Abstract
Despite the prevalence of transposable elements in the genomes of higher eukaryotes, what is virtually unknown is how they amplify to very high copy numbers without killing their host. Here, we report the discovery of rice strains where a miniature inverted-repeat transposable element (mPing) has amplified from ≈50 to ≈1,000 copies in four rice strains. We characterized 280 of the insertions and found that 70% were within 5 kb of coding regions but that insertions into exons and introns were significantly underrepresented. Further analyses of gene expression and transposable-element activity demonstrate that the ability of mPing to attain high copy numbers is because of three factors: (i) the rapid selection against detrimental insertions, (ii) the neutral or minimal effect of the remaining insertions on gene transcription, and (iii) the continued mobility of mPingelements in strains that already have >1,000 copies. The rapid increase in mPing copy number documented in this study represents a potentially valuable source of population diversity in self-fertilizing plants like rice.
Keywords: genome evolution, miniature inverted-repeat transposable element, transposon
Transposable element (TE)-mediated insertional mutagenesis has been recognized since McClintock's analysis of spotted corn kernels >60 years ago (1, 2). Although originally thought to be rare, genome-sequencing projects have revealed that most wild-type mammalian and plant genes harbor TE insertions in their introns and/or 5′ and 3′ untranslated regions (reviewed in ref. 3). Approximately 80% of human genes contain the long interspersed element L1, and >200,000 of the 1 million copies of the short interspersed element Alu are in our genes (4). At this time, very little is known about the actual role, if any, of TE sequences in the regulation of individual human genes. Furthermore, because most of the insertions occurred >5 million years ago, it is likely that their impact on gene expression will never be known because of the subsequent accumulation of additional mutations.
Many plant genes, in contrast, contain members of TE families that have recently proliferated and, as such, provide opportunities to observe the impact of TEs on gene expression and allelic diversification (5–7). Miniature inverted-repeat TEs (MITEs) are the predominant TE associated with plant genes. Like other plant class 2 TEs, MITEs are preferentially found in single-copy regions of the genome (3, 6, 7). What appears to distinguish MITEs from other class 2 elements is how they originate and amplify. The majority of characterized nonautonomous class 2 element families are >1 kb in length and can amplify to moderate copy number (usually <50 copies in a genome) after they arise by deletion from an autonomous element. In contrast, MITEs are short (≈100–500 bp) and appear to quickly amplify from one or a few elements to >1,000 (6, 7).
Plant genes have, on average, very short introns (≈200 bp, although some are >3 kb) when compared with their mammalian counterparts (≈2.5 kb on average) (8, 9). There is some evidence that plants cannot efficiently splice long introns: a requirement for short introns may be one reason short elements such as MITEs (and, less frequently, short interspersed elements) predominate in plant genes (8, 9). That is, there is a good chance that a MITE insertion into a plant gene will not disrupt gene expression. This is apparently the case, because there are hundreds of wild-type genes in databases with MITEs in their noncoding regions as well as alleles that differ, in part, by the presence or absence of MITEs (10). However, as with the TE insertions in human genes, the MITE polymorphism is one of several differences between alleles, making it virtually impossible to determine whether insertion altered gene expression.
To assess the impact of MITE insertions on gene and genome evolution, it first was necessary to identify MITEs that are still transposing. Such a family was recently found in rice (11–13). The rice genome is being sequenced because rice is the most important source for calories in humans and, fortuitously, because rice has the smallest genome among the cereals (≈430 Mb; maize ≈2,500 Mb; barley ≈5000 Mb) (14, 15). Whole-genome draft sequences are available for japonica and indica, two of the three subspecies of rice that have been independently domesticated from wild relatives (14, 15). Recent identification of the first active MITE, mPing, opened the door to addressing questions concerning the impact of MITE insertions into plant genes (11–13). However, although mPing is clearly an active MITE, its copy numbers were found to be relatively low, with <10 copies in the subspecies indica and from 1 to ≈50 copies in the subspecies japonica (11, 12). Thus, the available mPing-containing strains are not very useful in the design of experiments to understand how MITEs attain very high copy numbers and how they impact host gene expression.
