Abstract
Membrane proteins play vital roles in every aspect of cellular activities. To study diverse membrane proteins, it is crucial to select the right surfactants to stabilize them for analysis. Despite much effort, little progress has been made in elucidating their structure and function, largely because of a lack of suitable surfactants. Here we report the stabilization of a G protein-coupled receptor bovine rhodopsin in solution, using a new class of designer short and simple peptide surfactants. These surfactants consist of seven amino acids with a hydrophilic head, aspartic acid or lysine, and a hydrophobic tail with six consecutive alanines. These peptide surfactants not only enhance the stability of bovine rhodopsin in the presence of lipids and the common surfactants n-dodecyl-β-d-maltoside and octyl-d-glucoside, but they also significantly stabilize rhodopsin under thermal denaturation conditions, even after lipids are removed. These peptide surfactants are simple, versatile, effective, and affordable. They represent a designer molecular nanomaterial for use in studies of diverse elusive membrane proteins.
Keywords: lipid-like peptides, membrane proteins, self-assembly
Membrane proteins are involved in all aspects of vital cellular activities including energy conversion, photosynthetic electron transport, cell signaling, cell–cell interactions, cell adhesion, cell migration and movement, cytoskeletal organization, protein trafficking, viral fusion, information propagation, cellular secretory and neural synaptic activities, ion and metabolite transport, and respiratory transport. An approximate one-third of the genes in the human genome code for membrane proteins. Of that number, only a single human membrane protein structure (that of monoamine oxidase B) has been determined by x-ray diffraction at 3-Å resolution (1). To a lesser resolution (3.8 Å), the structure of human aquaporin 1 was determined by electron crystallographic diffraction (2). Thus, membrane protein structures largely remain elusive, primarily because of a lack of the right surfactants.
G protein-coupled receptors (GPCRs) comprise a large class of membrane proteins and play a crucial role in the signaling cascade (3, 4). Although they are vitally important in the pharmaceutical and biotechnology industries, medical science, and nanobiotechnology (3, 4), only a single bovine rhodopsin structure is known (5–8). To study the structure and function of diverse membrane proteins, including GPCRs, it is crucial to discover, search, design, synthesize, and select the right surfactants to stabilize membrane proteins.
It is estimated through extensive bioinformatics studies that approximately one-third of the total number of genes in sequenced organisms' genomes code for membrane proteins (9–13). Despite the importance of membrane proteins, dynamic studies of them and methods for their high-resolution structural analysis are rather limited. Although >35,000 soluble protein structures have been elucidated (Protein Data Bank, www.rcsb.org/pdb), only 215 membrane proteins, including 113 unique structures, have been determined as of July 2006 (http://blanco.biomol.uci.edu/membrane_proteins_xtal.html). Thus, membrane proteins pose a grand challenge that requires new tools, materials, and methods for systematic structural and other studies. Although numerous surfactants are available and have been used for many years in membrane protein studies, none have been completely satisfactory for use in stabilizing diverse membrane proteins, which quickly denature or aggregate in solution. New types of surfactants that can preserve membrane protein stability, structure, and function are prerequisites in tackling the problem and are urgently needed.
Previously, limited numbers of membrane proteins were solubilized, stabilized, and crystallized by using a variety of surfactants (12, 14–17). Most surfactants that have been successfully used contain a hydrophilic head and a hydrophobic tail that consists of 6–12 carbon atoms (12, 18–20). Surfactants with short hydrophobic tails prove to be the most useful for crystallization (21). In addition, several α-helical peptide surfactants and composite α-helix peptides with lipid tails have been reported to stabilize membrane proteins (22–24). However, these α-helical peptides are >30 residues long and expensive to obtain; they also require proper folding before they attain the properties of a surfactant.
