Abstract
The underlying mechanism by which the interspersed pattern of human segmental duplications has evolved is unknown. Based on a comparative analysis of primate genomes, we show that a particular segmental duplication (LCR16a) has been the source locus for the formation of the majority of intrachromosomal duplications blocks on human chromosome 16. We provide evidence that this particular segment has been active independently in each great ape and human lineage at different points during evolution. Euchromatic sequence that flanks sites of LCR16a integration are frequently lineage-specific duplications. This process has mobilized duplication blocks (15–200 kb in size) to new genomic locations in each species. Breakpoint analysis of lineage-specific insertions suggests coordinated deletion of repeat-rich DNA at the target site, in some cases deleting genes in that species. Our data support a model of duplication where the probability that a segment of DNA becomes duplicated is determined by its proximity to core duplicons, such as LCR16a.
Keywords: duplicons, LCR16 elements, lineage-specific duplications, segmental duplication
Based on the current sequenced animal genomes, human genomic architecture is unique in the abundance of large segmental duplications that are interspersed at discrete locations in the genome (1–5). Although recent duplications are common among other animal genomes, they are typically organized as clusters of tandemly arrayed segments (6). In humans and other great-ape genomes, ≈450 duplication hubs have been identified that have been the target of duplications from many different ancestral loci. This property has created regions of the genome that are complex mosaics of different genomic segments (7) where novel genes, fusion genes, and gene families have emerged (2, 8–13). Detailed studies of a few of the underlying regions (9, 11, 14) suggest that duplications have occurred in a stepwise fashion, involving subsequent larger segments of duplication as secondary events. The mechanism by which hundreds of kilobases of genomic sequence becomes duplicatively transposed to a new location on a chromosome is unknown.
Human chromosome 16 represents one of the most extreme examples of such recent segmental-duplication activity (15). More than 10% of the euchromatic portion of human chromosome 16p consists of segmental duplications known as LCR16 (low-copy repeat sequences on chromosome 16) (16, 17). During the initial sequence analysis of this chromosome, Loftus et al. (17) identified at least 20 distinct gene-rich LCR16 elements, ranging in size from a few kilobases to >50 kb in length, termed LCR16a–t. The majority of these were duplicated in an interspersed configuration throughout the chromosome. We subsequently identified a gene family, morpheus, within LCR16a that showed significant signatures of positive selection [Ka/Ks ratios up to 13.0 between humans and Old-World monkey (OWM) species]. The finished chromosome 16 sequence (18) provided the basis for a detailed analysis of these regions. We investigated the detailed organization of these regions among nonhuman primate species by sequencing large-insert clones from a diverse panel of primates to address questions regarding the mechanism of origin, the extent of structural variation among primates, and the relationship of these complex structures to the rapidly evolving LCR16a segment.
Results
Human LCR16 Genome Organization.
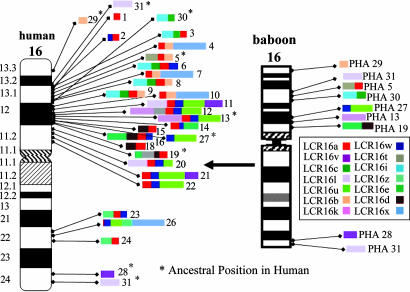
In humans, there are 17 complex blocks of LCR16 duplication (4.2 Mb of sequence) that contain 23 distinct copies of LCR16a with fewer copies of other flanking LCR16 segmental duplications (Table 4, which is published as supporting information on the PNAS web site, and Fig. 1 on human chromosome 16). Three blocks map to 16q22, whereas the remainder are distributed along the short arm of chromosome 16, where they occupy an estimated 11% of the euchromatin. The duplication blocks range in size from 604,376 bp (16p12.1/11.2) to solo copies of the LCR16a element (≈19,794 bp in length) (Table 5, which is published as supporting information on the PNAS web site, and Fig. 1). Of the 11 other LCR16 elements considered in this analysis (Table 1), all map within 109 kb of an LCR16a duplication. After excluding ancestral segments, we find only one exception where a block exists (LCR16uw, Fig. 1 and Table 6, which is published as supporting information on the PNAS web site) without a full-length (20 kb) LCR16a element. In contrast, two distinct “solo” LCR16a elements have been identified that are not associated with other duplicated segments (Table 5), including a single rogue segment that has been mapped outside of chromosome 16 to human 18p11.
Fig. 1.
