a need for understanding how new loci of natural alleles of known VTE genes positively impact tocopherol or vitamin E levels without detrimentally impacting fitness. The knowledge gained from the molecular characterization of QTL for tocopherol levels in Arabidopsis will contribute to our understanding in both of these areas.
Tocopherol levels in plant tissues fluctuate because of a variety of factors, both environmental and genetic, with leaf tocopherol levels being especially responsive to the environment. Levels and types of tocopherols also vary between and within species, cultivars, varieties, or accessions. Tocopherol QTL experiments in canola (18) and maize (19) have demonstrated that genetic variation in these crop plants can be mapped despite strong influence by environmental conditions (20, 21). Interestingly, no significant correlation was found between the levels of α- and γ-tocopherols in the genetic studies of canola (18, 20), but in maize several QTL for altered α/γ-tocopherol ratios were reported, although their cooccurrence with α-tocopherol QTL suggested they were due to an increase in α-tocopherol levels (19).
QTL analysis allows one to identify physiologically relevant genes for a trait that may not be readily identified by mutational studies. Additionally, the genes and alleles identified from QTL analysis are not constrained by preconceived ideas about the process and its regulation. Rather, QTL analysis utilizes the untapped genetic variation that exists among cultivars or, in the case of Arabidopsis, accessions to inform about a trait. Several studies have already used this method to identify nutritionally relevant QTL in Arabidopsis (22–25). Tocopherol QTL studies already have demonstrated an abundance of genetic variation in maize (19) and canola seed (18, 20); however, the molecular characterization of any identified locus has yet to be achieved. In this regard, the existing knowledge base concerning tocopherol synthesis in Arabidopsis combined with its genetic, genomic, and molecular tractability (26) should allow one to more readily identify the molecular basis of natural variation and determine whether the QTL identified affects known biosynthetic genes or identifies a novel locus that impacts the pathway. Here we report QTL analysis of two sets of Arabidopsis recombinant inbred lines that have identified several distinct QTL affecting tocopherol content and composition in seed. The construction of near-isogenic lines (NILs) and marker-assisted fine-mapping furthers our understanding of the regulation of tocopherol content in seed by determining the genes responsible for the QTL and has ramifications for breeding and metabolic engineering of tocopherols in crop plants.
Results
Arabidopsis Accession Variability.
The Arabidopsis thaliana accession Landsberg erecta (Ler) contained levels of tocopherols in seed lower than either the Columbia (Col) or Cape Verdi Islands (Cvi) accessions (Fig. 2), so the existing Col/Ler and Cvi/Ler recombinant inbred line (RIL) populations were chosen for QTL analysis. Harvested seed from each RIL population was analyzed by reversed-phase HPLC for individual and total tocopherols, and these data were used as quantitative values in QTL analysis.
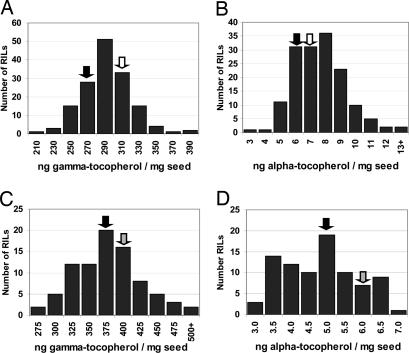
Fig. 2.
Range of tocopherol levels in seed from RILs indicates that a large genetic potential exists. Shown are frequency distributions of seed γ-tocopherol and α-tocopherol levels of Cvi/Ler RIL population lines (n = 2) (A and B) and Col/Ler RIL population lines (n = 1) (C and D). Levels of corresponding tocopherols for parental Ler, Cvi, and Col were grown in parallel with the corresponding RILs and are indicated by black, white, and gray arrows, respectively.
The range of seed tocopherol levels in both the Cvi/Ler and Col/Ler RIL populations exceeded that of the parental accessions (transgression), indicating that a large genetic potential exists for these traits (Fig. 2). Ler had lower tocopherol levels than either Cvi or Col (Fig. 2), and, for the majority of QTL, the Ler alleles conferred lower tocopherol levels. This joint contribution and relative proportion of parental accessions for the predicted QTL is in agreement with the transgressive nature of the tocopherol levels from the RIL populations. Although in the two independent RIL experiments the value for Ler differed, indicating influences of (unknown) environmental factors, it is important to emphasize that the relative rankings of different accessions, NILs, and recombinants between experiments do not. Standard deviations are typically <5% for γ-tocopherol and 10% for α-tocopherol. In this manuscript, all comparative seed tocopherol data reported were obtained from plants grown in the same experiment. The QTL identified were named QVE for quantitative vitamin E locus and numbered by location (Table 1). The major QTL for α-, δ-, and γ-tocopherols are described in detail below.
Table 1.