Actively transposing mPing elements were discovered independently in three laboratories working with three different sources of plant material: long-term cell culture (11), newly derived anther culture (12), and strains derived from the temperate japonica cultivar Gimbozu EG4 after γ-irradiation (13). Further analysis of one irradiated strain designated IM294 led to the isolation of an mPing element that had transposed into an exon of the Rurm1 gene (13). A second mutant allele isolated from another irradiated derivative of EG4 (HS110) was subsequently shown to contain an mPing insertion in an intron of Hd1, the rice homolog of the Arabidopsis floral-timing regulator, CONSTANS (16). In this instance, the insertion of mPing into the intron reduced, but did not abolish, gene activity.
The objective of this study was initially to understand the activation of mPing by irradiation of the rice strain Gimbozu EG4. In the course of this analysis, we found that mPing had amplified from ≈50 to >1,000 copies in EG4 and, independently, in several related landraces. Approximately 300 of the insertion sites in the rice genome were characterized with respect to their target sequence and proximity to a rice annotated gene. Most importantly, we determined that mPing activation did not require prior irradiation of EG4. That is, mPing was actively transposing and significantly increasing its copy number in EG4 and in related landraces, despite the fact that each of these strains contained ≈1,000 mPing elements.
Results
Determining mPing Copy Number
As a first step in understanding how irradiation may have activated mPing, we focused on Gimbozu EG4, the progenitor of the irradiated strain IM294. Gimbozu EG4 was derived from the cultivar Gimbozu, which, itself, was derived from another cultivar, Aikoku (in 1907), as the lone plant that remained erect after a storm caused all of the other plants in the field to lodge. The popularity of Aikoku and Gimbozu cultivars with Japanese farmers in the first half of the 20th century led to the use of these cultivars throughout the country and the breeding of many distinct but closely related strains and landraces.
In the course of analyzing the copy number of mPing elements among these landraces and in other rice cultivars, we discovered a dramatic amplification in four strains. The first indication of very high mPing copy number was the observation of smear hybridization when labeled mPing was used to probe DNA blots containing digested genomic DNA from 25 strains, including 21 landraces of Aikoku and Gimbozu (Fig. 1A). The very high copy numbers in these strains precluded the use of DNA blot hybridization to resolve individual copies. Instead, transposon display (TD) was used to provide a better estimate of copy number. TD is a modification of the amplified fragment-length polymorphism technique (17), where genomic DNA is digested, ligated to an adapter, and used as template for PCR with a labeled primer that anneals, in this case, near one end of mPing elements (11, 18, 19). All DNA fragments that are visualized on TD gels should have an adapter primer at one end and an mPing element at the other end.
Fig. 1.
Copy-number estimation of mPing elements in rice strains. (A) DNA blot of rice genomic DNAs digested with EcoRI and probed with digoxygenin-labeled mPing from the following strains: 1, Nipponbare; 2, Gimbozu EG4; 3, IM294 (irradiated mutant of EG4); 4, Kasalath (indica); 5, A126; 6, A127; 7, A135; 8, A161; 9, A183; 10, G193, 11, A103; 12, A105, 13, A106; 14, G172; 15, G175; 16, A176; 17, A101; 18, A102; 19, A104; 20, G185; 21, G190; 22, G174; 23, A157; 24, A119; 25, A123. Strains named with A are Aikoku-landraces and with G are Gimbozu-landraces. (B) Autoradiograph of a transposon display gel of mPing amplicons in Aikoku and Gimbozu strains. Each DNA sample was digested with MseI, ligated to an adapter, and used as template for PCR with an mPing primer and an adapter primer containing one (+G) selective base. Lane labels refer to the numbers in A and asterisks in A and B indicate the high-copy-number strains. (C) Copy-number estimates derived from transposon display gels. See Results for details.