LCR16 organization in human and baboon. The location, copy number, and structure of LCR16 duplications are depicted within the context of an ideogram for human (Left) and Papio hamadryas (PHA) (Right) based on the human genome reference sequence (hg16), BAC-end sequencing, and complete clone insert sequence of baboon clones. With the exception of the ancestral loci, duplication blocks are enumerated based on their position (p–q) on human chromosome 16 (Table 5).
Table 1.
LCR16 copy number among primates
| LCR16 | Human | Chimpanzee | Gorilla | Orangutan | Macaque | Baboon |
|---|---|---|---|---|---|---|
| a | 17 | 37 | 16 | 20 | 2 | 1 |
| t | 3 | 3 | 1 | 5 | 1 | 2 |
| b | 6* | 1 | 2 | 1 | 1* | 4 |
| w | 8 | 11 | 4 | 1 | 1 | 1 |
| u | 7 | 8 | 4 | 1 | 1 | 1 |
| c | 6* | 3 | 2 | 1 | 1* | 1 |
| k | 2* | 3 | 2 | 1 | 1* | 1 |
| d | 4* | 3 | 1 | 1 | 1* | 1 |
| e | 3* | 4 | 1 | 1 | 1* | 1 |
| i | 3* | 4 | 1 | 1 | 1* | 1 |
| l | 3* | 3 | 1 | 1 | 1* | 1 |
| v | 3 | 5 | 1 | 1 | 1 | 1 |
Copy number was based on experimental hybridization and BAC-end sequencing results (Methods). Bold type indicates a shift to single copy numbers.
*Copy-number estimate was based on the hg16 or rheMac2 sequence assembly.
Single-Copy Architecture of OWM Loci.
To investigate the evolutionary history of these complex genomic regions, we systematically recovered large-insert genomic clones corresponding to each human LCR16 segment from five nonhuman primate species including chimpanzee, gorilla, orangutan, macaque, and baboon. We designed a total of 12 probes, one corresponding to each of the LCR16 duplications, and hybridized each independently to available genomic BAC libraries (Methods). We identified 782 clones and estimated the copy number and cooccurrence of various LCR16 segments in these different species (Tables 1 and 6). The BAC hybridization revealed that the majority (11 of 12) of the LCR16 elements are single copy in OWM outgroup species (macaque and baboon) (Table 7, which is published as supporting information on the PNAS web site) and that copy number increases have occurred in a stepwise fashion, based on the inferred phylogenetic relationship of these species (Table 1). We observed a positional bias in the evolutionary order of these events. LCR16 segmental duplications located more distally from LCR16a, in general, are predicted to be more recent than those that map in closer proximity to human LCR16. Finally, we note that certain pairs of LCR elements (e.g., LCR16u and -w, as well as LCR16i and -c) consistently cohybridize to the same BACs, including the single-copy locus within OWM species, suggesting that these different duplicons originated from the same ancestral locus.
Duplication of Great Ape LCR16 Blocks into New Locations.
Mapping and sequencing of LCR16 segmental duplications within primate genomes has been problematic because the duplications are typically embedded in large duplication blocks that may exceed 100 kb in size. For example, in the chimpanzee genome, these regions are misassembled, are highly fragmented, or correspond to gaps (19). Large-insert genomic clones, such as BACs, can help circumvent this problem because BAC-end sequence (BES) may extend beyond the duplication blocks to anchor in unique sequence (20). Such sequence anchors provide information regarding the corresponding map position. We therefore selected 782 BACs for insert end-sequencing, generating 526 pairs of end-sequence that were informative for mapping purposes (Table 8, which is published as supporting information on the PNAS web site). Based on comparative mapping of macaque and baboon for each single-copy locus of LCR16, we unambiguously determined the most likely ancestral location of each segmental duplication, which mapped to nine distinct locations that were consistent between both outgroup species (Fig. 1; and see Fig. 4, which is published as supporting information on the PNAS web site). With the exception of LCR16t, LCR16a is not associated with any of these regions in OWM species (Fig. 1).
Using a similar strategy, we attempted to assign locations for corresponding loci within ape genomes using BES data. In contrast to OWMs, we identified multiple loci for each probe, the vast majority of which associated with LCR16a based on the hybridization results. We categorized ape loci as mapping to (i) an orthologous locus (based on the identification of LCR16 duplications at that position in human), (ii) an ancestral position (based on map positions of single copy loci in baboon and macaque), or (iii) a nonorthologous location (based on the absence of a corresponding duplication at that position in human) (Tables 2 and 5 and Fig. 4). We could assign 35 loci to one of these categories, whereas ≈27 were ambiguous (end sequences placed in duplicated sequences in humans or other primates, preventing accurate assignment; see Methods). We observed a spatial clustering of new insertions. Both sequence and BES data, for example, indicate that the distal portion of chromosome 16 has been the target of such LCR16 duplications, particularly within the chimpanzee lineage. Similarly, many of these orangutan insertions mapped to a 5-Mb region on human 13q12.1–13q12.3 (Fig. 4).
