Abstract
Mobile elements are important components of our genomes, with diverse and significant effects on phenotype. Not only can transposons inactivate genes by direct disruption and shuffle the genome through recombination, they can also alter gene expression subtly or powerfully. Currently active transposons are highly polymorphic in host populations, including, among hundreds of others, L1 and Alu elements in humans and Ty1 elements in yeast. For this reason, we wished to develop a simple genome-wide method for identifying all transposons in any given sample. We have designed a transposon insertion site profiling chip (TIP-chip), a microarray intended for use as a high-throughput technique for mapping transposon insertions. By selectively amplifying transposon flanking regions and hybridizing them to the array, we can locate all transposons present in a sample. We have tested the TIP-chip extensively to map Ty1 retrotransposon insertions in yeast and have achieved excellent results in two laboratory strains as well as in evolved Ty1 high-copy strains. We are able to identify all of the theoretically detectable transposons in the FY2 lab strain, with essentially no false positives. In addition, we mapped many new transposon copies in the high-copy Ty1 strain and determined its Ty1 insertion pattern.
Keywords: evolution, microarray, yeast, Ty1, integration
Transposable elements share one characteristic: they are able to physically move about their host genome, either by a cut-and-paste mechanism (most DNA transposons) or by a copy-and-paste process involving an RNA intermediate (retrotransposons). Occupying various and often substantial fractions of nearly every genome studied to date [human, 45% (1); chicken, 4.3% (2); mouse, 38% (3); yeast, 3% (4); maize, >60% (5), for example], transposons are under intense scrutiny as their complex contributions to evolutionary history are revealed through genome sequencing. It is clear that transposons have many effects on their host genomes: they can physically disrupt and potentially inactivate or alter genes upon transposition; mediate genome rearrangements once in place; and can affect gene expression in many ways, including enabling alternative splicing, triggering premature transcript termination, and facilitating gene breaking (for reviews see refs. 6 and 7). Importantly, transposon phenotypes do not require disruption of coding sequences. Defective or evolutionarily divergent elements such as the L1 element in humans (1, 8, 9) can also have profound effects.
The Saccharomyces cerevisiae Ty1 element is a well studied LTR-containing retrotransposon present in 20–30 copies in typical laboratory yeast strains (4, 10). This high copy number may result from the evolution of yeast and its population of retrotransposons under laboratory conditions; most wild yeast strains typically harbor lower Ty1 copy numbers (10–12).
Knowing all transposon insertion sites in a sample is very useful. First, such a method would be useful for studying transposon ecology, quickly addressing questions related to insertion site preference and the locations of transposon “hotspots” or “cold spots” in a genome. Second, studies of transposon evolution could benefit from a simple way to comprehensively scan the host genome for transposon locations. Third, individuals of the same species may carry varying transposon burdens; variations in transposon complement may be important factors in population dynamics and in phenotypes such as disease susceptibility.
We describe here a transposon insertion site profiling chip (TIP-chip), a custom tiling microarray-based strategy to search for transposons in either regions of interest or throughout an entire genome. By digesting sample genomic DNA, ligating to vectorettes, amplifying with a transposon-specific primer, fluorescently labeling the products, and hybridizing them to the TIP-chip, one can identify all sequences that flank the transposon being examined. Then, transposon profiles of different samples can be compared.
As a test of the TIP-chip strategy, we created a genomic tiling microarray for S. cerevisiae and used this to identify all Ty1 retrotransposons in two common lab strains and an experimentally derived Ty1 high-copy strain. We were able to correctly determine the locations of 94% of the known Ty1 elements in the S288C-derived FY2 strain, and identified 2 Ty1s not reported in the S288C DNA sequence. In addition, we examined the transposon profile of the L27-10 Ty1 high-copy strain. Comparing it with its parental strain GRF167, we observe at least 39 new Ty1 insertion sites, and we find that the population of new Ty1 insertions that occurred during the evolution of this strain is located largely (78%) within 2 kb of tRNA genes. Also, we found evidence in the high-copy strain for at least seven target regions in which multiple Ty1 elements were inserted, consistent with the existence of a limited number of high frequency target regions in the yeast genome (13).
Results
Supporting Information.
For further details, see Tables 2 and 3, Figs. 5–7, and Supporting Text, which are published as supporting information on the PNAS web site.
In Silico Design to Allow Comprehensive Amplification of the Yeast Genome.