Summary of the seed tocopherol QTL found in Arabidopsis
| RIL name | Trait | LG | Position, cM | Supporting interval, cM | lod score | % Explan | High parent | Known biosynthetic genes in interval |
|---|---|---|---|---|---|---|---|---|
| Cvi/Ler | ||||||||
| QVE3 | α-tocopherol | 1 | 88 | 86–90 | 3.15 | 9.0 | Cvi | VTE4, MECPS |
| QVE7 | γ-tocopherol | 3 | 0 | 0–4 | 2.88 | 8.3 | Cvi | IPP2 |
| QVE9 | γ-tocopherol | 5 | 2 | 0–6 | 4.39 | 12.9 | Ler | VTE5 |
| QVE10 | γ-tocopherol | 5 | 14 | 12–16 | 5.81 | 15.9 | Cvi | — |
| QVE12 | γ-tocopherol | 5 | 64 | 62–66 | 3.82 | 10.8 | Cvi | — |
| Col/Ler | ||||||||
| QVE1 | α-tocopherol | 1 | 10 | 8–12 | 9.46 | 40.5 | Col | HPPD |
| QVE2 | α-tocopherol | 1 | 50 | 46–52 | 5.42 | 25.4 | Ler | — |
| QVE4* | δ-tocopherol | 2 | 40 | 34–44 | 7.14 | 32.1 | Col | VTE2, TAT3 |
| QVE5 | γ-tocopherol | 2 | 44 | 40–46 | 5.44 | 25.5 | Col | VTE2, TAT3 |
| QVE6 | α-tocopherol | 2 | 50 | 46–54 | 4.33 | 20.9 | Ler | VTE2, TAT3, CMK |
| QVE8 | γ-tocopherol | 3 | 4 | 0–8 | 3.51 | 17.7 | Col | IPP2 |
| QVE11 | δ-tocopherol | 5 | 36 | 32–42 | 2.94 | 14.7 | Col | — |
| QVE13 | γ-tocopherol | 5 | 76 | 72–80 | 10.87 | 44.5 | Ler | — |
| QVE14* | δ-tocopherol | 5 | 92 | 88–96 | 7.63 | 33.9 | Ler | — |
All values were obtained by using PLABQTL composite interval mapping. LG, linkage group; % Explan, percentage of the phenotypic variance explained by a putative QTL (R2%); MECPS, 2-C-methylerythritol-2,4-cyclodiphosphate synthase; TAT3, tyrosine aminotransferase; CMK, 4-(cytidine-5′-diphospho)-2-C-methylerythritol kinase; —, no candidate.
*The only epistasis observed was between QVE4 and QVE14.
QTL Analysis.
α-tocopherol.
The highest contributing QTL for α-tocopherol (QVE1) was located on the top of chromosome 1 and contributed >40% of the explained phenotypic variation for the trait in the Col/Ler RIL population. Interestingly, a correlation between higher α-tocopherol levels and the presence of a Cvi allele in this region also was observed in the Cvi/Ler RIL population, but it was below the critical lod threshold (lod near 2; data not shown). Another major α-tocopherol QTL in Col/Ler (QVE2) explaining 25% of the phenotypic variation with an opposing allelic effect is loosely linked (40 cM apart) to QVE1. A third α-tocopherol QTL (QVE6) in Col/Ler is located on chromosome 2 near the erecta gene. None of these α-tocopherol QTL were found in the Cvi/Ler population, which contained only a single, unique α-tocopherol QTL (QVE3) on the lower arm of chromosome 1 in the region of the γ-tocopherol methyl transferase gene (VTE4).
γ-tocopherol.
More γ-tocopherol QTL than α-tocopherol QTL were detected: Chromosome 5 alone has four γ-tocopherol QTL. The three γ-tocopherol QTL on chromosome 5 in the Cvi/Ler population are located at 2, 14, and 64 cM (QVE9, QVE10 and QVE12, respectively), with the first two having opposite allelic effects. The Col/Ler population has a large-impact γ-tocopherol QTL located at 76 cM on chromosome 5 (QVE13). Additionally, there is a major γ-tocopherol QTL on chromosome 2 (QVE5) in Col/Ler and small γ-tocopherol QTL (QVE7 and QVE8) on the extreme top of chromosome 3 in both populations. Total tocopherol levels also were analyzed but are not discussed because this trait had a 0. 999 coefficient of correlation to γ-tocopherol, the predominant tocopherol (>90%) in Arabidopsis seeds, and the total and γ-tocopherol QTL consistently mapped to the same locations.
δ-tocopherol.
δ-Tocopherol levels were analyzed only in the Col/Ler RIL population and resulted in the identification of three δ-tocopherol QTL significant at P < 0.01. The two strongest δ-tocopherol QTL (contributing 34% and 32%, respectively) were found at 92 cM on chromosome 5 (QVE14) and just above the erecta region on chromosome 2 (QVE4). An epistatic interaction between QVE11 and QVE4 that contributes an additional 12% of phenotypic variation for δ-tocopherol was the only epistatic interaction detected among the 14 QVE loci identified. A third δ-tocopherol QTL at 36 cM (QVE11) contributed 20% of the phenotypic variation, with the Col allele contributing higher levels. Additional QTL with opposite allelic effects than QVE1 were detected on chromosome 5 (QVE12 at 64 cM and QVE14 at 92 cM). δ-Tocopherol levels were significantly correlated to γ-tocopherol levels in the Col/Ler population (r = 0.533).
Common QTL Between RIL Populations.