Table 2.
Primate segmental duplication BAC sequencing summary
| Species | Library | No. of BACs | Mbp, Mb | New insertion | Orthologous | Ancestral | Unknown |
|---|---|---|---|---|---|---|---|
| Chimpanzee | RPCI-43/CHORI-251 | 21 | 4.06 | 2 | 9 | 1 | 9 |
| Gorilla | CHORI-255 | 19 | 4.09 | 2 | 7 | 2 | 8 |
| Orangutan | CHORI-253 | 15 | 2.87 | 2* | 0 | 3 | 10* |
| Macaque | CHORI-250 | 3 | 0.46 | 0 | 0 | 3 | 0 |
| Baboon | RPCI-41 | 4 | 0.72 | 0 | 0 | 4 | 0 |
| Totals | 62 | 12.20 | 6 | 16 | 13 | 27 |
Sixty-two LCR16 BAC clones from five nonhuman primate species were sequenced and aligned to the human genome. Loci were classified as new insertion or orthologous based on the presence of unique anchors between human and nonhuman primate genomes. Ancestral loci correspond to the putative ancestral locus based on map position of single loci in baboon and macaque. For 27 clones, the precise map location could not be assigned because the entire insert consisted of segmental duplications.
*In the case of orangutan, most mapped to chromosome 13 and therefore were “new” insertions with respect to human and other apes, but the map location could not be further refined by orthologous anchors.
Sequence Structure of Nonorthologous Insertions and Lineage-Specific Duplications.
We sequenced 62 nonhuman primate LCR16-positive BAC clones, generating 12.2 Mb of genomic sequence from five nonhuman primate species (Table 2). For each sequence, we identified the best location in the genome based on alignment of unique flanking sequence (Methods). Sequence data generated from both the macaque and baboon unambiguously confirmed synteny and structure of the most likely ancestral position (Fig. 5, which is published as supporting information on the PNAS web site), including previously recognized distinct duplicons mapping to the same location (i.e., LCR16at and LCR16uw). We identified a minimum of six duplication blocks that were present at locations in ape genomes where there was no evidence of corresponding duplications in humans (Fig. 2 and Table 5). These insertions, which ranged in size from 33.4 kb to an estimated >200 kb, were always accompanied by a copy of LCR16a. Moreover, PCR breakpoint analyses (see below) and FISH analyses (data not shown) confirmed that these events occurred specifically within each lineage. We note that the sequenced insertions consist of both LCR16a and LCR16 elements flanking these regions, suggesting duplicative transposition, in some cases, of large (>100 kb), complex sequences into new locations.
Fig. 2.
Sequence alignment between human and nonhuman primate LCR16 loci. (a) Chimpanzee-specific insertion (AC097264) of 81,799 bp between genes DRF1 and FLJ31795 on chromosome 17q21.31. The new insertion consists of three LCR16 duplicons that are shared between human and chimpanzee (LCR16e, w and a) in addition to a flanking 16,820-bp lineage-specific duplication (LCR16a′, Table 9). The 5,962 bp of human sequence corresponding to the preintegration site are deleted in chimpanzee (Table 3). The extent of duplication of the underlying sequence based on WSSD analysis is shown for human (light blue), chimpanzee (pink), and orangutan (dark blue) for this and all subsequent images. (b) Chimpanzee-specific insertion (AC149436) of a segmental duplication mapping to chromosome 16p13.3. The insertion sequence (33,405 bp) consists of LCR16a and a chimpanzee-specific duplication (termed LCR16b′) of 7,403 bp, which is single-copy in human. A corresponding deletion of the integration site (16,100 bp) deletes the serine protease EOS gene in chimpanzee. (c) A 230-kb sequence in orangutan that is completely duplicated (dark blue bar). Two different segments flank the LCR16aw segmental duplication, including a 109-kb segment corresponding to human chromosome 13q34 (chr13:112701583–112831134) and a 99-kb segment from chromosome 16 (chr16:11526252–11625727). Both segments are unique in chimpanzee and human. (d) Orangutan genomic sequence (AC144879) shows the presence of an inserted duplication complex corresponding to human 13q12.11 (chr13:19666603–19839556). A 38,344-bp segment corresponding to the site of insertion in human has been deleted. Several orangutan-specific duplications are noted, including a 21-kb flanking duplication that maps to the corresponding region in human. This property shows that LCR16a and the ancestral locus (LCR16n′) were associated.
