Abstract
Androgens and the androgen receptor (AR) play important roles in the testes. Previously we have shown that male total AR knockout (T-AR−/y) mice revealed incomplete germ cell development and lowered serum testosterone levels, which resulted in azoospermia and infertility. However, the consequences of AR loss in particular types of testicular cells remain unclear. Using a Cre-loxP conditional knockout strategy, we generated a tissue-selective knockout mouse with the AR gene deleted in testis peritubular myoid cells (PM-AR−/y). Phenotype analyses showed that PM-AR−/y mice were indistinguishable from WT AR (AR+/y) mice with the exception of smaller testes size. PM-AR−/y mice have serum testosterone concentrations comparable with AR+/y mice. PM-AR−/y mice have oligozoospermia in the epididymis; however, fertility was normal. Although normal germ cell distribution ratio was found, total germ cell number decreased in PM-AR−/y mice. Further mechanistic studies demonstrated that PM-AR−/y mice have defects in the expression of Sertoli cells' functional marker genes such as tranferrin, epidermal fatty acid-binding protein, androgen-binding protein, and other junction genes including occludin, testin, nectin, zyxin, vinculin, lamininγ3, gelsolin, connection43, and N-cadherin. Furthermore, there were defects in peritubular myoid cell contractility-related genes such as endothelin-1, endothelin receptor A and B, adrenomedullin, adrenomedullin receptor, and vasopressin receptor 1a. Together, our PM-AR−/y mice provide in vivo evidence for the requirement of functional AR in peritubular myoid cells to maintain normal Sertoli cells function and peritubular myoid cell contractility, thus ensuring normal spermatogenesis and sperm output.
Keywords: knockout, transgelin, spermatogenesis
Peritubular myoid (PM) cells are mesenchymal cells that form the outer border of the seminiferous tubules and, in conjunction with Sertoli cells, produce the basement membrane required to maintain normal tubule morphology. The interactions between PM cells and Sertoli cells provide an example of a mesenchymal–epithelial cell interaction (1). PM cells contain a high percentage of the androgen receptor (AR) (2, 3) and provide a site for androgen signaling. It has been reported that the PM cells can synthesize several secretory products. Results from earlier in vitro studies indicated that androgen can act on PM cells to stimulate the production of a paracrine factor, termed P-Mod-S (peritubular factors that modulate Sertoli cell function), which modulates a number of Sertoli cell functions, such as the secretion of transferrin, inhibin, and androgen-binding protein (ABP), while providing nutritional interactions essential for germ cell development (4, 5). Another important function of PM cells is their ability to contract and induce peristalsis-like waves and impulses in the seminiferous tubule to aid the transport of spermatozoa through the tubular lumen and into the epididymis to control testicular output of both fluid and sperm; hence its regulation represents a key point in male fertility (6).
Androgens and the AR (7–9) play important roles in male fertility (10). The local actions of androgen on testis functions were initially demonstrated when testosterone alone, in the absence of the gonadotropins, leuteinizing hormone (LH), and follicle-stimulating hormone (FSH), could support spermatogenesis (11). Furthermore, AR has been detected in Sertoli, Leydig, PM, and spermatid cells (round and elongated) (2, 3), and mice lacking functional AR develop testicular feminization syndrome (12). Previously, we have shown that Sertoli cell-specific AR−/y mice revealed incomplete germ cell development and lowered serum testosterone levels, which resulted in azoospermia and infertility (13). However, the consequences of AR loss in other particular types of testicular cells, especially in PM cells, remain largely unknown.
Given the specific roles of PM cells in regulation of functional testes (14), we were interested in generating PM cell-specific AR knockout (PM-AR−/y) mice by using the Tagln (transgelin, smooth muscle protein 22-α)-Cre/loxP system (12, 15–17). By mating floxed AR mice (12) with a transgenic line possessing Tagln promoter-driven expression of the Cre recombinase (17), we obtained male PM-AR−/y mice with the AR gene deleted in PM cells.
Analyses of the PM-AR−/y mice revealed oligozoospermia but normal fertility. Mechanistic studies further suggest that Sertoli cell nursing function and PM cell contractility have defects. Together, these regulations might contribute to the slowdown or delay in the spermatogenesis and decreased sperm output, thus contributing to oligozoospermia.