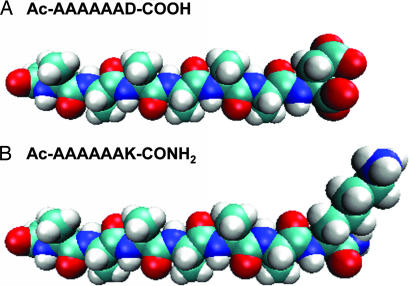
We previously reported the design of a class of self-assembling amphiphilic peptide surfactants that include G4D2, G6D2, G8D2, A6D, A6K, V6D, V6K, and L6D2 (25–27). These surfactants comprise ≈6–10 amino acid residues, are ≈2–3 nm in length, and have properties similar to those of common surfactants, such as n-dodecyl-β-d-maltoside (DM) and octyl-d-glucoside (OG). These peptides have one or two hydrophilic amino acids at one end, with either a negatively charged aspartic acid or a positively charged lysine followed by several consecutive hydrophobic amino acids such as glycine, alanine, valine, and leucine (Fig. 1). When the N terminus is acetylated, the surfactants A6D and V6D have two negative charges, one from the C terminus and the other from the aspartic acid side chain. On the other hand, A6K has one negative charge from the C terminus and a positive charge from the lysine side chain. When dissolved in water or ionic solutions, these peptide surfactants undergo self-assembly to form micelles, nanovesicles, or nanotubes (25–27).
Fig. 1.
Molecular models of peptide surfactants. (A) A6D. (B) A6K. Aspartic acid (D) bears negative charges, and lysine (K) bears a positive charge. Alanine (A) constitutes the hydrophobic tails with increasing hydrophobicity. Color code is as follows: cyan, carbon; red, oxygen; blue, nitrogen; white, hydrogen. Each peptide is 2.5 nm in length, similar to biological phospholipids.
Similar to common surfactants, these peptide surfactants have defined critical aggregation concentrations (CACs) in the submillimolar to millimolar range, depending on the hydrophobicity of the tails and the ionic concentration. For example, in water, A6D has a CAC of ≈1.6 mM, and A6K has a CAC of ≈1.5 mM; however, in phosphate-based saline (PBS) (10 mM sodium phosphate/150 mM NaCl, pH 7.4), A6D has a CAC of ≈0.25 mM, and A6K has a CAC of ≈0.23 mM. This difference is due to the charge screening effect in accordance with the Derjaguin–Landau–Verwey–Overbeek (DLVO) theory, which postulates that a critical coagulation concentration of counterions is required to allow assembly and that this concentration will be inversely proportional to the valence of the counterion raised to the sixth power (28). The peptide surfactant supramolecular structure is similar to that of phospholipids; namely, the formation of a polar interface sequesters the hydrophobic tails from water.
We asked whether the peptide surfactants are capable of stabilizing a well characterized membrane protein, such as GPCR bovine rhodopsin. Rhodopsin consists of seven transmembrane helices that form a binding pocket for the chromophore, 11-cis-retinal (29, 30). In a dark state, rhodopsin has an absorbance maximum of 500 nm. On absorption of a photon, the retinal isomerizes from 11-cis to all-trans and, concomitantly, rhodopsin undergoes conformational changes that result in an activated state, Meta II, which has a maximum visible absorbance of 380 nm (31–34). Rhodopsin can be thermally denatured, which leads to the loss of the 500-nm absorbing chromophore (A500). Here we report that the peptide surfactants stabilize GPCR bovine rhodopsin more effectively than the other common surfactants that have been tested so far. We also show that the designed peptide surfactants stabilize other membrane proteins, i.e., the photosystem I complex. Our studies suggest that short peptide surfactants may be promising material for further studies of membrane proteins.
Results
Peptide Surfactant A6D Enhances the Thermal Stability of GPCR Bovine Rhodopsin.
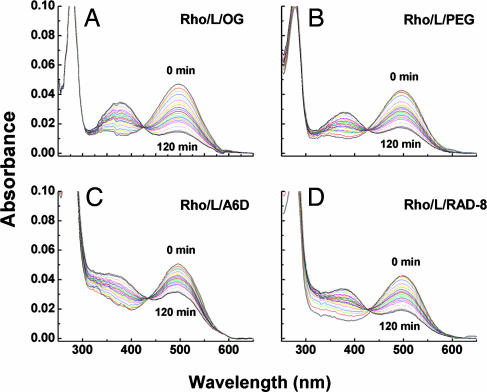
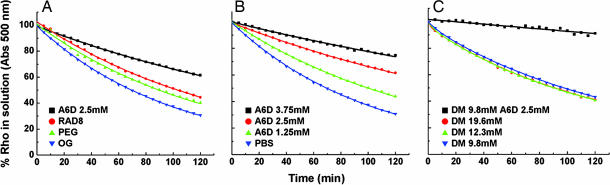
We examined the thermal stability of bovine rhodopsin in the presence of several surfactants. First, we extracted the rhodopsin from bovine retinae rod outer segments by using 2% OG (CAC of ≈25 mM) and obtained rhodopsin in phospholipid/OG mixed micelles. We then measured the thermal stability of rhodopsin in different surfactants and PBS solutions by following the absorbance decrease in A500 as a function of incubation time. We found that rhodopsin in a rhodopsin/phospholipid/OG complex displayed a thermal half-life of ≈71 min (Figs. 2 and 3A and B). When 2.5 mM A6D (≈10× CAC in PBS) was added to rhodopsin/phospholipid/OG mixed micelles, the half-life of rhodopsin increased to ≈173 min, representing a 2.5-fold increase in stability compared with the stability in OG alone (Figs. 2 and 3 A and B). To rule out nonspecific chromophore stabilization, we used another octapeptide, RAD8 (Ac-n-RADARADA-c-NH2), which is similar in size to A6D without significant surfactant properties. Rhodopsin in phospholipid/OG/RAD8 mix displayed a half-life of ≈99 min (Figs. 2 and 3A).