A TD gel of genomic DNA from 12 strains is shown in Fig. 1B. For all strains shown, an adapter primer with one selective base was used along with the mPing primer. Because there were too many bands for an accurate count in the high-copy-number strains (Fig. 1B, lanes 2 and 23–25), we used adapter primers with three selective bases for the high-copy-number strains and only a single selective base for the other strains (data not shown). The number of copies of mPing was estimated for the low-copy strains by counting all of the bands generated by using the four possible primer combinations. (Table 2, which is published as supporting information on the PNAS web site). The accuracy of this method for estimating copy number is evident by comparing the number of bands observed for the Nipponbare control (51 copies) with the actual number of elements in the virtually complete genome sequence (50 copies) (14). In contrast, the number of mPing elements in the high-copy-number strains (except EG4) was estimated by averaging the number of bands per primer by using 14 of the 64 possible primer combinations and multiplying this number by 64. For EG4, all 64 primer combinations were used, and all bands were counted. This direct method produced a copy number (1,163) that was not significantly different from the estimated copy number extrapolated from the results with 14 primers (1,198) (Table 3, which is published as supporting information on the PNAS web site). As shown in Fig. 1C, the high-copy-number strains harbor between ≈840 and ≈1,163 elements.
Clues to the relationships among these strains and the dynamics of mPing amplification can be deduced by comparing the patterns of bands on TD gels. For example, with two exceptions (A101 and A102; lanes 17 and 18 in Fig. 1B), all of the low-copy-number strains and Nipponbare (Fig. 1, lane 1) share many of their bands, thus revealing their very close genetic relationship. This finding is consistent with their breeding records; Gimbozu is a grandparent of a strain (Norin 22) that was used to generate Nipponbare. These data indicate that the ancestral Gimbozu genome had an mPing distribution much like what we see in extant Nipponbare. Thus, mPing elements have not been transposing in these lines for almost a century. In contrast, close examination of the TD band patterns of the high-copy-number strains revealed very few comigrating bands among the hundreds of bands examined in each strain (Fig. 1B, compare lanes 2 and 23–25 and data not shown). These data suggest that mPing amplification occurred independently in each of the high-copy-number strains. This scenario is consistent with the fact that these strains were cultivated in different geographically isolated regions of Japan.
mPing Insertion Sites in Rice.
The availability of most of the genomic sequence from two rice subspecies (14, 15), along with a large collection of full-length cDNAs (20), permitted a determination of the earliest stages of integration of a high-copy-number element family into or close to plant genes. To this end, we determined the flanking host sequences of a random subset of mPing insertions in the high-copy-number strains EG4, A119, and A123 by sequencing bands from TD gels after their recovery and reamplification. All of the bands recovered from the high-copy strains are referred to as “new insertions” to reflect the recent amplification of mPing in these strains. Recovered sequences were used to query the annotated rice genome database of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Like other Tourist-like MITEs, mPing targets the trinucleotide TAA (and its complement TTA), which is then duplicated upon insertion (Fig. 2). Because another Tourist-like MITE, the maize mPIF element, has a significant preference for a 9-bp target site (that includes and flanks TAA/TTA) (21), our analysis was extended to the three nucleotides upstream and downstream of the trinucleotide. A weak preference was evident for the sequence C/T T A/C T T/A A G/T A A/G (Fig. 2). Notably, the much stronger 9-bp preference of the mPIF element is the same as that of mPing.
Fig. 2.
Target-site preference of mPing insertions. A pictogram was constructed by using the nine nucleotides from 123 insertion sites. In this representation, the size of the letter is proportional to its frequency at a given position (generated at http://genes.mit.edu/pictogram.html by using default parameters). The gray rectangle indicates the trinucleotide duplicated upon insertion.
An extensive analysis of the genomic “neighborhoods” of 280 insertions was also undertaken, and the results are summarized in Table 1. An insertion site was considered to be in a single-copy region of the rice genome if the BLAST search hit only one BAC clone (with the exception of overlaps), whereas queries with more than two significant hits were considered to be in repetitive regions (22). In addition, the proximity of each insertion site to a transcribed region was assessed by extracting 5 kb flanking each mPing end and using these sequences to query the full-length cDNA collections in the Knowledge-Based Oryza Molecular Biological Encyclopedia (KOME, http://cdna01.dna.affrc.go.jp/cDNA) (see Materials and Methods) (20).
Table 1.