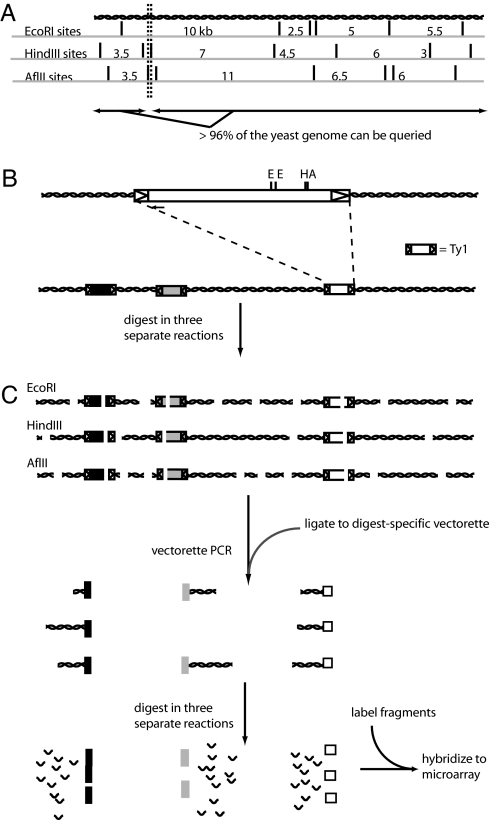
The DNA amplification protocol was designed based on the need to represent as much of the yeast genome as possible in the form of at least one fragment ≥1 kb long (allowing hybridization to multiple features on the TIP-chip, thereby increasing the statistical significance of positive signals) and <10 kb long (to maximize the yield of DNA amplified by the PCR). This was modeled by evaluating all possible two- and three-way mixtures of restriction digests of the actual yeast genomic sequence in silico chosen from a list of enzymes that generate sticky ends and cut Ty1 once or twice in appropriate regions, allowing the design of useful primers; enzymes also had to be efficient and cost-effective. Yeast genomic DNA was digested in three separate reactions with the single winning combination of enzymes (EcoRI, AflII, and HindIII) (Fig. 1A). With this combination of enzymes, 96.4% of randomly chosen insertion sites will yield detectable transposon flanks.
Fig. 1.
TIP-chip workflow. (A) Choosing restriction enzyme combinations for parallel digests of the yeast genome. The enzymes cut the DNA into overlapping pieces, so that each nucleotide of the yeast genome is contained in three separate restriction fragments, one from each enzyme. More than 96% of the yeast genome is contained in at least one fragment >1 kb and <10 kb; these somewhat arbitrary limits were chosen based on previous experience with PCR amplification and the proposed array design. (B) The Ty1 element, with LTRs shown as arrowheads. The small arrow at the 5′ end of the Ty1 denotes the position of the Ty1-specific primer used (JB8784; see supporting information). (C) Preparation of genomic DNA for hybridization to the TIP-chip. Genomic DNA is digested in three parallel reactions, with three restriction enzymes with 6-base recognition sequences. The digested fragments are ligated to digest-specific vectorettes and amplified by using vectorette PCR. Longer amplicons may not amplify well and may be underrepresented in the resulting mixture. The amplicons are then pooled and digested in three parallel reactions with three enzymes with 4-base recognition sequences. The resulting fragments are heat-inactivated, labeled, and hybridized to the microarray.
Once digested, fragments were amplified with vectorette PCR (14), a method that amplifies those restriction fragments containing the transposon-specific primer sequence (Fig. 1 B and C). This method has been used in mycobacteria to identify essential genes using transposons (15) and in Drosophila to screen for P-element insertions (16). The amplicons were digested, in three separate reactions, with three enzymes with 4-base recognition sites (MseI, MspI, and HpyCH4V), to produce small fragments suitable for microarray hybridization (Fig. 1C). Three enzymes were used in this step to minimize the effect of cutting in the middle of an already small fragment that would otherwise have hybridized to an array feature, leading to potential loss of signal; with three separate and subsequently pooled digests, sequences that could hybridize to an array feature are nearly all (44,229 of 44,290 or >99.9%) present at full length in at least one of the digests.
Construction of a Tiling Array with Complete Genome Coverage.