Fig. 2.
UV-visible absorption spectra of GPCR bovine rhodopsin (Rho) in PBS containing 1% OG (A), 0.9% PEG/1% OG (B), 2.5 mM A6D/1% OG (C), or 2.8 mM RAD8/1% OG (D). Spectra were recorded every 5 min, up to 120 min, at 40°C. L, lipid.
Fig. 3.
Stability kinetics of rhodopsin (Rho) in different surfactants. Thermal stability of rhodopsin was measured as a rate of the decay at A500. The A500 spectra at different time points are expressed as percentage of absorbance at the initial state of the experiment. (A) The half-life of rhodopsin in different surfactants (as described in the Fig. 2 legend) was as follows: 71 min in 1% OG, 90 min in 0.9% PEG/1% OG, 173 min in 2.5 mM A6D/1% OG, and 99 min in 2.8 mM RAD8/1% OG. (B) Stability of rhodopsin as a function of the concentration of the peptide surfactant A6D at 40°C. Half-life of rhodopsin was as follows: 277 min in 3.75 mM A6D/1% OG, 173 min in 2.5 mM A6D/1% OG, 101 min in 1.25 mM A6D/1% OG, and 71 min in 1% OG (PBS). (C) Stability of rhodopsin in DM with or without A6D at 50°C. Half-life of rhodopsin was as follows: not available in 2.5 mM A6D/9.8 mM DM, 96 min in 19.6 mM DM, 96 min in 12.3 mM DM, and 102 min in 9.8 mM DM.
It is known that proteins are usually more stable at higher concentrations. To rule out the concentration effect of A6D nonspecifically increasing rhodopsin stability, we added 0.9% PEG 1000 to the rhodopsin/lipid/OG complex. We observed that rhodopsin had a half-life of ≈90.0 min after a 2-h incubation; no significant increase in thermal stability was observed (Figs. 2 and 3A). These results suggest that enhanced stability is not due to higher concentrations of substance; rather, it is plausible that A6D formed mixed micelles with OG and phospholipid and effectively enhanced rhodopsin stability against thermal denaturation.
To evaluate the effectiveness of peptide surfactant A6D in stabilizing rhodopsin's structural integrity in the rhodopsin/phospholipid/OG/A6D complex, we carried out experiments by using various peptide surfactant A6D concentrations. We prepared rhodopsin/lipid/OG complex containing different concentrations of A6D, at 1.25 mM (≈5× CAC), 2.5 mM (≈10× CAC), and 3.75 mM (≈15× CAC), and then measured the loss of absorbance at 500 nm as a function of incubation time (Fig. 3B). We found that the stability half-life of rhodopsin increased from ≈71 min, without A6D, to ≈101, ≈173, and ≈277 min, respectively, as a function of A6D concentration increase. The nearly 4-fold increase in stability of rhodopsin was observed with the highest A6D concentration (3.75 mM) (Fig. 3B). These observations suggest that peptide surfactant A6D effectively stabilized GPCR bovine rhodopsin.