Characteristics of mPing insertion sites
| Strain | No. of analyzed insertions | Insert into | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Single copy, n(%)* | Exon | Intron | 0–1kb† | 1–3 kb† | 3–5kb† | >5 kb† | |||
| Nipponbare | 50 | 42 (84) | 1 | 3 | 9 | 7 | 2 | 20 | |
| EG4 | 221 | 201 (91) | 1 | 10 | 38 | 69 | 44 | 39 | |
| A119 | 26 | 24 (92) | 0 | 0 | 2 | 8 | 3 | 11 | |
| A123 | 33 | 31 (94) | 0 | 0 | 6 | 10 | 4 | 11 | |
| Total | 280 | 256 (91) | 1 | 10 | 46 | 87 | 51 | 61 | |
| Control | 300 | 189 (63) | 26 | 24 | 23 | 45 | 23 | 48 | |
| EG4 (de novo) | 23 | 23 (100) | 3 | 0 | 3 | 4 | 4 | 9 | |
* The number of insertions into single-copy sequences.
†Thes distance of the insertion sites from cDNA coding sequences.
The vast majority of the analyzed insertions were into single-copy regions and near transcribed DNA. For EG4, 201 of 221 mPing insertion sites (91%) were in single-copy regions (Table 1). Although fewer insertions from strains A119 and A123 were recovered, >90% of those analyzed were in single-copy regions. With regard to genic proximity, 70% of the new mPing insertions (195 of 256) are <5 kb from a cDNA coding sequence (Fig. 3), Table 1, and Table 4, which is published as supporting information on the PNAS web site), and of these 195 insertions, 1 is in an exon, 10 are in introns, and 46 are within 1 kb of a coding region. Although there are very few new insertions into genes, there are significantly more new insertions within 5 kb of a gene (70%) than the presumably older insertions in Nipponbare (44%; P = 0.002) (Tables 1 and 4; and see Table 5, which is published as supporting information on the PNAS web site).
Fig. 3.
mPing insertions in or near rice genes. mPing insertions are represented as black triangles, and exons and UTRs are gray and white boxes, respectively, that are connected by introns. Black arrowheads under the boxes represent the positions of primers used for PCR in (Fig. 4)
A control experiment was done to determine whether new insertions were preferentially in genic and/or single-copy regions. To this end, the genomic neighborhood of the new mPing insertions was compared with that of 300 randomly chosen control sequences, and the data were subjected to a contingency analysis (Tables 1 and 5). All rice strains analyzed showed significantly higher frequencies of mPing insertions into single-copy regions than the control. In contrast, control fragments were more frequently inside genes (Tables 1 and 5). The significant underrepresentation of genic hits among the new mPing insertion sites suggests that such insertions have been selected against in the very short time since mPing amplification. Rapid removal of genic insertions from the population is most likely explained by the fact that rice is a self-fertilizer and that most loci are homozygous. A PCR analysis of 50 randomly chosen new insertion sites revealed that five insertions were heterozygous (see Materials and Methods and data not shown). Such a situation could arise if an insertion allele is not yet fixed in the population and/or if mPing is still actively transposing (see below).
Transcripts from mPing-Containing Genes.
A previous study had shown that an mPing insertion in an intron of the rice homolog of the CONSTANS gene was responsible for a mutant phenotype, presumably because of aberrant alternative splicing of the mPing-containing intron (16). The impact, if any, of new mPing insertions into or near genes in the high-copy-number strains was explored by comparing transcripts produced by alleles with and without mPing. The alleles chosen represented what we believed were the best candidates for altered gene expression among all of the new insertions analyzed (Fig. 3). To this end, an RT-PCR analysis was undertaken for 13 genes with mPing insertions in exons or introns or within 200 bp of the cDNA hit and, using as template, RNA extracted from the leaves of EG4 and Nipponbare ((Fig. 3), see location of mPing insertions and PCR primers).