Two genomic regions potentially hold common QTL found in both RIL populations: QVE7 and QVE8 on the top of chromosome 3 and QVE12 and QVE13 in the middle of chromosome 5 (64 and 76 cM) (Table 1). Because the Ler allele contributes lower levels of γ-tocopherol for both QVE7 and QVE8, Ler may hold a common difference from both Cvi and Col. In contrast, for QVE12 and QVE13, different parents (Cvi from Cvi/Ler and Ler from Col/Ler) contributed the allele with the higher level of tocopherols. On chromosome 2, QTL for each of the three individual tocopherol components analyzed clustered to a central region near the erecta gene; the supporting interval of the γ-tocopherol QTL (QVE5) here overlaps with QTL for δ-tocopherol (QVE4) and α-tocopherol (QVE6) and may reflect a single QTL that affects all three traits because the position of the detected QTL is somewhat dependent on the selection of cofactors in composite interval mapping, especially in small populations. Interestingly, whereas QVE4 and QVE5 have Col as the contributing parent, Ler is the contributing parent for QVE6, suggesting an antagonism between γ- and α-tocopherol levels. Because QVE6 was not found in the Cvi/Ler RIL population, it is unlikely that the transcription factor ERECTA is the gene responsible for QVE6. It should also be noted that QVE4–QVE6 and QVE10 and QVE11 were located in regions showing segregation distortion.
Confirming the Impact of QTL in NILs.
NILs containing small regions of the contributing genetic background (Col or Cvi) introgressed into the Ler background were developed for several of the QVE. NIL G7 contains a 6-megabase (Mb) introgression of Cvi chromosome 1 in the Ler accession (Table 2). NIL G7 had an average 30% higher seed α-tocopherol level than its Ler counterpart, confirming the presence of QVE3 in this region on chromosome 1 (Table 2). The relative level of α-tocopherol is greater than initially estimated by QTL analysis and agrees better with estimates obtained from an additional experiment in which a subset of the Cvi/Ler RIL population was grown and evaluated again (data not shown). Additional recombinants of NIL G7 have narrowed QVE3 to a 0.8-Mb region that still encompasses VTE4.
Table 2.
Confirmation of QVE effects in NILs
| QTL locus | NIL name | Int | Chromosome no.: position, Mb | Relative γ-toco | Relative δ-toco | Relative α-toco |
|---|---|---|---|---|---|---|
| QVE3 | G7 | Cvi | 1:21–27 | 0.99 | 0.89 | 1.30 |
| QVE3 | G1A | Cvi | 1:21–24.4 | 0.99 | 1.13 | 1.53 |
| QVE3 | G4H | Cvi | 1:23.6–27 | 1.03 | 1.06 | 1.39 |
| QVE3 | G4G | Cvi | 1:21–23.5 | 0.98 | 0.95 | 0.92 |
| QVE3 | G3B | Cvi | 1:24.5–27 | 1.07 | 1.07 | 1.00 |
| QVE4/5/6 | 9Y | Col | 2:7.3–11.9 | 1.17 | 1.18 | 0.85 |
| QVE4/5/6 | 9E | Col | 2:8.1–11.9 | 1.17 | 1.11 | 0.92 |
| QVE4/5/6 | 9P | Col | 2:11.4–11.9 | 1.03 | 0.98 | 1.06 |
| QVE7 | LN26 | Cvi | 3:0–0.6 | 1.07 | 1.31 | 0.88 |
| QVE7 | 8E5 | Cvi | 3:0–0.101 | 1.14 | 1.43 | 0.97 |
| QVE7 | 4-212 | Cvi | 3:0.092–0.6 | 1.14 | 1.26 | 0.87 |
| QVE7 | 6A6 | Cvi | 3:0–0.088 | 1.01 | 1.12 | 1.05 |
Seed tocopherols levels are shown relative to Ler, which was set at 1.0 (N = 2–4). The bold numbers indicate that the NIL still encompasses the QTL. All introgressions (Int) were into the Ler accession. toco, tocopherol.
NIL 9Y maintains a 4.6-Mb portion of Col chromosome 2 introgressed into the Ler background. HPLC analysis confirmed the presence of both QVE4 and QVE5 in NIL 9Y; Col alleles in this region conferred an 18% increase in δ-tocopherol and a 17% increase in γ-tocopherol levels compared with Ler. NIL 9Y also has α-tocopherol levels 85% of those for Ler, indicating that QVE6 also is contained in this interval (Table 2). NIL 9E (8.1–11.9 Mb) has tocopherol levels nearly identical to NIL 9Y, whereas NIL 9P (11.4–11.9 Mb) restores Ler levels of α-, γ-, and δ-tocopherols. This finding suggests that QVE4–QVE6 are contained within the 3.3-Mb interval defined by NILs 9E and 9P (8.1–11.4 Mb). Further investigation is needed to characterize the additional recombinants across the interval to further refine the location of QVE4–QVE6 and possibly separate these QTL.
NIL LN26 contains a single Cvi introgression into the Ler background on the extreme top of chromosome 3 (22), with some additional introgressions in regions where no QVE are located (J. Keurentjes, L.B., and M.K., unpublished data). HPLC analysis showed that LN26 contained higher levels of both γ- and δ-tocopherols compared with the parental accession Ler, confirming the presence of QVE7 within the Cvi interval (Table 2).
Fine-Mapping of QVE7.