To determine whether the effect of A6D on stabilizing rhodopsin is specifically associated with OG, we combined A6D with DM (CAC of ≈0.15 mM) and measured rhodopsin half-life in rhodopsin/lipid/DM preparations. DM is one of the most common surfactants for solubilizing and stabilizing membrane proteins, and it has been shown to stabilize rhodopsin in the absence of lipids at room temperature in ≈2 days at 40°C. We first prepared samples containing different concentrations of DM to determine DM saturation concentration so that we could use peptide surfactant A6D to test whether the addition of A6D further enhances rhodopsin stability. We prepared samples containing different DM concentrations. We found that the half-life of rhodopsin in rhodopsin/phospholipid/9.8 mM DM (65× CAC), rhodopsin/phospholipid/13.3 mM DM (89× CAC), or rhodopsin/phospholipid/19.6 mM DM (130× CAC) was ≈102, ≈93, and ≈93 min, respectively (Fig. 3C).
The results showed that increasing DM concentration alone is insufficient to enhance bovine rhodopsin thermal stability. At a defined DM saturation concentration, we then added A6D to rhodopsin prepared in the DM concentration given above and monitored its thermal stability. Our results showed that, when the rhodopsin/phospholipid/DM sample was supplemented with an additional 2.5 mM A6D (10× CAC), the rhodopsin became extremely stable with little decay, even at 50°C (Fig. 3C). These results demonstrate that A6D significantly enhances rhodopsin stability against thermal denaturation when A6D and DM are combined to elevate surfactant effectiveness in protecting rhodopsin stability, possibly in a synergistic way.
Peptide Surfactant A6D Stabilized Rhodopsin in the Absence of OG.
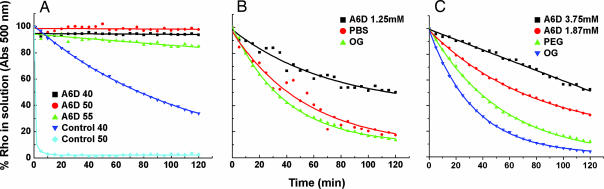
We then asked whether the peptide surfactant A6D alone, without OG or DM, could stabilize rhodopsin. We removed OG from the rhodopsin/lipid/OG mixture through extensive dialysis against buffers containing insignificant OG (defined as no OG) and then monitored rhodopsin thermal stability at 40°C, 50°C, and 55°C. Surprisingly, the rhodopsin/lipid/A6D complex without OG was very stable at all tested temperatures with little denaturation. In contrast, rhodopsin in control samples containing only OG, without A6D, had a half-life of ≈101 min at 40°C and <5 min at 50°C (Fig. 4A). These results suggest not only that A6D alone is sufficient to stabilize rhodopsin but also that peptide surfactant A6D can be used for protecting rhodopsin in solution without the common surfactants OG and DM. This observation opens the way to test the ability of A6D to stabilize and crystallize membrane proteins.
Fig. 4.
Kinetics of bovine rhodopsin under different conditions. (A) Stability of rhodopsin in the absence of OG at different temperatures. Half-life of rhodopsin was as follows: not available in 2.5 mM A6D at 40°C, 50°C, and 55°C; 101 min in control solution (1.25 mM A6D/1% OG) at 40°C; <5 min in control solution at 50°C. (B) Decay of A500 in delipidated rhodopsin in the absence of OG. Half-life of rhodopsin was as follows: 122 min in 1.25 mM A6D, 47 min in PBS, and 27 min in 1% OG (control). (C) Stability of delipidated rhodopsin at 40°C. Half-life of rhodopsin was as follows: 128 min in 3.75 mM A6D/1% OG, 76 min in 1.87 mM A6D/1% OG, 39 min in 0.9% PEG/1% OG, and 23 min in 1% OG.
Peptide Surfactant A6D Stabilized Rhodopsin in the Absence of Lipid and OG.
To determine whether A6D can substitute for phospholipids, we carried out experiments to test rhodopsin stability as a function of A6D concentration. We prepared four delipidated rhodopsin samples in (i) OG, (ii) 0.9% PEG in OG, (iii) 1.87 mM A6D in OG, and (iv) 3.75 mM A6D in OG. We then measured the thermal stability of these samples at 40°C. The delipidated rhodopsin in the OG/PEG mixture without A6D had the shortest half-life, ≈27 min. The stability of rhodopsin in A6D was increased as a function of the increasing concentration of A6D (Fig. 4B). Rhodopsin samples containing A6D at 1.87 mM (≈7× CAC) or 3.75 mM (≈15× CAC) displayed a half-life of ≈76 and ≈128 min, respectively. This result represents a 2.8- to 4.7-fold increase in stability compared with the stability in OG alone (Fig. 4B). These results suggest that A6D stabilizes rhodopsin effectively, even after lipid removal.