Of the 13 genes, 4 were not expressed in the leaves of either strain (Fig. 4). Transcripts were detected for 7 genes, but no significant difference in transcript size or abundance was observed between EG4 and Nipponbare (Fig. 4). For 2 genes (Fig. 4, Fig. 1) and 9), reproducibly lower levels of product were detected in EG4 than in Nipponbare, whereas gene 8 showed reproducibly more product in EG4 than in Nipponbare. Despite these apparent quantitative differences, we were surprised that all of the mPing-containing alleles that could be analyzed were transcribed and that no significant splicing alterations were observed. Additional experiments using RNA isolated from different tissues or from plants grown under a variety of regimens would be required to see whether any of the insertions are responsible for more subtle alterations in transcription.
Fig. 4.
Analysis of transcription from mPing-containing rice genes. (A) RT-PCR products resolved on agarose gels. Lane numbers refer to genes 1–13 in Fig. 3, where the position of PCR primers is shown for each, whereas M is a 100-base ladder. The source of leaf RNA was Nipponbare (N) or Gimbozu EG4 (G). (B) PCR with genomic DNA from Nipponbare (N) or Gimbozu EG4 (G) using the same primer sets as in A.
Transposition of mPing in High-Copy-Number Strains.
Before this study, transposition of mPing had been detected only in cell and anther culture and in the irradiated progeny of strain EG4 (11–13) to our knowledge. However, the different banding patterns of high-copy-number strains observed in this study (Fig. 1B) suggested independent bursts of transposition, whereas the finding that some of the insertion sites in these lines are heterozygous was consistent with continued activity of mPing.
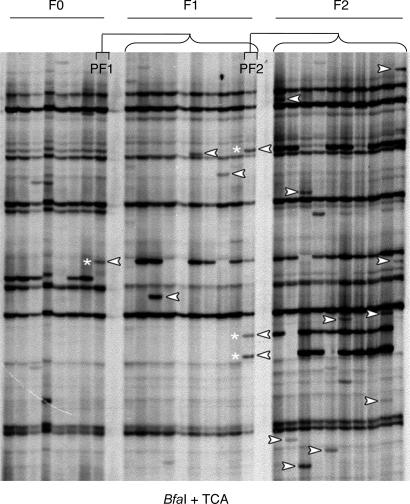
To determine whether mPing was active before irradiation, DNA was isolated for TD analysis from three generations of strain EG4 by using plants propagated by single-seed descent (Fig. 5). Specifically, TD was performed with mPing and adapter primers and, as template, DNAs isolated from eight progeny (F0) of a single selfed parent, the 10 progeny (F1) from one selfed F0 plant (PF1), and the 10 progeny (F2) from one selfed F1 plant (PF2) (Fig. 5). A new mPing insertion should appear as a band that is in one individual but not in its siblings or its parent (Fig. 5). white arrowhead). Such bands are herein referred to as “de novo insertions” to clearly distinguish them from the previously described new insertions (see Table 1). To verify that such bands were de novo insertions of mPing and not artifacts of PCR, a two-step procedure was devised. First, a random subset of putative de novo bands was chosen, reamplified, and sequenced. Next, the flanking sequences were used to guide the design of primers to amplify the insertion site in the parent (PF1) and progeny (F2) (Fig. 6, which is published as supporting information on the PNAS web site). Finally, the proximity of the de novo insertions to rice genes was determined and compared with the mPing-insertion sites in strains EG4, A119, and A123 (the previously described new insertions) (Table 1; and see Table 6, which is published as supporting information on the PNAS web site).
Fig. 5.
Transposon display analysis of mPing insertions of generational material from strain EG4. See Results for details.
Recall that the lower-than-expected number of mPing insertions in exons and introns of the high-copy-number strains (Table 1) suggested that negative selection had already acted to remove most insertions in rice genes (in both exons and introns). If this interpretation were correct, then some of the de novo insertions, which have not been exposed to selection, would be expected to be in rice genes. Analysis of the insertion sites from the 10 progeny (Fig. 5, plants designated F1) of a single parent (Fig. 5), plant PF1) led to sequences at 23 sites that were verified as de novo insertions by the two-step procedure described above (Fig. 5 and Table 1). Among these 23 insertions were 3 into exons of rice genes (Table 6). In addition, PCR analysis revealed that all of the de novo insertions were heterozygous (Fig. 6).