Identification of transposon insertion sites by microarray is limited by the fact that most microarrays cover only exons, whereas transposons are often targeted to intergenic regions. The TIP-chips are simple tiling arrays, constructed as custom arrays on the 44K 60mer Agilent platform. First, the yeast genome was masked according to the Saccharomyces Genome Database (SGD) annotation: repeats were omitted from the sequences used for feature selection. Using a combination of sequences identified by Primer3 and evenly spaced oligonucleotides falling between the former, the features are placed, on average, every 280 nt, and give nearly 25% direct coverage of the masked yeast genome.
Because the tiling features are so closely spaced, each transposon flank of >600 bp will hybridize to at least two and typically more adjacent array features, giving an unambiguous, readily visible signal in the form of a line of spots (as our tiling features are not randomized but are placed in reading order across the array, in chromosomal order). This method enables easy visual differentiation of sporadic background feature hybridization from actual transposon flank hybridization; however, it does increase the potential effects of spatial artifacts.
FY2 Strain.
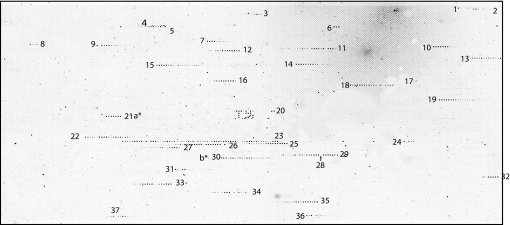
We first hybridized FY2 genomic DNA to the TIP-chip. FY2 is an S288C derivative closely related to the strains used for the S. cerevisiae genome sequencing project. With the Ty1-specific primer used, 31 of the 32 known Ty1 elements (31 annotated in SGD in addition to one known FY2-specific insertion) were expected to hybridize to the array. Two of the elements are present in tandem orientation, and the 3′ element is undetectable because it lacks nonrepetitive sequences flanking its 5′ end; this leaves 30 elements that should be visible on the array. Fig. 2 shows the FY2 array in grayscale, with numbers marking each putative transposon flank identified. This experiment was performed in duplicate, and the same 37 lines were seen on the second array. Table 1 gives details for the sequences associated with each line of spots, and documents the successful capture of 30 of 30 detectable Ty1 elements, giving a true positive rate of 100%. Table 1 also gives the distance from the nearest end of each line on the array to the central base of the target site duplication of the transposon that it identifies; for all but one case where the distance is >1 kb, the apparently large distance is due to intervening repetitive, masked features. For the rest of the inserts, the mean distance is 408 nt and many lines terminate very close to their transposons, often within 50 nt.
Fig. 2.
Typical hybridization of FY2 amplicons to a TIP-chip. Each putative transposon flank appears as a line on the array. The bound features are numbered; these numbers correspond to Table 1. The numbers are placed so that they are nearest the endpoint of the linear signal closest to the Ty1 element and thereby indicate the orientation of the Ty1 element. Ty1 hybridization controls (features spanning the LTRs) in the middle of the array produce the “TY” pattern. Interruptions in the lines of spots represent intervening hybridization negative controls.
Table 1.
FY2 insertions
| No. | Chr | Start | Stop | Nearest Ty1 | Distance to Ty1 | Ty1 name | Ty1 orientation |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 166210 | 166517 | 166161 | 49 (l) | YARCTy 1-1 | − |
| 2 | 2 | 214467 | 215596 | 221042 | 5446 (r) | YBLWTy 1-1 | + |
| 3 | 2 | 259092 | 259575 | 259578 | 3 (r) | YBRWTy 1-2 | + |