QTL analysis predicted QVE7 to be located above 3 cM on the Ler/Cvi chromosome 3 map (Table 1 and Fig. 3). This interval does not contain any known tocopherol biosynthetic genes (VTE1-VTE5) (Fig. 1), and the only gene related to plastidic isoprenoid synthesis in the interval is isopentenyl disphosphate isomerase 2 (IPP2; locus At3g02780; 0.602 Mb). Because of the early availability of NIL LN26 and the promise of a novel candidate gene, QVE7 was chosen for isolation by fine-mapping. HPLC analysis of homozygous recombinants between LN26 and Ler was used to phenotypically classify individual recombination events within the ≈600-kb Cvi interval of NIL LN26. This analysis delineated the region of interest to a 10-kb interval located between 92 and 102 kb of chromosome 3 (Fig. 3) and eliminated IPP2 as a candidate gene for QVE7. The DNA across this 10-kb interval was fully sequenced in the Ler and Cvi accessions and compared with flanking sequences from the most informative NILs. This analysis identified the nearest polymorphisms internal to the recombination events at base pairs 92,662 and 101,091, further delineating the interval of interest to 8.5 kb.
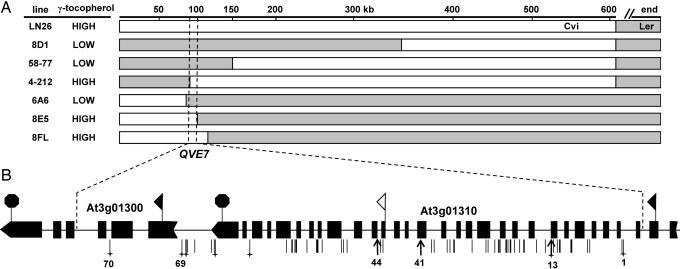
Fig. 3.
Fine-mapping of QVE7 to an 8.5-kb region of chromosome 3. (A) Summary of the most informative NILs used to fine-map the location of the γ-tocopherol QTL, QVE7. The Cvi allele of QVE7 confers increased γ-tocopherol levels; Ler DNA in recombinants is indicated by shaded regions. The Cvi genomic region common to all lines that have an increased γ-tocopherol phenotype is between 92,660 and 101,100 bp on the top of chromosome 3. The top scale is in kilobases. (B) Predicted gene structures and polymorphisms of At3g01300 and At3g01310. Sequence data were obtained for Ler and Cvi and were aligned to the published Col sequence. The black rectangles indicate predicted exons, and the ribbon tail and arrow indicate the predicted first and final exons of each gene, respectively. Black flags indicate predicted translational start sites; the black octagons indicate stop codons. The white flag indicates the alternate start site for At3g01310 modeled by the Munich Information Center for Protein Sequence. Vertical lines below the diagram indicate the position of polymorphisms between Ler and Cvi; those where Ler differs from both Col and Cvi are marked with a cross. Bold arrows indicate polymorphisms conferring amino acid changes or splice-site-junction alterations that are most likely to confer a phenotypic change. The associated numbers correspond to the polymorphisms numbered in Tables 3 and 5.
The 8.5-kb region delineating QVE7 contains only two predicted genes, the promoter and first three exons of At3g01300 and virtually the entire At3g01310 gene. At3g01300 encodes a 491-aa putative protein kinase with predicted serine/threonine kinase activity and ATP-binding motifs. No differences between accessions were detected in the At3g01300 coding region, and only a single 2-bp insertion was found in the second intron in Ler (Table 3). A homozygous mutant with a T-DNA insertion in the fourth exon of At3g01300 in the accession Col, SALK039402 (27), did not affect γ-tocopherol levels compared with wild type (Table 4), indicating that the At3g01300 gene is not involved in controlling seed tocopherol levels in Col. There are, however, several polymorphisms in the predicted promoter region of At3g01300, including an 11-bp deletion in Ler ≈60 bp before the TATA box (Table 3; see also Table 5, which is published as supporting information on the PNAS web site). Although we cannot exclude the possibility that At3g01300 promoter polymorphisms might impact seed tocopherol levels in Ler or Cvi, the combined data for At3g01300 suggest that it is unlikely to encode QVE7.
Table 3.
Select sequence polymorphisms between accessions within QVE1
| Polymorphism no. | Polymorphism Ler→Cvi | Position relative to Col sequence, bp | Details of position | Col resembles |
|---|---|---|---|---|
| 1 | A→G | 101,091 | Intron At3g01310/cT22N4w CAPS | Cvi |
| 13 | A→G | 99,884 | Ile→Thr, amino acid 219, At3g01310 | Cvi |
| 41 | A→T | 97,694 | Asp→Glu, amino acid 490, At3g01310 | Ler |
| 44 | G→T | 96,943 | Intron, fourth base 3′ from exon, At3g01310 | Ler |
| 69 | — →GTAGTTTTTTA | 93,687–93,697 | Promoter region, At3g01300 (−95 bp) | Cvi |
| 70 | TA→ — | 92,662 | Intron, At3g01300 | Cvi |
Shown are select sequence polymorphisms between Ler and Cvi accessions within the 91,910–101,450 bp region of chromosome 3. Each polymorphism, its position relative to the published Col chromosome 3 sequence (www.arabidopsis.org), and the Col allele are listed, along with placement and inferred effect on the associated gene. Polymorphism no. corresponds to numbered arrows in Fig. 3B. CAPS, cleavage-amplified polymorphic sequences.