Results
Generation of Mice with Conditional Knockout of AR in PM Cells
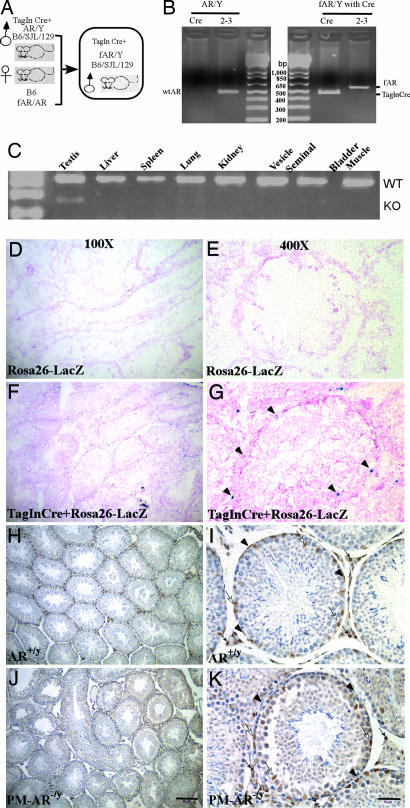
Using a Cre-loxP conditional knockout strategy, we mated female heterozygous floxed AR mice with male Tagln-Cre mice to generate PM-AR−/y and WT AR (AR+/y) littermates (Fig. 1A). We detected Cre and floxed AR DNA fragments in tail genomic DNA of 21-day-old PM-AR−/y mice (Fig. 1B).
Fig. 1.
Generation of mice with conditional knockout of AR in PM cells (PM-AR−/y). (A) Mating strategy to generate PM-AR−/y mice. (B) Identification and confirmation of PM-AR−/y mice. Genomic DNA was isolated from tail snips and used as template for PCR with primers select and 2–3. The detailed method and primer sequences have been described (12, 15). The expression of floxed AR and Tagln-Cre in the tail genomic DNA of PM-AR−/y male mouse was confirmed by PCR. (C) RT-PCR of various tissues harvested from PM-AR−/y mice. Only the mRNA from the testes of the PM-AR−/y mice shows a knockout band (KO) and a WT AR band when primers exon 1 and exon 3 are used; other tissues have only a WT band. (D–G) X-Gal staining to confirm the Cre recombination. (D and E) No positive staining was found in ROSA-26 LacZ control mice testis. (F and G) Positive blue signal was found in PM cells of double transgenic (Tagln-Cre and ROSA26-LacZ) mice. Neutral fast red was used to counterstain the nuclei. (H–K) Immunostaining of AR protein with primary rabbit polyclonal antibody N20 in testicular sections from AR+/y and PM-AR−/y mice, which are all counterstained with hematoxylin to reveal the location of cell nuclei. (H and I) In AR+/y testis, AR staining was found in PM cells (arrowheads), Sertoli cells (white arrows), and Leydig cells (black arrows). (J and K) The PM-AR−/y testis shows no AR staining in PM cells; however, Sertoli cells and Leydig cells retain positive staining signals. (Scale bar: D, F, H, and J, 160 μm; E, G, I, and K, 40 μm.)
In addition, various tissues were harvested from 14-wk-old PM-AR−/y and AR+/y mice. Only testes showed deletion of AR exon 2 with a 180-bp signal from RT-PCR using primers for exon 1 and exon 3. In contrast, none of the other tissues examined, including liver, spleen, lung, kidney, seminal vesicle, bladder, and muscle, showed deletion of AR exon 2 (Fig. 1C). This finding suggests that selective disruption of AR occurred in the testes.