During our analysis of these insertions, we observed segments that were duplicated specifically within each species (gray and black bars in, which is published as supporting information on the PNAS web site). These lineage-specific duplications ranged in size from a few kb to >80 kb in length and were frequently associated with genic regions (Table 9, which is published as supporting information on the PNAS web site). There are two important structural properties regarding these lineage-specific duplications associated with these insertions. First, these lineage-specific duplications most frequently map at the periphery of duplicated segments that are shared between great apes (Figs. 2 and 6). These findings are consistent with hybridization (Table 1) and phylogenetic results (see below), which show evolutionarily younger duplicons accreting at the edges. Second, most of these peripheral lineage-specific segmental duplications originate from chromosomal regions where LCR16a activity can be documented as having recently occurred (Fig. 2d and see Fig. 5u). These associations with ancestral loci suggest that lineage-specific LCR16 segments originate in regions of prior LCR16a integration.
As a more direct test of association with LCR16a, we performed a series of independent hybridization experiments with each of the eight lineage-specific duplications in orangutan that were not identified as duplicated in chimpanzee or human. We estimated the copy number of each duplication and then cross-referenced positive clones by PCR to determine whether they were associated with LCR16a in the orangutan (Table 10, which is published as supporting information on the PNAS web site). Seventy-two percent (100 of 139) of clones detected by using a lineage-specific probe were also positive for LCR16a. When BES data were used to eliminate the ancestral locus, we found that 94% (17 of 18) of the duplicated loci were in association with LCR16a. We found only one exception where an orangutan-specific duplication had occurred without LCR16a. These data indicate that different intrachromosomal euchromatic duplications have emerged at different locations in a different lineage but focused, once again, around the LCR16a core. Interestingly, both copy number and sequence divergence decrease in a gradient-like fashion as distance from LCR16a increases (Fig. 7, which is published as supporting information on the PNAS web site). Thus, even though the chromosome, the location, and the content of the segmental duplication differ, we observe a virtually identical complex mosaic pattern of segmental duplications and polarity vis-à-vis LCR16a in different primate species.
Recurrent and Independent Duplications of LCR16a.
Sequencing of the baboon and macaque genomes confirmed the ancestral location of each LCR16 segment (Fig. 1). Using noncoding primate genome sequences, we constructed a neighbor-joining phylogenetic tree for each of the 14 human LCR16 duplicons (Fig. 8, which is published as supporting information on the PNAS web site). The tree topology and corresponding branch lengths were remarkably consistent with the evolutionary order of events predicted from the initial hybridization results. The LCR16a phylogenetic analysis reveals two distinct clades, one monophyletic origin with respect to human/African ape sequences and a second monophyletic origin for the orangutan loci (Fig. 6). This finding is consistent with molecular clock data, which indicate that LCR16a expansions have occurred independently in each of the two lineages. It is interesting that, when the duplication architecture is superimposed over the LCR16a phylogeny, similar block architectures cluster. For example, in the case of human, three distinct groups can be recognized based largely on the presence of flanking LCR16 duplicons (LCR16b, d, or k/l). These associations supersede relationships predicted based on orthology, suggesting large-scale genetic exchanges since speciation of humans and great apes (21).
The finding of so many independent, recurrent duplications of the LCR16a segment prompted us to investigate whether there might be evidence for additional, more ancient copies of LCR16a that were not originally identified as a result of our threshold for detection (i.e., >90% sequence identity). Five additional loci were discovered, including three nearly full-length copies on chromosome 10q22.3 as well as two partial copies on chromosomes Xp11.22 and 11p15.4 (Fig 9, which is published as supporting information on the PNAS web site). Three of these five homologous LCR16a structures were embedded within complex duplication blocks flanked by chromosome-specific segmental duplications. The extensive substitutions (≈0.2–0.3 substitutions per site) suggest that these duplications of LCR16a occurred much earlier during primate evolution (>40 million years) (22). Analysis of the recent rhesus macaque genome assembly confirmed the presence of Xp11.22, 11p15.4, and one of the 10q22.3 loci at syntenic positions to these human copies, confirming duplication of these before the divergence of the macaque/human lineages.
Junction Analysis.