Table 4.
Seed γ-tocopherol levels in T-DNA insertion lines
| Line | Nearest gene | Position of T-DNA | γ-Tocopherol, ng per mg of seed | % WT |
|---|---|---|---|---|
| SALK039402 | At3 g01300 | Exon | 269 ± 6 | 98.5 |
| SAIL244D05 | At3 g01310 | Promoter | 265 ± 9 | 96.9 |
| Col-0 | — | None | 274 ± 10 | 100 |
Samples were run in duplicate for seed from nine different homozygous T-DNA lines or from three wild-type (Col) plants. —, not applicable.
Polymorphisms Within At3g01310.
The predicted At3g01310 gene is 7.8 kb in length and contains 31 predicted exons that would encode a 119.5-kDa protein of 1,057 aa (www.arabidopsis.org). Most of the predicted At3g01310 ORF except for the initial two exons is contained within the 8.5-kb QVE7 region, defined by recombination events. At3g01310 is currently categorized as encoding an expressed protein with similarity to a predicted acid phosphatase and to an unknown human protein GB:BAA24863 (www.arabidopsis.org). A histidine acid phosphatase signature is present at amino acids 370–384 (LrcViaVvRHGdRtP; http://ca.expasy.org/prosite). There is a paralog to At3g01310 on chromosome 5, At5g15070, which has a similar predicted gene structure and 88% predicted protein identity.
Sequence comparison of the Cvi and Ler alleles of At3g01310 across the 8.5-kb QVE7 interval identified 63 polymorphisms between the two accessions (Fig. 3B and Table 5), including two nucleotide substitutions that would lead to alterations in the At3g01310 coding region: Asp-490 and Ile-219 present in Ler are converted to Gln-490 and Thr-219 in Cvi. The other 61 At3g01310 polymorphisms are either within introns or are silent coding substitutions. However, two of these intron changes are within 4 bp 5′ to exon boundaries and may cause a change in splicing frequency. CTAG/is converted to ATAG/in Cvi before exon 21; the A at the −4 position in Cvi has a 33% usage rate compared with only 6% for the C in Ler. Before exon 23, the consensus ACAG/in Ler is converted to ATAG/in Cvi, but this change results in only a slight change in usage rate (59% to 33%) (www.bio.net/bionet/mm/arab-gen/1992-October/000750.html).
As mentioned earlier, the similar locations and phenotypes of QVE7 and QVE8 suggested that they might correspond to the same locus in the two RIL populations. If this were the case, polymorphisms in Ler relative to both Cvi and Col could be a common molecular mechanism underlying the altered tocopherol levels in both QVE7 and QVE8. The majority of the 63 polymorphisms identified in At3g01310 were unique to the Cvi parent: Only 3 of the 63 polymorphisms were alike between Cvi and Col. Included among these polymorphisms is the Ile-219 to Thr-219 substitution in Ler and two other polymorphisms in noncoding regions (Fig. 3B and Table 5).
Public gene expression data (Max Planck Institute of Developmental Biology, http://csbdb.mpimp-golm.mpg.de/index.html) and studies in our own laboratory (data not shown) using the Affymetrix (Santa Clara, CA) ATH1 GeneChip indicate that At3g01310 is expressed in a number of tissues, including developing siliques and seeds, where gene expression would be most likely to impact tocopherol levels in mature seed. However, the ATH1 array element probe set for At3g01310 corresponds only to the last three exons of the predicted 31 exon gene and, as described in Discussion, this probe set may not accurately reflect gene expression from the entire 7.8-kb region of At3g01310. A SAIL transgenic line (SAIL244D05) with a T-DNA insertion 215 bp upstream of the predicted protein start codon of At3g01310 did not significantly affect seed γ-tocopherol levels (96.9%; see Table 4). However, it is not currently known whether this particular mutation affects transcription of the At3g01310 gene in developing seed. Other T-DNA insertions in the coding region of At3g01310 available from the GABI-Kat collection are currently under investigation.
Discussion
We have undertaken a quantitative genetic approach in A. thaliana to identify the molecular basis of heritable natural variation in seed tocopherol levels between three different accessions. Fourteen QVE loci have been identified that impact tocopherol content and composition in seed in two Arabidopsis RIL populations. The location, heritability, and impact of five of these QVE loci have been confirmed in NILs, and in one case fine-mapping has identified an 8.5-kb interval containing two genes, At3g01310 and At3g01300, that had not been previously implicated in controlling seed tocopherol levels.
Most of the QVE loci identified predominantly affect levels of a single tocopherol and in general do not impact multiple tocopherols. Possible exceptions are QVE4, QVE5, and QVE6, which impact δ-, γ- and α-tocopherols, respectively, and overlap as a complex QTL on chromosome 2. Additional fine-mapping will be required to determine whether QVE4–QVE6 are due to the activity of one or more genes. Although δ-tocopherol was not evaluated in the Cvi/Ler RIL population, δ-tocopherol levels also appear to be affected by the γ-tocopherol QTL, QVE7 (Table 2).