We then asked whether removing residue phospholipids from the preparation, namely, A6D alone without endogenous residue phospholipids, could stabilize delipidated rhodopsin. We carried out experiments to purify delipidated rhodopsin, using anti-rhodopsin-1D4 Ab immunoaffinity chromatography to remove the endogenous phospholipids from the rhodopsin preparation by extensive washing with the peptide surfactant A6D and replacing the phospholipids with A6D. This method has been successfully used to remove phospholipids from rhodopsin purified from bovine retinae rod outer segments. After affinity purification to obtain delipidated rhodopsin, we tested rhodopsin stability in the absence of both OG and phospholipids. We found that delipidated rhodopsin in the absence of both phospholipids and OG had a short half-life of ≈47 min at 40°C. Furthermore, adding OG to the delipidated rhodopsin did not have a significant stabilization effect. On the other hand, delipidated rhodopsin stabilized by A6D alone had a half-life of ≈122 min at 40°C, a 2.6-fold increase in stability (Fig. 4C).
Discussion
Surfactants play a vital role in our understanding of the structure and function of membrane proteins. Numerous attempts have been made to discover, synthesize, and select a variety of surfactants to facilitate the study of membrane proteins, but little progress has been made so far. It is widely known that obtaining stable membrane proteins for high-resolution structures requires the right surfactants. The designer short peptide surfactants reported here belong to a class of the simplest surfactants, which are easily adapted for molecular engineering for individual membrane proteins.
We have found that this class of designed peptide surfactants can stabilize not only the function of GPCR bovine rhodopsin in solution but also multiple subunits of the photosystem I (PSI) protein complex (35) on a dry surface (36, 37). These peptide surfactants represent a promising approach to membrane protein crystallization. V6D was used to solubilize the integral membrane protein glycerol 3-phosphate dehydrogenase in solution, when assayed through protein gel electrophoresis, retaining its functional enzymatic activities (38).
This class of short peptide surfactants, including A6D, V6D, A6K, and others, can be very useful. We show here that A6D not only can synergistically interact with the common surfactants OG and DM and lipids to enhance rhodopsin stability, but it also can effectively protect rhodopsin function against thermal denaturation in the absence of both lipid and common surfactants. This observation suggests that peptide surfactants interact with membrane proteins to stabilize them. Indeed, our previous results showed that peptide surfactants undergo self-assembly to form micelles, nanovesicles, and nanotubes (25–27).
These short peptide surfactants may have several advantages in studying diverse membrane proteins. (i) Their biochemical properties resemble common surfactants with similar CACs and seem not to denature membrane proteins and membrane protein complexes, as shown by our studies and those of other researchers. (ii) They are chemically and structurally simple and can be adapted quickly for a wide variety of uses. (iii) They are short with high purity, soluble in water, and stable for long periods at ambient temperature. (iv) They are affordable worldwide because peptide manufacturing is a mature industry, and the price is decreasing steadily. (v) They can readily be used with other common surfactants in a combinatorial manner.
Short Peptide Surfactants Stabilize Other Membrane Proteins.
We used green plant PSI (35) to demonstrate that these designer short peptide surfactants can stabilize membrane proteins (36, 37). PSI is a chlorophyll-containing membrane protein complex that is the primary reducer of ferredoxin and the electron acceptor of plastocyanin. We isolated the complex from the thylakoids of chloroplasts by using a common surfactant, Triton X-100. The chlorophyll molecules associated with the PSI complex provide an intrinsic steady-state emission spectrum between 650 and 800 nm at 77 K that reflects the organization of the pigment–protein interactions. In the absence of surfactants, a large blue shift of the fluorescence maxima from ≈735 nm to ≈685 nm indicates a disruption in light harvesting subunit organization, thus disrupting chlorophyll–protein interactions. The commonly used membrane protein-stabilizing surfactants, DM and OG, did not stabilize the ≈735-nm complex with the ≈685-nm spectroscopic shift. However, before drying the sample, addition of the peptide surfactant Ac-AAAAAAK (A6K) at an increasing concentration significantly stabilized the PSI complex (36). Moreover, in the presence of the A6K peptide surfactant, the PSI complex is stable in a dried form at room temperature for at least 3 weeks (36, 37). Another peptide surfactant, Ac-VVVVVVD (V6D), also stabilized the complex, but to a lesser extent. These observations suggest that peptide surfactants may stabilize membrane protein complexes on a dry surface.