Two types of junctions could be identified based on our comparison of nonhuman primate and human sequences: (i) those that traversed lineage-specific duplications that had not been observed in humans (termed accretion boundaries) and (ii) those corresponding to the sites of new insertions (i.e., unique-duplication transitions where the LCR16 duplications were not present at that locus in human). The latter, termed insertion boundaries, provided the opportunity to study the architecture of the integration sites before duplicative transposition.
We generated precise sequence alignments and examined the repeat content for a total of 12 insertion and 23 accretion boundaries. As a control for the quality of sequence and assembly, subsets of these boundaries were tested and validated by junction-PCR amplification and sequencing of the PCR product (Fig. 3). Overall, ≈55% (19 of 35) of the junctions showed the presence of an Alu repeat mapping precisely at the accretion or insertion boundary. Of these, ≈95% (18 of 19) corresponded to younger subfamilies (AluS and AluY) (Table 11 and Fig. 10, which are published as supporting information on the PNAS web site). This threefold enrichment confirms previous findings that younger Alu repeat elements are significantly enriched at the breakpoints of segmental duplication (23, 24). Because of the lineage-specific nature of the duplications, donor and acceptor relationships in most cases could be readily defined. We noted 11 examples where the transition between donor and acceptor sequences occurred within homologous (although not identical) repeat elements.
Fig. 3.
Breakpoint resolution of a chimpanzee insertion. The schematic depicts a segmental duplication insertion of 82 kb and the corresponding deletion of 6.0 kb at the preintegration site with respect to the human reference sequence. PCR breakpoint analysis shows that repeat sequences were present in common ape ancestors but that insertion was specific to chimpanzee and bonobo. Variability in PCR products is due to insertion and deletion of Alu repeats, which are common in repeat-rich regions (22, 42). The preintegration locus consists of 92.7% common repeats.
Interestingly, for six examples where a new segmental duplication was clearly documented at a new location in a nonhuman primate species, we observed a corresponding genomic deletion of the preintegration site (Figs. 2 and 3). These deletions ranged in size from 3.4 to 80.1 kb in length (median length = 5.9 kb) and were remarkably repeat-rich (77.3%) (Table 3). The evolutionary age of the corresponding repeat subfamilies and junction PCR indicate that these complex retroposon repeat structures represent the ancestral state. In one case, the corresponding segmental duplication was associated with the deletion of an entire serine protease gene in chimpanzee (Fig. 2b). This gene deletion was previously shown to be specific to the chimpanzee lineage (25), and our results clearly indicate a previously undescribed mechanism underlying its excision. Although the number of sites is still limited, these data suggest that coordinated deletion of repeat-rich DNA is a hallmark feature of de novo segmental duplication.
Table 3.
Composition of preintegration sites based on human reference genome (hg16)
| Species | GenBank accession no. | Chr | Human coordinates | Size, kb | Repeats, % | Percent LTR | Percent LINE | Percent SINE | Percent unique |
|---|---|---|---|---|---|---|---|---|---|
| PTR | AC097264 | 17 | Chr17:43247305–43253267 | 5.9 | 92.7 | 0.0 | 8.3 | 70.6 | 7.3 |
| PTR | AC149436 | 16 | Chr16:2832391–2848391 | 16.1 | 61.8 | 17.3 | 13.4 | 23.7 | 38.2 |
| GGO | AC145025 | 16 | Chr16:27163482–27169307 | 5.8 | 83.7 | 30.0 | 26.3 | 22.8 | 16.3 |
| GGO | AC145240 | 16 | Chr16:23281866–23285303 | 3.4 | 95.8 | 0.0 | 23.6 | 71.3 | 4.2 |
| PPY | AC144877 | 13 | Chr13:22779969–22852281 | 80.1 | 55.6 | 5.0 | 23.1 | 22.0 | 44.4 |
| PPY | AC145239 | 13 | Chr13:23733424–23739125 | 5.7 | 74.1 | 2.2 | 55.5 | 15.2 | 25.9 |
| Average | 19.5 | 77.3 | 9.1 | 25.0 | 37.6 | 22.7 |
*Sequence content of deleted regions in human genome that contain a previously undescribed, lineage-specific segmental duplication within a nonhuman primate species (PTR, Pan troglodytes; GGO, Gorilla gorilla; PPY, Pongo pygmaeus).
Discussion
Our detailed sequence and evolutionary analysis of a subset of primate segmental duplications reveals unexpected properties regarding their origin and expansion. We summarize these properties and the supporting data and put forward a model for LCR16 segmental duplication and associated structural variation of primate genomes.