Because plants do not set aside a germ line early in development, insertions that occur during plant growth can be inherited if the cell lineage with the insertion contributes to reproductive structures. To determine whether de novo insertions were inherited, we analyzed the single-seed descent material in 16 separate TD reactions that used 1 of 16 primers with three selective bases (of 64 possible primers). Only 1 of the 16 reactions (BfaI + TCA) is shown in Fig. 5. To estimate the number of de novo insertions in generations F1 and F2, bands observed in only one progeny and not in the parent or siblings were counted, and the (per primer) average (from the 16 reactions) was multiplied by 64. The results of this analysis led to an estimate of 63.2 and 49.2 de novo insertions per plant per generation in the F1 and F2 generations, respectively (Tables 7 and 8, which are published as supporting information on the PNAS web site).
The number of de novo mPing insertions that were inherited from the F1 generation to the F2 generation was estimated by counting the de novo bands appearing in one parent (Fig. 5), PF2) that segregated in the F2 progeny (Fig. 5), asterisk). Based on this analysis, >80% (35 of 43) of the de novo bands in parent PF2 were inherited in the F2 progeny (Tables 7 and 8).
Finally, TD analysis of one parent and 10 progeny from each of two other high-copy-number strains, A119 and A157 (Table 1), also revealed de novo bands when parent and progeny were compared (Fig. 7, which is published as supporting information on the PNAS web site). Verification that a subset of these bands were bona fide de novo insertions indicated that mPing was also actively transposing in these lines (Fig. 6).
Discussion
The ability to analyze the earliest events in the amplification of mPing elements in rice has begun to answer several questions about how MITEs attain high copy numbers despite their preference for insertion into genic regions. Our results indicate that mPing has amplified in two discrete stages: from 1 to ≈50 copies in tropical vs. temperate japonica strains (11), then from ≈50 to 1,000 and more copies in a few related temperate varieties. To our knowledge, this second stage of amplification has not been observed for any other DNA transposable element and may explain, in part, the exceptionally high copy number of MITEs.
The first plateau at 50 copies appears very stable, as evidenced by the apparent lack of mPing mobility in Nipponbare and related strains over the past century (Fig. 1). Stable inactivation of mPing could be caused by the absence of a necessary component in these strains (e.g., an autonomous element) or effective host repression. Activation of mPing in Nipponbare anther culture demonstrates that this strain is capable of supporting mPing amplification (that is, it has all necessary genetic components) (12) but that transposition is normally repressed, probably by epigenetic mechanisms.
However, although mPing can be activated in the laboratory by anther or cell culture, our results demonstrate that mPing was also activated in the farmer's field. Transposon display analysis of DNA from individual parent and progeny plants from three of the four high-copy-number strains (EG4, A119, and A157) demonstrate that mPing is still actively transposing, because new insertions are readily detected (Figs. 5–7 and Tables 6–8). To our knowledge, these strains had not been subjected to mutagenic treatment before their analysis. Furthermore, our data (and breeding records) indicate that the high-copy-number strains and Nipponbare shared a common ancestor and that this ancestor had many of the same mPing insertion sites as Nipponbare (Fig. 1B and data not shown). To account for these data, we suggest that repression of mPing transposition was overcome independently in the high-copy-number strains in the course of their cultivation. In addition, once host repression is overcome, it cannot be easily reestablished, as evidenced by the continued movement of mPing elements in three strains that already have ≈1,000 copies (Fig. 5 and 7). Furthermore, the rate of accumulation of mPing insertions per generation is staggering, with dozens of heritable insertions documented in the single-seed decent EG4 plants (Fig. 5 and Tables 7 and 8).
Clearly, an understanding of how TEs amplify to very high copy numbers also requires knowledge of how a host can sustain the onslaught of hundreds, perhaps thousands, of TEs into and near its genes. As with element amplification, our results indicate that selection also occurs in stages. The first stage is the rapid elimination of most exon and intron insertions from rice genes in the high-copy-number strains (Table 1, compare Total with Control, and Table 5). Evidence that mPing can readily insert into exons and introns is provided by the identification of mPing insertions in rice exons and introns among the de novo events (Table 1 and 6). A later, second stage of selection is suggested by a comparison of the insertions within 5 kb of rice genes in high-copy-number strains (Table 1 and 5, “total” 195 insertions within 5 kb of 256 insertions in single-copy regions, or 76%) vs. the older insertions in Nipponbare (22 insertions within 5 kb of 42 insertions in single-copy regions, or 52%). The apparent selection against mPing elements close to rice genes over time suggests that some fraction of this very large group of insertions is impacting host fitness, perhaps by causing subtle changes in rice gene expression.