| 4 | 3 | 81701 | 82002 | Ty2 cross-match? | |||
| 5 | 3 | 146051 | 148410 | 148613 | 203 (r) | FY2-specific Ty1* | + |
| 6 | 3 | 168649 | 169309 | Misannotated LTR† | |||
| 7 | 4 | 651589 | 653289 | 651414 | 177 (l) | YDRCTy 1-1 | − |
| 8 | 4 | 802394 | 802746 | 803192 | 446 (r) | YDR170W-A | Ty1 ORF + |
| 9 | 4 | 884651 | 888630 | 884213 | 438 (l) | YDRCTy 1-2 | − |
| 10 | 4 | 992810 | 993967 | 992634 | 176 (l) | YDRCTy 1-3 | − |
| 11 | 4 | 1093276 | 1095636 | 1095764 | 128 (r) | YDRWTy 1-4 | + |
| 12 | 4 | 1203843 | 1206693 | 1206696 | 3 (r) | YDRWTy 1-5 | + |
| 13 | 5 | 111456 | 116123 | 116290 | 167 (l) | ura3-52 insertion‡ | + |
| 14 | 5 | 449762 | 452131 | 449314 | 448 (l) | YERCTy 1-1 | − |
| 15 | 5 | 498549 | 501233 | 498414 | 135 (l) | YERCTy 1-2 | − |
| 16 | 7 | 532701 | 535606 | 535766 | 160 (r) | YGRWTy 1-1 | + |
| 17 | 7 | 568112 | 568432 | 567762 | 350 (l) | YGRCTy 1-2 | − |
| 18 | 7 | 823457 | 826056 | 823309 | 148 (l) | YGRCTy 1-3 | − |
| 19 | 8 | 549666 | 551790 | 549634 | 32 (l) | YHRCTy 1-1 | − |
| 20 | 10 | 203885 | 203926 | LTR§ | |||
| 21 | 10 | 470739 | 472376 | 472379 | 3 (r) | YJRWTy 1-1 | First of two tandem + elements |
| 22 | 12 | 221324 | 224876 | 218910 | 2418 (l) | YLR035C-A | Ty1 ORF − |
| 23 | 12 | 459949 | 460576 | ND¶ | |||
| 24 | 12 | 489581 | 490388 | 481896 | 7685 (l) | YLRCTy 1-1 | − |
| 25 | 12 | 584882 | 593097 | 593149 | 52 (r) | YLRWTy 1-2 | + |
| 26 | 12 | 645906 | 650793 | 650828 | 35 (r) | YLRWTy 1-3 | + |
| 27 | 12 | 815946 | 817995 | 818034 | 39 (r) | FY2-specific Ty1* | + |
| 28 | 13 | 183810 | 184138 | 184172 | 34 (r) | YMLWTy 1-1 | + |
| 29 | 13 | 191099 | 196331 | 196334 | 3 (r) | YMLWTy 1-2 | + |
| 30 | 13 | 378622 | 385148 | 378619 | 3 (l) | YMRCTy 1-4 | − |
| 31 | 14 | 102522 | 103628 | 102519 | 3 (l) | YNLCTy 1-1 | − |
| 32 | 14 | 517252 | 518954 | 519164 | 210 (r) | YNLWTy 1-2 | + |
| 33 | 15 | 114236 | 117701 | 117704 | 3 (r) | YOLWTy 1-1 | + |
| 34 | 15 | 590985 | 594106 | 594822 | 716 (r) | YORWTy 1-2 | + |
| 35 | 16 | 52854 | 55959 | 62377 | 6418 (r) | YPLWTy 1-1 | + |
| 36 | 16 | 810602 | 811799 | 810560 | 46 (l) | YPRCTy 1-2 | − |
| 37 | 16 | 843409 | 857882 | 844410 (+) and 856552 (−) | Internal‖ | YPRWTy 1-3 and YPRCTy 1-4 | Two tail-to-tail elements |
For each apparent transposon flank seen on the array in Fig. 3A, a detailed analysis of the chromosomal coordinates spanned by the bound features, along with the known coordinates, SGD name, and orientation of the nearest Ty1 element (all from the SGD feature table), is shown. Also shown is the distance to the nearest known Ty1 element, with (l) indicating that the transposon is located nearest to the left end of the line and (r) marking transposons nearest the right ends of the lines.
*These Ty1 elements are not reported in the original S288C isolate genome sequence and are thus inferred to represent insertions that occurred during strain construction or subsequent laboratory subculture.
†This sequence contains an LTR that was not annotated in the SGD database version used to design the array. Because all amplicons contain Ty1 LTR sequences, this region is in fact expected to hybridize.
‡This insertion is known to be present in strain FY2 and not in the strains used for the genome sequencing project.
§LTR unintentionally left unmasked.
¶This insertion is in a repetitive portion of the rDNA region of chromosome 12, and we did not attempt to ascertain the exact position of new Ty1 insertion site.
‖Two inverted tail-to-tail Ty1s are expected to lie internal to the hybridization line, rather than at one endpoint.