Recurrent Duplications.
We show that LCR16a has duplicated independently in each of the great-ape lineages to new euchromatic locations (Fig. 2). Most of the complex duplication blocks on human chromosome 16 are or have been associated with a full-length copy of LCR16a. Human and orangutan LCR16a map to different locations in the two genomes (Fig. 4). More ancient, full-length copies of the LCR16a element have been identified on different chromosomes, once again associated with complex regions of duplication. These data indicate that LCR16a duplications have occurred independently multiple times, and this 20-kb sequence has an inherent proclivity to duplicate to new locations.
Duplication Polarity.
Other LCR16 elements have accumulated in a stepwise fashion focused around LCR16a to form complex duplication blocks (Fig. 6). Unlike LCR16a, solitary duplications (i.e., not associated with another LCR) are rarely identified for these (in the one clear case in human, analysis of the structure showed it to be a deletion of LCR16a) (Fig. 4v). Based on outgroup sequence data (macaque and baboon), most of these LCR16 elements originate from ancestral single-copy sequences (Fig. 1). We show that younger and less abundant duplications accumulate at the periphery of LCR16a (Fig. 7). In the case of orangutan, a completely analogous structure of flanking duplications (independent in origin) has emerged flanking LCR16a (Figs. 2 and 3). These data suggest polarity of duplication around LCR16a.
Ancestral Associations.
Our hybridization and sequencing (Tables 1 and 2) data indicate that several of the ancestral loci of intrachromosomal segmental duplication on chromosomes 13 and 16 have been associated with LCR16a. In gorilla, for example, we find LCR16a in close proximity to LCRl (although at least in humans, such an association no longer exists). Two other examples (Figs. 2d and 4u) indicate that ancestral positions of LCR16 in chimpanzee and orangutan map in close proximity with LCR16a and are associated with lineage-specific duplications in these species. We propose these associations with LCR16a have served to prime lineage-specific duplications from these regions.
Coordinated Deletion.
Our detailed analysis of six new insertions have shown that, in all six cases, the newer insertions involved the coordinated deletion of sequences. The preintegration sequences are highly enriched for common repeat sequences and may be prone to double-strand breakage events. The coordinated deletion of target site nucleotides has been observed for several atypical L1 integration events (26, 27) and may implicate single-strand annealing (SSA) and/or synthesis-dependent annealing (SDSA) (28, 29) as part of the pathway of segmental duplication.
Core Duplicon-Flanking Transposition Model.
We have shown that LCR16 segmental duplications change in copy number, composition, and, more remarkably, location among humans and great apes. These regions of the genome may be loosely classified as a form of mobile DNA. Unlike typical common repeats (30), however, this process has moved and juxtaposed large gene structures, frequently in a lineage-specific manner, into new genomic contexts. The complex set of data presented here argues that LCR16a has played an active role in creating the duplication architecture on human chromosome 16 and orangutan chromosome 13. We propose that other LCR16 duplications have been duplicated passively, essentially as genetic hitchhikers as part of this process. The association of LCR16a elements with ancestral loci, especially younger duplication events, suggests that a property of the sequence itself has the potential to duplicate sequences to new locations. This has occurred independently and at different times during human–great ape evolution. These events are associated with both deletions and other rearrangement events that have subtly restructured human and great ape chromosomes during evolution. Core duplicons, similar to LCR16a, have recently been identified for other chromosomes with an overabundance of intrachromosomal duplications (31, 32). It is possible that this characteristic may represent a general property of the human/great ape genome.
There are at least two possible explanations for our observations. First, the LCR16a sequence may have evolved mechanistically as a preferred template for gene conversion events to new locations in the human genome. In this model, LCR16a would serve as a source for the directional repair of a double-strand breaks in the genome, perhaps similar to yeast mating-type switching (33). The Alu-repeat richness of the LCR16a cassette would provide the homology to promote single-strand annealing and/or SDSA. These findings might explain the coordinated deletion of preintegration sites, the enrichment of Alu repeats at breakpoints, and the finding that sequences flanking LCR16a become duplicated. If the LCR16a sequence carries an inherent enhancer of gene conversion, it is unclear how the process could be so processive (hundreds of kilobases) or why it, as opposed to other Alu-rich repeat regions of the genome, is the preferred source.