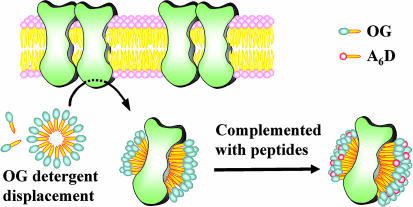
Proposed Model of How Peptide Surfactants Interact with Membrane Proteins.
We now wish to introduce a plausible model to explain how simple peptide surfactants interact with membrane proteins, particularly rhodopsin or other GPCRs (Fig. 5). In this model, the peptide surfactants form micelles and other nanostructures in the absence of proteins. When membrane proteins, e.g., rhodopsin, are present, these small peptide surfactants surround rhodopsin and act to protect it from thermal denaturation, similar to the action of chaperones, lipids, and other surfactants.
Fig. 5.
A proposed model of rhodopsin stabilization using peptide surfactants. GPCR bovine rhodopsin was extracted with OG from the membrane. After surfactant exchanges, the hydrophobic alanine tail of A6D forms the rhodopsin–surfactant complex only on the belt area. Small peptide surfactants surround rhodopsin and act to protect it from thermal denaturation, similar to the action of chaperones. This action may be similar to that of lipids and other surfactants. The proposed dimeric GPCR bovine rhodopsin is embedded in the cellular membrane. The lipids (shown with pink heads) of the membrane form bilayers. The surfactants (shown with gray heads) are OG, which is used for the initial purification, and peptide surfactant A6D (shown with red heads). All tails are shown in yellow. The lipids have two tails, and both OG and A6D have a single tail.
Perspective on Designer Lipid-Like Peptides for Membrane Protein Studies.
The field of designer short and simple peptide surfactants is in its infancy. However, several observations described here indicate that this class of surfactants will be very useful. We have designed a small number of variations from 20 natural l-amino acids, not all in mirror image, of 20 d-amino acids, as well as an increasing number of unnatural amino acids. All of these amino acids can be incorporated into the class of short peptide surfactants. A combinatorial approach can be readily applied to produce peptides with a wide range of properties for membrane protein study. So far, we have only focused on peptide surfactants containing homogenous hydrophobic tails. One possibility for the specialization of molecules is the exploration of mixtures of short and long tails; heterogeneous tails; tails with many hydrophobic residues including valine, leucine, methanine, isoleucine, proline, phenylalanine, tyrosine, tryptophan; and different head groups including sugars and other polar molecules. Peptides that further enhance stabilization and crystallization of membrane proteins might then be identified. Another possibility is the exploration of mixing several different surfactants together as cocktails. Naturally, proteins can select the most appropriate peptide components from such a mixture for use in stabilization and crystallization to uncover their structure and function.
Methods
GPCR Bovine Rhodopsin Source.
Frozen bovine retinae were obtained from J.A. Lawson (Lincoln, NE). Cell culture media and supplements were from Irvine Scientific (Santa Ana, CA) and Sigma (St. Louis, MO).
Chemicals, Surfactants, and Peptide Surfactants.
DM was purchased from Anatrace (Maumee, OH). OG was purchased from Roche Molecular Biochemicals (Mannheim, Germany). CNBr-activated Sepharose was purchased from Amersham Pharmacia (Little Chalfont, U.K.). All designed peptide surfactants were custom synthesized and characterized by the Biopolymers Laboratory at the Massachusetts Institute of Technology and Synpep (Dublin, CA).
Purification Materials.
Peptides corresponding to the C terminus (T340–A348) of rhodopsin were synthesized by the Biopolymers Laboratory at the Massachusetts Institute of Technology and purified by HPLC. Rhodopsin nonapeptide T340–A348 was used to elute rhodopsin from rhodopsin-1D4 Sepharose beads at a concentration of 100 μM throughout.
mAbs Coupled to Sepharose Beads.
Anti-rhodopsin mAb rhodopsin-1D4 (39) was prepared by the National Cell Culture Center (Minneapolis, MN). The mAb rhodopsin-1D4 was coupled to CNBr-activated Sepharose beads as described (40, 41), except that 10 mg of the purified mAb proteins was bound per 1 ml of rehydrated beads. The resulting mAb rhodopsin-1D4 Sepharose beads had a capacity to bind ≈1 mg of rhodopsin per milliliter of settled beads.
Solubilization of Rhodopsin in Different Surfactants.