In addition to four matches to Ty1 or LTR sequences that were not excluded from the array design due to annotation problems, there are five signals in the FY2 array that did not correspond to annotated Ty1 elements or LTRs. Line 4 is most likely a spurious cross-match to a Ty2 element that happens to contain a sequence with a high-scoring 22 of 24 exact, yet gapped match to our primer. Line 23 represents binding to very repetitive features in the rDNA region of chromosome 12; this potentially FY2-specific insertion is not easily confirmed; in fact, any insertion into repetitive DNA cannot be localized with complete certainty. The other three signals have biologically interesting explanations. One of these, number 13, is the known ura3-52 allele, consisting of a Ty1 insertion into the URA3 ORF, not present in the S288C sequence but known to exist in the FY2 strain (17, 18), and the other two, numbers 5 and 27, were verified by PCR and sequencing and found to represent two previously unknown Ty1 insertions, and thus are additional true positives. One of the new insertions (number 5) occurs on chromosome 3, between two tRNA genes, and the other (number 27) is on chromosome 12, very close to a tRNAArg. Our false positive rate is therefore essentially zero (with the possible exception of lines 4 and 23).
Inverted Ty1 elements, if oriented tail-to-tail, will appear as a single line of spots (with the Ty1 elements positioned inside rather than at one endpoint of the line); we observed this in the FY2 array (number 36) and, knowing the true position of all of the Ty1 elements, were able to correctly interpret these signals. In analyzing an unknown strain, any given linear signal may therefore represent more than one insertion, and PCR or other verification techniques are necessary if pinpointing the location of all transposons is required. This can be done by designing two PCR primers outside each endpoint of the line of spots, to amplify potential transposon junctions in both directions. Two known Ty1 elements were not expected to be detected by our array for technical reasons, shown as letters in Fig. 2. One of these, YJRWTy1–2, on chromosome 10 (a* in Fig. 2), is part of a tandem Ty1 duo and is therefore undetectable using the set of primers used in this experiment, as it has no unique sequences flanking its 5′ end, only Ty1 sequences. The other element, YMRCTy1–3, (b* in Fig. 2) is somewhat degenerate at the site matching our primer, with two internal mismatches out of 24 nucleotides, and is apparently undetectable with the primers used. In a more comprehensive version of the TIP-chip strategy, one could design several transposon-specific primers, and pool the resulting amplicons before labeling and hybridization. However, with our array design, we cannot currently recover Ty1 insertions into preexisting Tys.
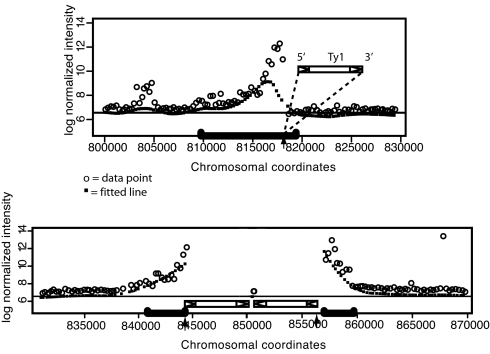
In Fig. 2, many of the lines appear less intense at one end than at the other, and the more intense end corresponds to the end nearest the transposon. This phenomenon is a layering effect, due to the cumulative fluorescence of overlapping restriction fragments from the transposon flanks. The features nearest the transposon will be bound by subfragments from each of the three restriction fragments, the next set of features by only two, whereas the furthest features will be bound by subfragments only from the longest restriction fragment. Furthermore, the longer amplicons are likely to be amplified less efficiently, magnifying this effect. This creates a directional intensity gradient in each line, with the most intensely fluorescent features nearest the endpoint of the line identifying the site of the transposon insertion, as evident in Fig. 2. As can be seen (Fig. 3), this directionality can be inferred computationally. We first normalized and smoothed the data as described in Materials and Methods and then scanned for regions of five or more features in a row with Z scores above a predefined cutoff. The slope of each line of features was calculated and this slope correlates perfectly with the position of the transposon insertion site relative to the endpoints of the line. In 33 of 33 (100%) cases where the line found by this method corresponded to a known Ty1 insertion site, the correct position and orientation of the Ty1 could be inferred from the slope of the line (Fig. 3A). This method correctly identified the tail-to-tail element insertion in line 37 (Fig. 3B).
Fig. 3.
Two Ty1 insertion sites from the FY2 strain, shown as graphs of log normalized intensity versus chromosomal coordinates. The top graph, from chromosome 12, shows a new Ty1 insertion site (line 26 in Fig. 2), in which the Ty1 lies on the right side of the line (downstream in chromosome coordinates, confirmed by PCR), giving the line a positive slope. The bottom graph displays the same information for two known tail-to-tail Ty1 insertions on chromosome 16 (line 37 in Fig. 2). The gap in the line is due to masking of the 6-kb Ty1 elements; there are no features spanning this region. Arrowheads mark positions of confirmed and known Ty1 elements. Blue brackets mark regions for which the Z score is >2.5 for each spot (P value ≈ 0.01 for each spot, therefore much lower for the entire line).