An alternative explanation for the apparent strong association of new duplications with LCR16a may be as an indirect consequence of intense selection. We have shown previously that the gene family encoded by LCR16a shows among the strongest signatures of positive selection among humans and African ape genes (8). It is possible that the complex pattern of duplication is simply a consequence of the pressure to produce more divergent copies of LCR16a at distinct locations. We do not favor this model completely, because positive selection of the morpheus gene family occurred only among humans and African apes (Ka/Ks = 10–13 for exon 2 when compared with the OWM outgroup). Our data indicate that complex duplicated blocks have emerged completely independently in the orangutan lineage, where there is no strong evidence of positive selection (Ka/Ks ≈ 1.0). Moreover, we have identified more ancient copies of LCR16a on chromosomes 10 and 11, and the X chromosome, suggesting that this piece of genetic material was inherently unstable and duplicating before positive selection. We therefore favor a duplication-driven model of DNA transposition. This dynamic model for genomic duplication helps to explain the nonrandom spatial–temporal distribution of segmental duplications in human and great apes.
Methods
Genomic Library Hybridization and BAC End-Sequencing.
Large-insert genomic BAC libraries (minimum 6-fold coverage) from chimpanzee (RPCI-43), gorilla (CHORI-255), the orangutan (CHORI-253), the olive baboon (RPCI-41), and the rhesus macaque (CHORI-250) were probed by hybridization for each individual LCR16 duplication (Table 12, which is published as supporting information on the PNAS web site) as described (20). A total of 782 LCR16-positive BACs were selected, and the inserts were end-sequenced. Repeat-masked BES was rescored for quality and mapped against the human genome (MEGABLAST 12PATCh–d BES–D 3–p 93–F m–UT–s 150–R T).
BAC Sequencing.
BACs were subjected to shotgun sequencing at the National Institutes of Health Intramural Sequencing Center (34) and the Baylor College of Medicine Human Genome Sequencing Center to (35) at least 6-fold sequence redundancy. A subset of clones (n = 25) corresponding to potential new insertions were selected for ordered and oriented sequence assembly.
Sequence Annotation.
Nonhuman primate BAC sequence was compared with human genome sequence by using Miropeats (36), two_way_mirror (J. Bailey, personal communication), and ALIGN (37), using parameters optimized for global alignment of primate sequences (22). The best map location was defined as one where human and nonhuman primate sequences align within nonduplicated flanking unique sequences. If the entire BAC was duplicated, the most significant correspondence by BLAST sequence homology was used, and the location was classified as “ambiguous.” We examined the extent of recent duplication (>94%) for each clone using the whole-genome shotgun sequence-detection strategy for human (2), chimpanzee (38), and orangutan (E.E.E., unpublished data). FISH hybridization was used to assess duplication/unique status in gorilla (M.V., unpublished data). For simplicity, human chromosome designations are used for nonhuman map descriptions (39).
PCR Breakpoint Analysis.
A subset of breakpoints associated with lineage-specific insertions were validated by designing PCR assays across the breakpoint junctions (Table 12) and amplification of genomic DNA from a panel of primate lymphoblast-derived DNAs. The dense repeat content of many of the breakpoints precluded design of assays across all insertion breakpoints.
Phylogenetic Analysis.
We extracted overlapping sequences corresponding to each of the human segmental duplications from nonhuman primate sequences and generated multiple sequence alignments using ClustalW (40) and corresponding neighbor-joining phylogenetic trees (MEGA). We considered only noncoding sequences by processing the multiple sequence alignments for corresponding cDNA using MAM software. We used Kimura's two-parameter method (41) for all estimates of genetic distance.
Supplementary Material
Acknowledgments
We are grateful to Baishali Maskeri, Robert Blakesley, Andy Sharp, and Eray Tuzun for technical assistance. We thank the large-scale sequencing centers (Baylor College of Medicine and the Washington University Genome Sequencing Center) for access to genome assembly data (macaque) and trace sequence data from the chimpanzee and orangutan before publication. This work was supported in part by National Institutes of Health (NIH) Grant GM58815 (to E.E.E.) and by the Intramural Research Program of the National Human Genome Research Institute, NIH. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Abbreviation
- OWM
Old-World monkey.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos: AC092562, AC07264–AC07266, AC097268, AC097270–AC097271, AC097326–AC097328, AC097330–AC097334, AC117273, AC118583, AC119422, AC144359–AC144362, AC144462, AC144590, AC144875–AC144877, AC144879–AC144881, AC145000, AC145025, AC145040, AC145177, AC145239–AC145240, AC145242–AC145243, AC145295, AC145353–AC145354, AC145356, AC145400–AC145403, AC146492, AC146844, AC146898, AC146952, AC147576, AC148534, AC148537– AC148538, AC148619, AC148838–AC148839, AC148841, AC148882, AC149436, AC150449, AC153733, and AC154112).