Bovine retinae rod outer segment membranes were prepared (40, 41) and urea-stripped (42). Samples of the membranes containing 1.4 nmol of rhodopsin were solubilized in 0.5 ml of PBS containing 0.5 mM phenylmethylsulfonyl fluoride with 1% DM or 2% (vol/vol) OG. The suspensions were centrifuged (100,000 × g for 15 min), and the supernatants were kept in the dark at 4°C.
Thermal Stability Study of Rhodopsin in Peptide Solution in the Presence of the Surfactant.
Rhodopsin solubilized (32–34) in PBS containing 2% (vol/vol) OG was added to the same volume of PBS containing 0, 2.5, 5.0, or 7.5 mM A6D, 1.8% PEG, or 5.4 mM RAD8. In another experiment, rhodopsin solubilized in PBS containing 1% (19.8 mM) DM was added to the same volume of PBS containing 5.0 mM A6D or 0, 5.0, or 19.8 mM DM. A decrease in A500 was recorded every 5 min up to 120 min at 40°C for the samples containing OG and at 40–55°C for those containing DM (Figs. 2 and 3). UV-visible absorption spectroscopy was performed with a Lambda 6 spectrophotometer (Perkin-Elmer, Wellesley, MA) equipped with a temperature-regulated cuvette holder with a slit width of 2 nm, 480 nm/min scan speed, and a response time of 1 second. The A500 remaining at different time points was expressed as the percentage of absorbance present at the start of the experiment. The half-life of rhodopsin in each solution was calculated by using a single exponential curve fit. The plot obtained from the percentage of absorbance at different time points was fitted to a single exponential curve with two parameters (Eq. 1).
The plot obtained from the A500 values at different time points was directly fitted to a single exponential curve with three parameters (Eq. 2) when the plot from the percentage of absorbance could not be fitted to Eq. 1.
The half-life (t1/2) was calculated by using Eq. 3.
Thermal Stability Study of Rhodopsin in Peptide Solution in the Absence of OG.
Rhodopsin solubilized in PBS containing 2% (vol/vol) OG was added to the same volume of PBS containing 2.5 mM A6D. Then, 0.5 ml of the mixture was poured into the tube. The tube was covered with 10 kDa of cutoff membrane and dialyzed against 4.5 ml of PBS containing 1.25 mM A6D for 40 h in the dark at 4°C. A resultant sample decrease in A500 was recorded every 5 min for 120 min at 40°C. A decrease in A500 of the predialysis mixture containing 1% OG and 1.25 mM A6D was also monitored as a control (Fig. 4A).
Removal of Phospholipids from Rhodopsin.
Samples of the membranes containing 4.7 nmol of rhodopsin were solubilized in 1,000 μl of PBS containing 2% (vol/vol) OG. The suspension was mixed with 250 μl (settled beads, concentration of 50%) of rhodopsin-1D4 Sepharose and rotated for 3 h. The 1D4 Sepharose beads were then packed into a minicolumn (7-mm inner diameter) and washed with PBS containing 1% OG by using 500 bed volumes (total of 62.5 ml). The rhodopsin was then eluted with 1.5 ml of PBS containing 1% OG and the C-terminal nanopeptide.
Thermal Stability Study of Rhodopsin in Peptide Solution in the Absence of Phospholipids.
The delipidated rhodopsin solution was added to the same volume of 1% OG in PBS containing 0, 3.75, or 7.5 mM A6D, or 1.8% PEG (Fig. 4B). In another experiment, the delipidated rhodopsin solutions were poured into tubes. Each tube was covered with 10 kDa of cutoff membrane and dialyzed against 4.5 ml of PBS or PBS containing 1.25 mM A6D or 1% OG (Fig. 4C). For all samples, a decrease in A500 was recorded every 5 min, up to 120 min a205t 40°C.
Acknowledgments
We thank Steve Yang, Alexander Rich, and members of S.Z.'s laboratory for helpful and stimulating discussions. This work was supported in part by grants from the National Science Foundation to the Center for Bits and Atoms at the Massachusetts Institute of Technology. Y.N. gratefully acknowledges generous support from Menicon Co., Ltd., Japan.
Abbreviations
- GPCR
G protein-coupled receptor
- DM
n-dodecyl-β-d-maltoside
- OG
octyl–d-glucoside
- CAC
critical aggregation concentration.
Footnotes
The authors declare no conflict of interest.
