When transposable elements (TEs) were discovered in maize by Barbara McClintock >50 years ago they were regarded as a curiosity; now they are known to be the most abundant component of probably all eukaryotic genomes. They account for almost 50% of the human genome and >70% of the genomes of some grass species, including maize. As such, they make up the vast majority of the output of genome sequencing projects. The availability of so much new information has fueled a revolution in their analysis and studies of their interaction with the host.
In addition to discovering TEs, McClintock also uncovered disparate ways that TEs can alter genetic information. At one end of the spectrum she found that TEs could restructure genomes through element-mediated chromosomal rearrangements. At the other end she and others found they could generate new alleles by inserting into and around genes and altering their expression. Thus, the presence and extraordinary abundance of TEs in eukaryotic genomes promote a myriad of genome-altering events.
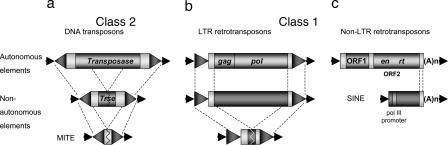
TEs are fragments of DNA that can insert into new chromosomal locations, and they often make duplicate copies of themselves in the process. Eukaryotic TEs are divided into two classes, according to whether their transposition intermediate is RNA (class 1) or DNA (class 2) (Fig. 1). For all class 1 elements, the element-encoded transcript (mRNA) forms the transposition intermediate. In contrast, with class 2 elements, the element itself moves from one site to another in the genome.
Fig. 1.
Structural features and classification of eukaryotic TEs. Elements are divided into two classes, according to whether their transposition intermediate is RNA (class 1) (b and c) or DNA (class 2) (a). Class 1 elements are further divided into two groups on the basis of transposition mechanism and structure: LTR retrotransposons (b) and non-LTR retrotransposons (c). Each class contains autonomous and nonautonomous elements. Autonomous elements encode proteins required for transposition (gag, capsid-like protein; pol, reverse transcriptase; ORF1, a gag-like protein; en, endonuclease; rt, reverse transcriptase). Nonautonomous elements do not encode these proteins but retain the cis sequences necessary for transposition. TSDs are the black arrowheads flanking each element, and the inverted repeats at the termini of class 2 elements (a) and the direct repeats at the ends of LTR retrotransposons (b) are represented by large gray arrowheads. A color version of this figure originally appeared in box 1 of ref. 1.
Each group of TEs contains autonomous and nonautonomous elements. Autonomous elements have ORFs that encode the products required for transposition. In contrast, nonautonomous elements do not encode transposition proteins but are able to transpose because they retain the cis sequences necessary for transposition. Integration of almost all TEs results in the duplication of a short genomic sequence (called a target site duplication, or TSD) at the site of insertion.
Eukaryotic DNA (class 2) transposons usually have a simple structure with a short terminal inverted repeat (TIR) (≈10–40 bp, but can be up to ≈200 bp) and a single gene encoding the transposase. Transposase binds in a sequence-specific manner to the ends of its encoding element and to the ends of nonautonomous family members. Once bound, transposase initiates a cut-and-paste reaction whereby the element is excised from the donor site (generating an “empty site”) and inserted into a new site in the genome. The elements studied by McClintock, including the Ac/Ds and Spm/dSpm families, are DNA transposons capable of insertion and excision.
Class 1 retroelements can be divided into two groups on the basis of transposition mechanism and structure. LTR retrotransposons have long terminal repeats (LTRs) in direct orientation that can range in size from ≈100 bp to several kilobases. Autonomous elements contain at least two genes, called gag and pol. The gag gene encodes a capsid-like protein, and the pol gene encodes a polyprotein that is responsible for protease, reverse transcriptase, RNase H, and integrase activities. An element-encoded transcript that initiates from a promoter in the 5′ LTR and terminates in the 3′ LTR is transported to the cytoplasm. There it serves as both mRNA and template for double-stranded cDNA that is transported into the nucleus where it can then integrate into the genome. The host can mitigate this increase in genome size by mediating homologous recombination between the identical or near-identical LTRs of full-length elements, generating a much shorter solo LTR. LTR retrotransposons compose the largest fraction of most plant genomes, where they appear to be the major determinant of the tremendous variation in genome size.
Non-LTR retrotransposons are divided into the autonomous long interspersed elements (LINEs) and the nonautonomous short interspersed elements (SINEs). LINEs encode two ORFs, which are transcribed as a bicistronic mRNA composed of ORF1 (an RNA binding protein) and ORF2 (endonuclease and reverse transcriptase activities). Both LINEs and SINEs terminate by a simple sequence repeat, usually poly(A). LINE transcripts initiate at a promoter within the 5′ end of the element and terminate at or often downstream of the simple repeat sequence. SINEs are characterized by an internal RNA pol III promoter near the 5′ end. SINEs are a heterogeneous group of elements that range in length from 90 to 300 bp and are derived either from a variety of tRNA genes or from 7SL RNA.
An unexpected finding from the analysis of genome sequences is that TE content varies from species to species in two important ways: by the classes of TEs present and their fractional representation in the genome and by the level of TE activity. The yeast Saccharomyces cerevisiae has only LTR retrotransposons (called Ty elements), whereas class 1 non-LTR retrotransposons predominate in mammalian genomes, with class 2 DNA transposons making up <5% of the TE fraction. The genomes of flowering plants, including both monocots (e.g., grasses such as rice and maize) and dicots (e.g., Arabidopsis and tomato), have a rich collection of both class 1 and class 2 elements, with LTR retrotransposons comprising the largest fraction of most characterized genomes.
Genome-wide activity of TEs also varies from species to species. Given the rich genetic analysis of TE-mediated mutations in maize, Drosophila melanogaster, and Caenorhabditis elegans, it is not surprising that these genomes were found to contain many young, active TE families. Flowering plants, especially members of the grass clade (e.g., rice, maize, barley, and wheat), are in an epoch of TE-mediated genome diversification, with the participation of many families of active class 1 and class 2 elements. In contrast, extant mammalian genomes have many fewer active TE lineages; however, TE activity appears to vary significantly between species.
The eight research articles in this Special Feature by no means cover the breadth of current TE research. Rather, they illustrate several prevalent themes of TE analysis. One theme is the integration of computer-assisted searches of genome sequence databases into the analysis of TEs and their impact on the genome. A second theme that goes hand-in-hand with the expanding databases is the development of new experimental methodologies that permit genome-wide analysis of the dynamic relationship between TEs and their host. Another theme is the multidisciplinarity of modern TE research. Four sections of this journal, Biochemistry, Evolution, Genetics, and Plant Biology, are represented among the eight research papers in this Special Feature.
Footnotes
The author declares no conflict of interest.
References
- 1.Feschotte C, Jiang N, Wessler SR. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]