In this study, we have documented a burst of MITEs during rice domestication. A valid question is whether our results are also applicable to natural plant populations. We believe that the answer is yes, based on the fact that prior evolutionary analyses of plant genomic sequences revealed that MITE families arise from the burst of only a few elements over a very short period (6, 10, 11). Thus, although the dynamics of selection clearly differ in domesticated vs. wild plant species, both populations contain collections of repressed TEs that are poised to amplify under suitable environmental conditions. These bursts of amplification are a potentially valuable source of population diversity in normally selfing plants like rice. In addition, the continued movement of mPing in strains with >1,000 mPing elements makes this a potentially valuable source of tagging populations that can be used in rice gene discovery.
Materials and Methods
Plant Material and DNA Extraction.
Aikoku and Gimbozu cultivars and landraces were obtained from the GenBank project of the National Institute of Agrobiological Science, Ibaraki, Japan. Genomic DNA was extracted from 3-week-old seedlings by the cetryltrimethylammonium bromide method (23) or by using DNA easy plant mini kit (Qiagen, Valencia CA).
DNA Blot Hybridization.
Genomic DNA (100 ng) was transferred to Hybond N+ nylon membranes (Amersham Pharmacia Biotech, Piscataway, NJ) after EcoRI digestion and electrophoresis through a 1% agarose gel with 1× Tris–acetate–EDTA buffer and probed with mPing (1–430 nt, GenBank accession no. AB087615) labeled with digoxygenin by using PCR DIG Probe Synthesis kit (Roche Applied Science, Indianapolis, IN) (using primer sequences forward, 5′-TCGTCAGCGTCGTTTCCAAGT-3′, and reverse, 5′-TGGAGGGGTTTCACTTTGACG-3′).
Copy-Number Determination.
mPing copy number was determined by TD and performed as described (18, 19). Final annealing temperature for selective amplification was 58°C with the 33P-labeled primer. Primer sequences were MseI + 0, 5′-GACGATTGAGTCCTGAGTAA-3′ and MseI + NNN, 5′-GACGATTGAGTCCTGAGTAANNN-3′ (NNN stands for AAA, AAT,……,CCC); mPing P1, 5′-TGTGCATGAGACACCAGTG-3′; mPing P2, 5′-CAGTGAAACCCCCATTGTGAC-3′. Copy-number estimates are described in Results.
Annotation of mPing Flanking Sequences.
DNA fragments from TD gels were excised, reamplified, and cloned as described (18, 19). Sequences of cloned fragments were determined by the Molecular Genetics Instrumentation Facility (University of Georgia). The mPing flanking sequences were annotated by using a PERL script named GB_1020;mPing_1020;annotator (available on request). Bioperl modules including Bio::Seq, Bio::SeqIO, Bio::SearchIO, Bio::LiveSeq::DNA, Bio::Tools::Run::StandAloneBlast, Bio::SearchIO::blast, and Bio::Index::Fasta were used (24). The sequences obtained from TOPO cloning vectors (Invitrogen, Carlsbad, CA) containing PCR-amplified DNA from TD gel bands were used as input. Additional input parameters including “genomic database for BLAST,” “flanking size of the insertion site,” and “cDNA database name” were used to locate nearby full-length cDNA. The vector sequences (TGGAATTCGCCCTT… AAGGGCGAATTCTGCAGA) were filtered and the insert sequences oriented according to mPing end sequences. Each of these oriented sequences was used as query for a BLAST search of the Nipponbare genomic DNA database (word size = 7, filter = F, e = 1e-3). Database hits that share >95% similarity were considered matches.