Few of the QVE identified were shared between the two RIL populations, despite the fact that Ler is a common parent, which is not uncommon and has been previously reported with these two RIL populations (28). For the majority of the QVE, this classification will be helpful in distinguishing which polymorphisms are relevant as the underlying cause of the QTL, because only those specific to the accession in which the QTL is present should be of consequence. It is also possible that QTL detection sensitivity may not have been equivalent in the two RIL populations or that environmental/genotype interactions contributed to differences in resolving QTL in the two RIL populations.
One major advantage of studying the natural variation of tocopherols in Arabidopsis is that, by integrating the genetic, genomic, and molecular tractability of the organism with prior work on tocopherol biosynthetic genes, one can readily determine whether a known biosynthetic gene is a candidate for a specific QVE. This is the case for 6 of the 14 QVEs identified, QVE1, QVE3, QVE4–QVE6, and QVE9, which contain known tocopherol biosynthetic pathway genes in their intervals [hydroxyphenylpyruvate dioxygenase (HPPD), VTE4, VTE2, and VTE5, respectively]. NILs and initial fine-mapping of QVE3, QVE4, QVE5, and QVE6 has confirmed association of the tocopherol phenotype with the VTE4 and VTE2 candidate genes in these intervals. One can readily rationalize how allelic variation at VTE4, which encodes γ-tocopherol methyltransferase, could impact α-tocopherol levels in QVE3 given that seed-specific overexpression of VTE4 led to an 80-fold increase in seed α-tocopherol and a corresponding decrease in its substrate, γ-tocopherol (9). It is less straightforward to understand how allelic differences at the HPPD locus in the QVE1 interval could be the basis of altered seed α-tocopherol levels; overexpression of HPPD modestly increased total seed tocopherol levels but had little effect on α-tocopherol content (29). Determining whether allelic variation at known tocopherol biosynthetic loci is indeed responsible for the phenotypes of QVE1, QVE3–QVE6, and QVE9 must remain the subject of future analysis.
Eight other QVE loci localize to areas of the genome where no known tocopherol biosynthetic genes occur and suggest that there are several additional genes that impact tocopherol levels in seed. This identification of loci affecting seed tocopherol levels demonstrates a key aspect of QTL analysis: It allows researchers to cast a wide net that is complemented, rather than constrained, by prior work in the area. The QVE locus with the most advanced fine-mapping, QVE7, has delineated the responsible gene to an 8.5-kb interval on the top of chromosome 3. Two genes are contained within this interval, the 5′ half of At3g01300 and almost all of At3g01310. Although recombination, sequence data, and At3g01300 mutational data are most consistent with the At3g01310 gene being QVE7, determining the molecular mechanism of the accession alleles is not straightforward.
The predicted At3g01310 gene is extremely large (7.8 kb and 31 exons), and the data are still ambiguous as to whether it encodes a single gene. The last 12 exons are represented by abundant 5′ and 3′ ESTs, and the first 4 exons also are represented by several ESTs, but no transcripts have been identified that encompass the entire predicted 1,057-aa coding region (http://signal.salk.edu/cgi-bin/tdnaexpress and www.arabidopsis.org). Interestingly, the Munich Information Center for Protein Sequence predicts that the At3g01310 gene starts at position 97,083, which would encode a 498-aa protein (56.3 kDa) (http://mips.gsf.de/proj/thal/db). A full-length transcript, U23068, initiates here, giving support to this model, and there are no known transcripts in this region that extend beyond the predicted 5′ end of the shorter gene. This shorter predicted gene structure would eliminate amino acids 1–558 of the 1,057 aa predicted for the At3g01310 protein, including the two coding-sequence differences between Ler and Cvi and the histidine acid phosphatase signature at amino acids 370–384. The only nucleotide change likely to be of consequence in the shorter predicted gene structure is polymorphism 44 (Table 3), which would affect a predicted intron splice junction in Cvi. The only currently available T-DNA knockout mutant in the interval, 215 bp upstream of the start codon of the predicted 7.8-kb At3g01310 gene, does not impact the levels of seed tocopherols in the Col accession when homozygous. However, several additional T-DNA insertional mutants at various locations internal to the predicted 7.8-kb (and shorter) At3g01310 transcript should help to illuminate the functionality of the locus. Finally, tiling array data (http://signal.salk.edu/cgi-bin/atta) do not show a clear-cut unidirectionality of transcription over the At3g01310 gene, and, because only exons 28–31 are represented on the Affymetrix ATH1 chip, we cannot use public expression data for expression in other regions of the predicted 7.8-kb At3g01310 gene. Detailed transcript analysis of this region will need to be undertaken to identify the length, orientation, and abundance of transcripts in this region in developing seeds of Ler and Cvi.
Plant seeds are the major food source for the world's population, especially in developing countries where, because of necessity, the seed of a single crop is often the primary dietary food component. However, such over-reliance on a single food crop can have devastating impacts on human health, because no single crop provides the necessary balance of essential macro- and micronutrients, resulting in the prevalence of severe nutrient deficiencies. An important target of research in plant metabolism in the coming decade is to understand at a fundamental level the biosynthesis, acquisition, transport, and storage of essential macro- and micronutrients, with the goal of using this information to drive the development of world crops that provide more balanced nutrition. The benefit of QTL analysis in Arabidopsis is that the experimental tractability of the system allows relatively rapid progress in understanding the molecular basis of variation for the trait. Such studies have significant potential to increase our understanding of how the metabolism of essential micronutrients is regulated and should yield novel targets for metabolic engineering and, in this case, for engineering increased vitamin E in crop plants.