L27-10 Ty1 High-Copy Strain.
We also used the TIP-chip to profile the Ty1 composition of a Ty1 high-copy strain and its immediate parent strain. This high copy strain has undergone ten cycles of retrotransposition and thus is expected to carry numerous additional copies of Ty1 elements in its genome (ref. 19, and L.Z.S., C. J. Cost, M. L. Zupancic, E. M. Caputo, and J.D.B., unpublished data). The TIP-chip should provide an excellent method for mapping these insertions comprehensively; this was tested in L27-10, a yeast strain derived from GRF167 (MATα, ura3-167, his3Δ200).
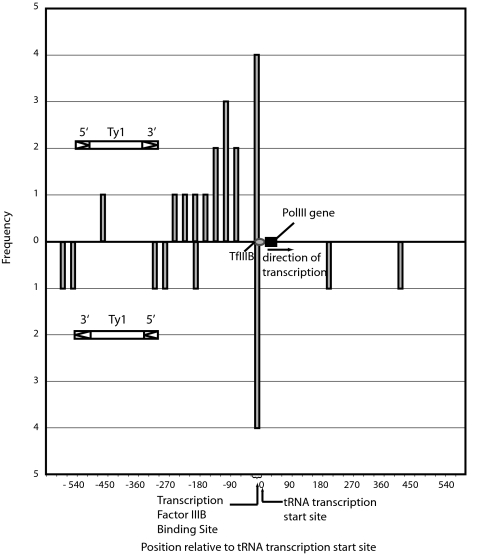
We identified 66 lines hybridizing to the L27-10 TIP-chips that were not seen in GRF167, and two lines for the GRF167 strain that were not seen in any of the L27-10 TIP-chips. The latter class may represent a new insertion in GRF167 or a deletion in L27-10. A virtual overlay of the data from the L27-10 and GRF167 TIP-chips shows the signals that appear in one array and not the other (Fig. 5). Each signal in L27-10 not seen in GRF167 was examined in detail by PCR and sequence analyses (see supporting information for detail). In total, 66 insertions were identified, this finding is in good agreement with real-time PCR experiments that predict this strain harbors ≈70 new Ty1 elements (data not shown). The 66 insertions fell into three classes, sequence confirmed (24 or 36%), likely true positive (29 or 44%) and likely false positive (20%). Although it is not possible to definitively determine the false positive rate without a complete genome sequence from this strain, the data suggest a true positive rate of 80%. Ty1 elements insert near RNA polymerase III (polIII) transcripts (4, 20, 21); this is also true of the sequence-confirmed new copies of Ty1 that accumulate in the high copy strain, as 92% of these are within 2 kb of a polIII gene. Interestingly, one of these insertions hit SNR52, the only snoRNA transcribed by polIII in yeast (22). Table 2 details the locations of all 66 new insertions. Fig. 4 displays all sequence-confirmed insertions within 600 nt of polIII-transcribed target genes; nearly all insertions fall upstream of these targets.
Fig. 4.
Histogram showing confirmed new Ty1 insertion positions relative to transcription start sites of tRNA genes (at 0). Bin size is 150 nucleotides; orientation of Ty1s is indicated.
Seven of the previously unidentified PCR-amplified insertions were actually positioned in the middle of the line seen on the array; those signals are presumed to represent flanks from multiple Ty1 elements inserted in close genomic proximity, evidence for such clusters was found by sequence analysis (Table 2). Additional analysis will be needed to comprehensively pinpoint every single Ty1 element in this strain. The TIP-chip, however, gives a very rapid and complete “big picture” of the positions of the new insertions, and it is visible at a glance that the new Ty1 copies are inserted in a dispersed manner throughout the genome.