Target-site preference of mPing in the rice genome was investigated by first eliminating the mPing sequence from sequences recovered from TD. Similarity searches of the trimmed sequences were carried out by using the BLASTN program (filter = F, e = 1e-20) against the Nipponbare genomic DNA database (25). Database hits were considered when pairwise alignment length was equal to the length of query sequences within 10% error. Twenty bases of genomic sequence preceding the first site of each query were obtained. As a consequence, flanking sequences of both ends of 123 mPing insertion sites were identified.
To identify genes near mPing insertion sites, 10 kb of flanking genomic sequence (5 kb on each side of an mPing insertion site for each of the genomic hits) was extracted from the Nipponbare genomic DNA database and used as query for a BLAST search against the full-length cDNA database (http://cdna01.dna.affrc.go.jp/cDNA) (20). High-scoring pairs (HSPs) >50 bp and >98% identity were considered matches. Positions (upstream, downstream, or inside) and distances of cDNA sequences relative to mPing insertion sites were determined based on position information of the hits in HSPs. If a cDNA sequence hit spans an mPing site (either contiguous or with a gap), the mPing insertion was considered to be inside of the cDNA sequence. If the start of a cDNA is downstream of a mPing insertion site, the insertion was considered to be upstream of the cDNA, and the distance between the insertion site and the cDNA start was calculated. The distance of a downstream insertion from a cDNA was calculated from the 3′ end of the cDNA sequence.
Computer Simulation (Control).
For the control experiment (Table 1), fragments of up to 10 kb were randomly generated from the annotated Nipponbare genome database and used as queries for BLAST searches of the same database to determine copy number. BLAST searches of the cDNA database were also performed. The middle of each 10-kb sequence was taken as the insertion site and used in cDNA distance determinations.
Expression Analysis.
RNA was extracted from leaves of Nipponbare and EG4 by using the RNeasy Plant Mini kit (Qiagen). Synthesis of cDNA was with Invitrogen SuperScript RTII (Invitrogen). PCR was then performed by using primers designed to amplify respective genes. Primer sequences that were used for these PCRs are available on request.
Supplementary Material
Acknowledgments
We thank the GenBank project of the National Institute of Agrobiological Science in Japan for providing seeds of Aikoku and Gimbozu; Deep Shah, Mika Morita, and Nami Ueda for assistance with experiments; and Dr. Jeff Bennetzen for advice regarding data analysis. The study was funded by a grant from the National Science Foundation Plant Genome Program (to S.R.W.) and Research Project Grants-in-Aid for Scientific Research 14360005 and 15380006.
Abbreviations
- TD
transposon display
- TE
transposable element
- MITE
miniature inverted-repeat TE.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.McClintock B. Cold Spring Harbor Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig N, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: Am Soc Microbiol; 2002. [Google Scholar]
- 4.Oshima K, Okada N. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 5.Kwase M, Fukunaga K, Kato K. Mol Gen Genom. 2005;274:131–140. doi: 10.1007/s00438-005-0013-8. [DOI] [PubMed] [Google Scholar]
- 6.Feschotte C, Jiang N, Wessler SR. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Arbuckle J, Wessler SR. Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorkovic ZJ, Kirk DA, Lambermon MHL, Filipovicz W. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 9.Varagona MJ, Purugganan M, Wessler SR. Plant Cell. 1992;4:811–820. doi: 10.1105/tpc.4.7.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bureau TE, Wessler SR. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang N, Bao Z, Zhang X, McCouch SR, Eddy SR, Wessler SR. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi K, Terauchi K, Wada M, Hirano HY. Nature. 2003;421:167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- 13.Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. Nature. 2003;421:170–172. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- 14.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varmam H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al. Science. 2002;296:79–92. [Google Scholar]
- 16.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler SR. Proc Natl Acad Sci USA. 2000;97:10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casa AM, Nagel A, Wessler SR. Methods Mol Biol. 2004;260:175–188. doi: 10.1385/1-59259-755-6:175. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. Science. 2003;301:376–379. [Google Scholar]
- 21.Zhang X, Jiang N, Feschotte C, Wessler SR. Genetics. 2004;166:971–986. doi: 10.1534/genetics.166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Murray MG, Thompson WF. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al. Nucleic Acids Res. 2006;34:741–744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.