Materials and Methods
Plant Growth and Materials.
A population of 154 Cvi/Ler RILs (30) and 58 Col/Ler RILs (31) were grown on soil in growth chambers, and seed was analyzed for tocopherol content. Plants were grown on soil in 16:8-h light/dark, 22°C/18°C cycles, with ≈100 μE·m−2·s−1 of light. RIL population plants were grown in pairs in 3-in pots (1 in = 2.54 cm). Genotyping and selection of homozygous recombinants were conducted early on seedlings such that only one plant reached rosette stage per pot, with parental controls being treated similarly. Harvested seeds were allowed to dry in a desiccator at least two weeks before extraction for HPLC analysis.
NILs G7, G1A, G4H, G4G, and G3B were created by backcrossing the recombinant inbred line CVL6 (CS22005) to Ler three times, with selective genotyping at each generation to maintain Cvi alleles in the region of the predicted QTL (21–27 Mb on chromosome 1) and Ler elsewhere in the genome. NILs 9Y, 9E, and 9P were similarly created with the RIL CL386 (CS1992) and maintain a portion of Col chromosome 2 introgressed into the Ler background. NILs LN26, 4-212, 4-214, and 58-77 were developed as described in ref. 22. NILs 8D1, 6A6, 8E5, 8FL, and others not shown were developed by backcrossing LN26 to Ler and screening for F2 recombinants within the Cvi region. Fine-mapping situated the Cvi/Ler boundary of NIL LN26 between 599 and 640 kb at the top of chromosome 3 (data not shown) (22).
Tocopherol Extraction and HPLC Analysis.
Tocopherols were extracted from dried mature Arabidopsis seed as described (32). Reversed-phase HPLC separation of the tocopherols and quantitative analysis of tocopherol composition were performed as described in ref. 33 with the following modifications. The extract was resuspended in acetonitrile and injected on a C18 Sherisorb ODS-2, 250- × 4.6-mm, reversed-phase HPLC column (Column Engineering, Ontario, CA), and the 0–66.6% B portion of the gradient was performed for 0–13 min. Tocopherols were detected and quantified by fluorescence at 290-nm excitation and 325-nm emission. Tocol was used as an internal standard to correct for sample recovery. Tocopherol levels were quantified relative to known standards by using five-point standard curves, with regression curves generally >0.995.
QTL Mapping.
Individual and total tocopherol levels from each RIL population were used as quantitative values in QTL analysis. MapQTL 4.0 (Plant Research International, Wageningen University and Research Centre, Wageningen, The Netherlands) was used for initial interval mapping for both Cvi/Ler and Col/Ler populations. Permutation (1,000) analyses were performed to calculate the critical lod score for each trait in each population; the significant lod scores (P = 0.05) varied from 2.4 to 2.7 for the different traits. Locus data for the Cvi/Ler RIL population (30) and for the Col/Ler RIL population (31) (http://arabidopsis.info/new_ri_map.html) were used for the analysis. PLABQTL was used for composite interval mapping (34). Because of the high density of the markers in the Col/Ler RIL population (31), loci with less complete scoring in highly dense regions were omitted to reduce nonoptimal exclusion by PLABQTL due to colinearity. Cofactors used for calculation were automatically chosen by the PLABQTL program, and a critical lod score of 2.5 was preset. Any QTL that failed to exceed a critical lod score determined to be >2.5 for that trait by permutation analysis have been omitted.
Genotyping.
PCR template DNA was prepared as in ref. 35 with following modifications. Fresh leaf or floral tissue was ground with 4-mm glass beads in 0.25 M sodium hydroxide in 1.1-ml microtubes (Dot Scientific, Inc., Burton, MI) by a paint shaker. The majority of markers used were simple sequence-length polymorphisms obtained from the Arabidopsis Information Resource database or designed around insertions/deletions between Ler and Col obtained from the Monsanto (St. Louis, MO) polymorphism collection. Amplified products were resolved on a TBE (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8) agarose gel containing 2% ultrapure agarose and 2% NuSieve GTG agarose (Cambrex, Rockland, ME). Cleavage-amplified polymorphic sequence markers were designed according to ref. 22 or were designed from polymorphisms identified by sequencing. Homozygous T-DNA insertion lines were identified by PCR using the T-DNA LB primer and flanking primers designed from wild-type Columbia sequence.
Sequencing.
DNA from Ler, Col, and NIL 8E5 was isolated by using a modified version of the “shorty” protocol described by the University of Wisconsin Arabidopsis Knockout Facility (www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/FindingYourPlant.asp) as follows: leaf tissue was ground at room temperature with an Eppendorf pestle in the described extraction buffer, extracted twice in phenolchloroform, precipitated, and washed once before resuspension in water. Equal amounts of DNA from three amplification PCRs were pooled and then treated with ExoSAP-IT (Amersham Biosciences/USB, Cleveland, OH). DNA sequencing was performed by the Research Technology Support Facility at Michigan State University (East Lansing, MI). Sequences were aligned to the published Col backbone (www.arabidopsis.org) by using the SeqManII program from DNASTAR, Inc. on a Macintosh computer.