References
- 1.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Nat Struct Biol. 2002;9:922–926. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 2.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 3.Watson S, Arkinstall S. The G-Protein Linked Receptor Facts Book. London: Academic; 1994. [Google Scholar]
- 4.Haga T, Berstein G. G Protein-Coupled Receptors. Raton, FL: CRC, Boca; 1999. [Google Scholar]
- 5.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 9.Wallin E, von Heijne G. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kall L, Sonnhammer EL. FEBS Lett. 2002;532:415–418. doi: 10.1016/s0014-5793(02)03730-4. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Engelman DM, Gerstein M. Genome Biol. 2002 Sep 19; doi: 10.1186/gb-2002-3-10-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loll PJ. J Struct Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 14.Gennis RB. Biomembranes: Molecular Structure and Function. New York: Springer; 1989. [Google Scholar]
- 15.Michel H. Crystallization of Membrane Proteins. Boca Raton, FL: CRC Press; 1990. [Google Scholar]
- 16.Ostermeier C, Michel H. Curr Opin Struct Biol. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 17.le Maire M, Champeil P, Moller JV. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 18.Ringler P, Heymann B, Engel A. Practical Approach Series. In: Baldwin SA, editor. Membrane Transport. Oxford: Oxford Univ Press; 2000. pp. 229–268. [Google Scholar]
- 19.Howard TA, McAuley-Hecht KE, Cogdell RJ. Practical Approach Series. In: Baldwin SA, editor. Membrane Transport. Oxford: Oxford Univ Press; 2000. pp. 269–307. [Google Scholar]
- 20.Garavito RM, Ferguson-Miller S. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 21.Hauser H. Biochim Biophys Acta. 2000;1508:164–181. doi: 10.1016/s0304-4157(00)00008-3. [DOI] [PubMed] [Google Scholar]
- 22.Schafmeister CE, Miercke LJ, Stroud RM. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]
- 23.McGregor CL, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Prive GG. Nat Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 24.Soomets U, Kairane C, Zilmer M, Langel U. Acta Chem Scand. 1997;51(3 Suppl):403–406. doi: 10.3891/acta.chem.scand.51-0403. [DOI] [PubMed] [Google Scholar]
- 25.Vauthey S, Santoso S, Gong H, Watson N, Zhang S. Proc Natl Acad Sci USA. 2002;99:5355–5360. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoso S, Hwang W, Hartman H, Zhang S. Nano Lett. 2002;2:687–791. [Google Scholar]
- 27.von Maltzahn G, Vauthey S, Santoso S, Zhang S. Langmuir. 2003;19:4332–4337. [Google Scholar]
- 28.Verwey EJW, Overbeek JTG. Theory of the Stability of Lyophobic Colloids. Amsterdam: Elsevier; 1948. pp. 106–115. [Google Scholar]
- 29.Gerber GE, Gray CP, Wildenauer D, Khorana HG. Proc Natl Acad Sci USA. 1977;74:5426–5430. doi: 10.1073/pnas.74.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mogi T, Stern LJ, Marti T, Chao BH, Khorana HG. Proc Natl Acad Sci USA. 1988;85:4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grobner G, Burnett IJ, Glaubitz C, Choi G, Mason AJ, Watts A. Nature. 2000;405:810–813. doi: 10.1038/35015604. [DOI] [PubMed] [Google Scholar]
- 32.Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 33.Reeves PJ, Thurmond RL, Khorana HG. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Shem A, Frolow F, Nelson N. Nature. 2003;426:630–635. doi: 10.1038/nature02200. [DOI] [PubMed] [Google Scholar]
- 36.Das R, Kiley PJ, Segal M, Norville J, Yu A, Wang L, Trammell S, Reddick LE, Kumar R, Stellacci F, et al. Nano Lett. 2004;4:1079–1083. [Google Scholar]
- 37.Kiley P, Zhao X, Bruce BD, Baldo M, Zhang S. PLoS Biol. 2005;3:1180–1186. doi: 10.1371/journal.pbio.0030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh JI, Du S, Tordajada A, Paulo J, Zhang S. Biochemistry. 2005;44:16912–16919. doi: 10.1021/bi051357o. [DOI] [PubMed] [Google Scholar]
- 39.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papermaster DS. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 41.Papermaster DS. Methods Enzymol. 1982;81:240–246. doi: 10.1016/s0076-6879(82)81037-9. [DOI] [PubMed] [Google Scholar]
- 42.Shichi H, Somers RL. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]