Cre recombination was analyzed by breeding the Tagln-Cre mice with the ROSA26-LacZ reporter line (18), which harbors a bacterial β-galactosidase reporter gene, in which the expression relies on the deletion of a loxP-flanked “stop” sequence that separates the ROSA26 promoter and the LacZ gene (18). Thus the LacZ gene is expressed where Cre is expressed and activated. Peritubular cells show efficient recombination and intensive staining accordingly, in contrast to nonsmooth-muscle cells that do not undergo recombination in double transgenic mice (Tagln-Cre and ROSA26-LacZ) (Fig. 1 F and G). Furthermore, no positive staining was found in ROSA26-LacZ control mice (Fig. 1 D and E). Immunohistochemical staining was applied to confirm that the loss of AR was specific to PM cells. As a control, AR-positive staining in PM cells was found in AR+/y testes (Fig. 1 H and I). As shown in Fig. 1 J and K, AR-positive staining was also observed in Sertoli and Leydig cells of PM-AR−/y, whereas no AR staining was found in PM cells that surround the seminiferous tubles of PM-AR−/y mice. This finding suggests that AR expression was lost selectively in PM cells. All together, results from genomic DNA genotyping, Cre recombination staining, tissue mRNA RT-PCR detection, and immunohistochemical staining reveal that the AR gene was selectively knocked out in PM cells of PM-AR−/y mice.
Oligozoospermia with Normal Fertility in PM-AR−/y Mice.
We observed spermatozoa number decreased significantly in epididymal semen of PM-AR−/y mice compared with AR+/y mice (n = 10; P < 0.05). There was a 57% reduction in sperm count for PM-AR−/ycompared with AR+/y littermates. The total epididymal semen sperm count in PM-AR−/y mice was 12.1 ± 3.4 × 106 while that in AR+/y mice was 28.3 ± 7.0 × 106. Despite the reduction in sperm count, we found normal sperm motility in PM-AR−/y mice at 57.0 ± 9.0% compared with AR+/y mice at 56.5 ± 3.7%.
Interestingly, normal fertility was found in PM-AR−/y mice despite the oligozoospermia. To evaluate fertility in PM-AR−/y mice, three 8-wk-old male mice of each genotype, AR+/y, PM-AR−/y, and AR−/y, were mated with 8-wk-old C57BL/6 female mice. In three successive sets of 2-wk pairings, PM-AR−/y mice were mated with six different female mice. Vaginal plugs were found on the morning after mating, and total pup numbers were recorded. As a negative control, total AR knockout (T-AR−/y) male mice failed to impregnate their mates, and vaginal plugs were not found after mating. Together, results from Table 1 show that PM-AR−/y mice are oligozoospermic, but have normal fertility.
Table 1.
Fertility assessment and epididymal content analysis
| Mice | Mate no. |
Vaginal plug | Sperm count (106)/epididymis | Motility, % | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| AR+/y | 9.2 ± 2.6 | 8.6 ± 1.5 | 8.5 ± 2.6 | + | 28.3 ± 7.0 | 56.5 ± 3.7 |
| PM-AR−/y | 9.0 ± 2.6 | 8.6 ± 1.5 | 8.7 ± 2.1 | + | 12.1 ± 3.4* | 57.0 ± 9.0 |
| T-AR−/y | 0 | 0 | 0 | − | No epididymis | |
*,P < 0.05 vs. AR+/y.
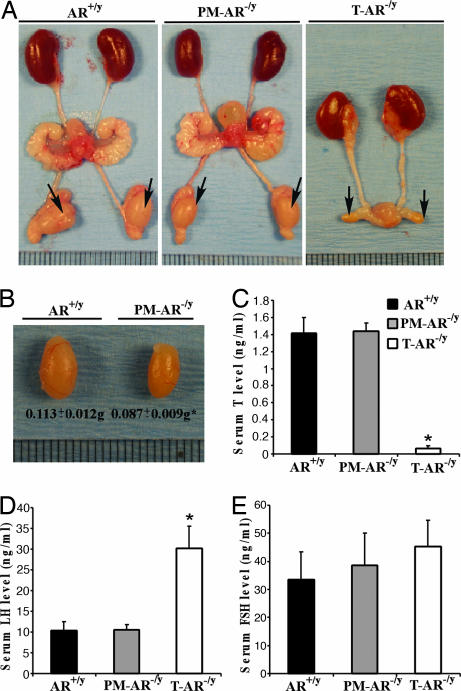
Decreased Testis Size and Normal Ranges of Serum Testosterone, LH, and FSH Levels in PM-AR−/y Mice
Phenotype analyses show there were no significant differences in most of the genitourinary organs or in total body weight (AR+/y, 29.0 ± 3.7 g vs. PM-AR−/y, 28.4 ± 2.3 g) between 10 pairs of 14-wk-old AR+/y and PM-AR−/y mice (Fig. 2A). However, the size of the testis in PM-AR−/y mice significantly decreased nearly to 76% of AR+/y (0.087 ± 0.009 vs. 0.113 ± 0.012 g for 14-wk-old PM-AR−/y vs. AR+/y mice) (Fig. 2 B). In contrast, T-AR−/y mice have even smaller testes with no seminal vesicles, epididymis, or prostate (Fig. 2A) (13). Serum hormone assays show that PM-AR−/y mice have normal ranges of serum testosterone, LH, and FSH levels compared with AR+/y mice; in contrast, T-AR−/y mice have significantly lower serum testosterone and higher LH levels, whereas only marginal changes in FSH levels were found compared with AR+/y mice and PM-AR−/y mice (Fig. 2 C–E).