Supplementary Material
Acknowledgments
We thank Kamal Swarup (University of Nottingham, Nottingham, United Kingdom) and Narasimham Upadyayula, Torbert Rocheford, and Martin Bohn (University of Illinois at Urbana–Champaign) for assistance with MapQTL and PLABQTL. We also thank Michael Grusak and members of his laboratory (Baylor College of Medicine, Houston, TX) and Scott Sattler (Michigan State University, East Lansing) for assistance with RIL populations. This work was supported by grants (to D.D.P.) from the U.S. Agency for International Development, HarvestPlus, and the Grand Challenges in Global Health initiative.
Abbreviations
- QTL
quantitative trait loci
- NIL
near-isogenic line
- RIL
recombinant inbred line
- Cvi
Cape Verde Islands accession
- Col
Columbia accession
- Ler
Landsberg erecta accession
- Mb
megabase.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “From Functional Genomics of Model Organisms to Crop Plants for Global Health,” held April 3–5, 2006, at The National Academy of Sciences in Washington, DC. The complete program is available on the NAS web site at www.nasonline.org/functional_genomics.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Hossain T, Rosenberg I, Selhub J, Kishore G, Beachy R, Schubert K. Proc Natl Acad Sci USA. 2004;101:5158–5163. doi: 10.1073/pnas.0401342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinon V, Ravanel S, Douce R, Alban C. Plant Physiol. 2005;139:1666–1676. doi: 10.1104/pp.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellapenna D, Pogson B. Annu Rev Plant Biol. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- 4.Ajjawi I, Shintani D. Trends Biotechnol. 2004;22:104–107. doi: 10.1016/j.tibtech.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Diaz de la Garza R, Quinlivan EP, Klaus SM, Basset GJ, Gregory JF, III, Hanson AD. Proc Natl Acad Sci USA. 2004;101:13720–13725. doi: 10.1073/pnas.0404208101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, Drake R. Nat Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 7.Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 8.DellaPenna D. Trends Plants Sci. 2005;10:574–579. doi: 10.1016/j.tplants.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Shintani D, DellaPenna D. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- 10.Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porfirova S, Bergmuller E, Tropf S, Lemke R, Dormann P. Proc Natl Acad Sci USA. 2002;99:12495–12500. doi: 10.1073/pnas.182330899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D. Plant Cell. 2003;15:2343–2356. doi: 10.1105/tpc.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi Q, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, Gruys KJ, Last RL. Plant Cell. 2006;18:212–224. doi: 10.1105/tpc.105.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DellaPenna D, Last RL. Physiol Plant. 2006;126:356–368. [Google Scholar]
- 15.Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, et al. Plant Cell. 2003;15:3007–3019. doi: 10.1105/tpc.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collakova E, DellaPenna D. Plant Physiol. 2003;133:930–940. doi: 10.1104/pp.103.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanwischer M, Porfirova S, Bergmuller E, Dormann P. Plant Physiol. 2005;137:713–723. doi: 10.1104/pp.104.054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marwede V, Gul MK, Becker HC, Ecke W. Plant Breeding. 2005;124:20–26. [Google Scholar]
- 19.Wong JC, Lambert RJ, Tadmor Y, Rocheford TR. Crop Science. 2003;43:2257–2266. [Google Scholar]
- 20.Marwede V, Schierholt A, Mollers C, Becker HC. Crop Science. 2004;44:728–731. [Google Scholar]
- 21.Galliher HL, Alexander DE, Weber EJ. Crop Science. 1985;25:547–549. [Google Scholar]
- 22.Bentsink L, Yuan K, Koornneef M, Vreugdenhil D. Theor Appl Genet. 2003;106:1234–1243. doi: 10.1007/s00122-002-1177-9. [DOI] [PubMed] [Google Scholar]
- 23.Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. Plant Physiol. 2003;131:345–358. doi: 10.1104/pp.102.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vreugdenhil D, Aarts MGM, Koornneef M, Nelissen H, Ernst WHO. Plant Cell Environ. 2004;27:828–839. [Google Scholar]
- 25.Zhang ZY, Ober JA, Kliebenstein DJ. Plant Cell. 2006;18:1524–1536. doi: 10.1105/tpc.105.039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jander G. Methods Mol Biol. 2006;323:115–126. doi: 10.1385/1-59745-003-0:115. [DOI] [PubMed] [Google Scholar]
- 27.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 28.Ungerer MC, Halldorsdottir SS, Purugganan MD, Mackay TF. Genetics. 2003;165:353–365. doi: 10.1093/genetics/165.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsegaye Y, Shintani DK, DellaPenna D. Plant Physiol Biochem. 2002;40:913–920. [Google Scholar]
- 30.Alonso-Blanco C, El-Assal SED, Coupland G, Koornneef M. Genetics. 1998;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 32.Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D. Plant Cell. 2003;15:1320–1332. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L, DellaPenna D. Plant Mol Biol. 2001;47:379–388. doi: 10.1023/a:1011623907959. [DOI] [PubMed] [Google Scholar]
- 34.Utz HF, Melchinger AE. J Agric Genomics. 1996;2:1. [Google Scholar]
- 35.Gilliland LU, McKinney EC, Asmussen MA, Meagher RB. Genetics. 1998;149:717–725. doi: 10.1093/genetics/149.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.