References
- 1.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE. Am J Hum Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW. Genome Biol. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Lu HH, Chung WY, Yang J, Li WH. Mol Biol Evol. 2005;22:135–141. doi: 10.1093/molbev/msh262. [DOI] [PubMed] [Google Scholar]
- 5.She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, et al. Nature. 2004;430:857–864. doi: 10.1038/nature02806. [DOI] [PubMed] [Google Scholar]
- 6.She X, Liu G, Ventura M, Zhao S, Misceo D, Roberto R, Cardone MF, Rocchi M, Green ED, Archidiacano N, et al. Genome Res. 2006;16:576–83. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichler EE. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 8.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 9.Courseaux A, Nahon JL. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- 10.Paulding CA, Ruvolo M, Haber DA. Proc Natl Acad Sci USA. 2003;100:2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR. Genome Res. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccarelli FD, von Mering C, Suyama M, Harrington ED, Izaurralde E, Bork P. Genome Res. 2005;15:343–351. doi: 10.1101/gr.3266405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandepoele K, Van Roy N, Staes K, Speleman F, van Roy F. Mol Biol Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 14.Eichler EE, Johnson ME, Alkan C, Tuzun E, Sahinalp C, Misceo D, Archidiacono N, Rocchi M. J Hered. 2001;92:462–468. doi: 10.1093/jhered/92.6.462. [DOI] [PubMed] [Google Scholar]
- 15.Stallings R, Doggett N, Okumura K, Ward D. Genomics. 1992;7:332–338. doi: 10.1016/0888-7543(92)90249-r. [DOI] [PubMed] [Google Scholar]
- 16.Stallings R, Whitmore S, Doggett N, Callen D. Cytogenet Cell Genet. 1993;63:97–101. doi: 10.1159/000133509. [DOI] [PubMed] [Google Scholar]
- 17.Loftus B, Kim U, Sneddon V, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby M, Barnstead M, Cronin L, et al. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- 18.Martin J, Han C, Gordon LA, Terry A, Prabhakar S, She X, Xie G, Hellsten U, Chan YM, Altherr M, et al. Nature. 2004;432:988–994. doi: 10.1038/nature03187. [DOI] [PubMed] [Google Scholar]
- 19.Chimpanzee Sequencing Analysis Consortium (CSAC) Nature. 2005;437:69–87. [Google Scholar]
- 20.Yohn CT, Jiang Z, McGrath SD, Hayden KE, Khaitovich P, Johnson ME, Eichler MY, McPherson JD, Zhao S, Paabo S, Eichler EE. PLoS Biol. 2005;3:1–11. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson MS, Oliver K, Loveland J, Humphray S, Dunham I, Rocchi M, Viggiano L, Park JP, Hurles ME, Santibanez-Koref M. Am J Hum Genet. 2005;77:824–840. doi: 10.1086/497704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey JA, Giu L, Eichler EE. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurka J, Kohany O, Pavlicek A, Kapitonov VV, Jurka MV. Proc Natl Acad Sci USA. 2004;101:1268–1272. doi: 10.1073/pnas.0308084100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puente XS, Gutierrez-Fernandez A, Ordonez GR, Hillier LW, Lopez-Otin C. Genomics. 2005;86:638–647. doi: 10.1016/j.ygeno.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert N, Lutz S, Morrish TA, Moran JV. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert N, Lutz-Prigge S, Moran JV. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 28.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugawara N, Haber JE. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 31.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zody MC, Garber M, Sharpe T, Young SK, Rowen L, O'Neill K, Whittaker CA, Kamal M, Chang JL, Cuomo CA, et al. Nature. 2006;440:671–675. doi: 10.1038/nature04601. [DOI] [PubMed] [Google Scholar]
- 33.Haber JE. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 34.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 35.Scherer SE, Muzny DM, Buhay CJ, Chen R, Cree A, Ding Y, Dugan-Rocha S, Gill R, Gunaratne P, Harris RA, et al. Nature. 2006;440:346–351. doi: 10.1038/nature04569. [DOI] [PubMed] [Google Scholar]
- 36.Parsons J. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 37.Myers EW, Miller W. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 39.McConkey EH. Cytogenet Genome Res. 2004;105:157–158. doi: 10.1159/000078022. [DOI] [PubMed] [Google Scholar]
- 40.Higgins DG, Thompson JD, Gibson TJ. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 41.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 42.Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.