Discussion
Transposable elements occupy a very important niche in the biology of most, if not all, organisms. Surprisingly few tools exist for comprehensively mapping the genomic distribution of transposons in any given sample and, in particular, their variation between different individuals of the same species (a question of potential medical as well as biological significance). The TIP-chip microarray methodology meets this need. We have used the TIP-chip to successfully profile the transposons in the FY2 yeast strain, identifying 100% of the detectable transposons as well as two previously unknown insertions. With some modifications, such as using multiple transposon-specific primers in the PCR and amplifying and separately labeling both transposon flanks (so that head-to-head insertions and solo LTR insertions can be recovered), this success rate can increase and additional information can be extracted from these arrays. Our unique strategy for finding and quantifying lines and their slopes, and thereby determining more precisely the location of the insertion site, is also extremely informative. We have profiled the transposons in a high-copy Ty1 strain and have uncovered a large number of new insertions, in agreement with previous predictions. Even in the face of very complex multiple transposon insertions in close proximity, the TIP-chip is a very valuable first-pass tool, because it quickly identifies most or all of the transposon insertion sites in any given sample, and for many applications, including polymorphism studies, knowing the rough location of the transposon insertions is sufficient.
Although our studies were done in yeast, we expect that the transition to more complex genomes will not be an insurmountable challenge. Work done in Drosophila (16) and in the banana (23) using similar techniques shows that vectorette PCR is easily adaptable to complex genomes. Furthermore, we performed TIP-chip analysis on a yeast–human DNA mixture in which the yeast genome was mixed with a 100-fold excess of human genomic DNA by weight, mimicking a human genome experiment. The TIP-chip data were qualitatively similar to the control chip done with only a small amount of yeast DNA, although background was slightly higher (Fig. 7). Thus, the basic technique described is readily applied to more complex samples.
The TIP-chip is an important step forward for transposon studies, because it is a simple, yet effective method for examining the transposon terrain in any given sample, allowing profiling of biologically and medically relevant transposons in a high-throughput manner.
Materials and Methods
Design of the Microarray.
A total of 41,995 60-bp features were chosen from the yeast genome in a three-step process. First, the yeast genome was masked according to the SGD annotation; retrotransposons, LTRs, telomere repeats, and X and Y′ elements were excluded from the sequences used for feature selection. Second, Primer3 (ref. 24; www-genome.wi.mit.edu/cgi-bin/primer/primer3_1020;www.cgi) was used to choose oligonucleotides with the lowest likelihood of conformational problems (parameters: optimal size, 60; Tm min, 72; Tm opt, 76; Tm max, 80; otherwise default); however, this process did not yield enough oligonucleotides spaced at the required high density: some oligos were spaced up to 10 kb apart, and only 38,455 were chosen along the yeast genome. Finally, the remaining oligonucleotides were placed evenly across any gaps with complete disregard for sequence properties. The 60-mers were arranged in sequence order on the microarray such that hybridization to adjacent features would produce visible lines. Custom 44K 60mer Agilent microarrays (AMADID 013306) were used.
Amplification of Transposon Flanking Fragments.
We followed the basic vectorette protocol first described in Riley et al. (14). Yeast genomic DNA, prepared as described by Yuan et al. (25), was treated with RNase, if necessary, and 20 μg of gDNA was immediately digested with EcoRI, AflII, and HindIII in three separate 250-μl reactions. After digestion, the fragments were heat-inactivated at 65°C for 20 min and then ligated to the annealed vectorette primers (JB9408, common to all reactions, JB9409 for the EcoRI fragments, JB9487 for the AflII fragments, and JB9488 for the HindIII reaction). See supporting information for primer sequences. After ligation, the fragments were amplified by using the vectorette primer, JB9410, and also the Ty1-specific primer, JB8784, complementary to sequences adjacent to the 5′ LTR.
The amplified Ty1-adjacent fragments were pooled and digested in three parallel reactions with MseI, MspI, and HpyCH4V. The digests were heat inactivated and then pooled and labeled for use on the microarray. The products were purified and concentrated on a Microcon column (Amicon, Millipore, Bedford, MA), boiled, and spotted onto microarrays and covered with coverslips. The microarrays were hybridized overnight and washed in 2× SSC, 0.03% SDS for 5 min at 65°C, then in 1× SSC for 5 min at room temperature, and finally in 0.2× SSC for 5 min at room temperature. Microarrays were allowed to air dry and then were scanned in a GenePix 4000B scanner from Axon Instruments (Sunnyvale, CA), using GenePix Pro 5.1 software.
Microarray Analysis: Finding and Quantifying Lines.
Two methods (outlined in more detail in supporting information) were used to define lines of spots that were above the background. In analysis method 1, we simply looked at the F635 median–B635 median difference and empirically set a cutoff defining hybridized vs. unhybridized features. We then scanned the data in order of ID (which is the same as chromosomal coordinates) and looked for three or more features in a row above the cutoff, with fewer than two intervening features below the cutoff. In analysis method 2, we first normalized the data to minimize spatial effects. We took advantage of the fact that there should be no lines of features with high intensity in the vertical dimension and estimated spatial biases by fitting a loess curve to the log intensity versus column number (horizontal dimension) scatterplot. We did this for each row and used the residuals as the normalized data. Because amplified probes are expected only in the horizontal dimension, features related to amplified regions will appear as outliers in the log-intensity versus column number plots and thus ignored by loess (a robust procedure). We added back the median log intensity of the original data to keep it in the original scale.
The features were naturally segmented across chromosomes by the repeat masking performed during the construction of the array. To reduce noise, we smoothed the data in each chromosomal segment (in the horizontal dimension) by using a running window of ten features and averaging each window using loess to remove outliers.
Empirical densities of the log intensity smoothed data (not shown) showed that the log-intensity data were normally distributed with the exception of a few outliers. These outliers, of course, are related to the feature of interest. Therefore, we assumed that log-intensities associated with unamplified regions followed a normal distribution. We refer to this as the null distribution. Because of the outliers, we estimated the mean and variance of this distribution with the robust summary statistics: the median and MAD (median absolute distance) of the log intensities. With the null-distribution properly estimated, we were then able to covert the smoothed log-intensity data into Z scores (subtract the mean and divide by the standard deviation).
Scanning the data once more, we looked for regions of five or more features in a row with Z scores above a predefined cutoff (we used 2.5, which roughly corresponds to a marginal P value of 0.01). The slope of each line of features was then calculated; positive slopes correspond to Ty1 elements on the plus strand, negative slopes correspond to Ty1 features on the minus strand, and near-zero slopes indicate tail-to-tail inverted Ty1 pairs.
Supplementary Material
Acknowledgments
S.J.W. was supported by National Institutes of Health (NIH) Training Grant CA009139. L.Z.S. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-1858-05). This work was supported in part by NIH Grants GM36481 and CA16519 (to J.D.B.).
Abbreviations
- TIP
transposon insertion site profiling
- SGD
Saccharomyces Genome Database.
Note Added in Proof.
A similar method for mapping transposon insertion sites was independently developed by Gabriel and colleagues (A. Gabriel, J. Dapprich, M. Kunkel, D. Gresham, S. Pratt, and M. Dunham, personal communication).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5646).
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Wicker T, Robertson JS, Schulze SR, Feltus FA, Magrini V, Morrison JA, Mardis ER, Wilson RK, Peterson DG, Paterson AH, et al. Genome Res. 2005;15:126–136. doi: 10.1101/gr.2438005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 5.Messing J, Dooner HK. Curr Opin Plant Biol. 2006;9:157–163. doi: 10.1016/j.pbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Han JS, Boeke JD. BioEssays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 7.Kazazian HH., Jr Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 8.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissinot S, Chevret P, Furano AV. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 10.Liti G, Peruffo A, James SA, Roberts IN, Louis EJ. Yeast. 2005;22:177–192. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- 11.Eibel H, Gafner J, Stotz A, Philippsen P. Cold Spring Harbor Symp Quant Biol. 1981;45:609–617. doi: 10.1101/sqb.1981.045.01.079. [DOI] [PubMed] [Google Scholar]
- 12.Sniegowski PD, Dombrowski PG, Fingerman E. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 13.Bachman N, Eby Y, Boeke JD. Genome Res. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sassetti CM, Boyd DH, Rubin EJ. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggert H, Bergemann K, Saumweber H. Genetics. 1998;149:1427–1434. doi: 10.1093/genetics/149.3.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winston F, Dollard C, Ricupero-Hovasse SL. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 18.Rose M, Winston F. Mol Gen Genet. 1984;193:557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- 19.Boeke JD, Eichinger DJ, Natsoulis G. Genetics. 1991;129:1043–1052. doi: 10.1093/genetics/129.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 21.Devine SE, Boeke JD. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 22.Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Hernandez JB, Swennen R, Sagi L. Transgenic Res. 2006;15:139–150. doi: 10.1007/s11248-005-2544-5. [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 25.Yuan DS, Pan X, Ooi SL, Peyser BD, Spencer FA, Irizarry RA, Boeke JD. Nucleic Acids Res. 2005;33:e103. doi: 10.1093/nar/gni105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.