Fig. 2.
Morphology of testes from 14-wk-old AR+/y, PM-AR−/y, and AR−/y male mice. (A and B) Decreased testes size in PM-AR−/y mice. PM-AR−/y mice have genitourinary organs of similar size as AR+/y mice except for smaller testes, which were 76% of T-AR−/y mice (n= 10; ∗, P < 0.05 vs. AR+/y). In contrast, there are small testes but no seminal vesicle, epididymis, or prostate in T-AR−/y mice. Arrows indicate testes (each unit of the rule equals 1 mm). (C–E) Serum testosterone, LH, and FSH levels in 14-wk-old male AR+/y, PM-AR−/y, and T-AR−/y mice (n= 8, 8, and 5, respectively). PM-AR−/y mice have normal range of serum testosterone, LH, and FSH levels compared with AR+/y mice; in contrast, T-AR−/y mice have significantly lower serum testosterone and higher LH and only a marginal change in FSH level compared with the age-matched AR+/y and PM-AR−/y mice (∗, P < 0.05 vs. AR+/y).
Normal Germ Cell Distribution Ratio in PM-AR−/y Mice.
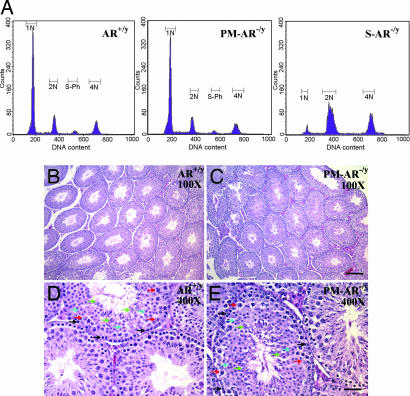
We analyzed spermatogenesis by determining the relative distribution of germ cell populations in the testes of PM-AR−/y, AR+/y, and Sertoli cell-specific AR knockout (S-AR−/y) mice. Using flow cytometric scanning of propidium iodide-labeled AR+/y germ cells, we detected four main histogram peaks of DNA content that correspond to haploid (1N; round and elongated spermatids and spermatozoa), diploid (2N; spermatogonia, preleptotene, and diplotene primary spermatocytes, secondary spermatocytes, and somatic cells), tetraploid cells (4N; G2 spermatogonia, leptotene, zygotene, and pachytene primary spermatocytes), and S-phase cells between peaks 2N and 4N (spermatogonial cells and preleptotene spermatocytes synthesizing DNA). The proportion of somatic cells has been shown to comprise <3% of the total testicular cells in the mouse testis (19). PM-AR−/y mice show normal germ cell distribution ratios (Fig. 3A) that are consistent with H&E staining results (Fig. 3 B). In comparison, we found that S-AR−/y mice with knockout of AR only in Sertoli cells (13) show an increase in tetraploid cells and cells with haploid DNA content were barely detectable. The diploid-to-tetraploid ratio found in S-AR−/y mice suggests that spermatogenic arrest occurs mainly before the first meiosis, in agreement with our previous findings (13).
Fig. 3.
Normal germ cell distribution in PM-AR−/y mice. (A) Analyses of germ cell DNA content of testes from AR+/y, PM-AR−/y, and S-AR−/y mice by flow cytometry. 1N represents haploid cells, 2N represents diploid cells, and 4N represents tetraploid cells. S-phase represents spermatogonial cells and preleptotene spermatocytes synthesizing DNA. PM-AR−/y mice show the normal germ cell distribution ratio (A), which is consistent with the H&-staining data (B–E). All types of germ cells can be found in PM-AR−/y mice (D and E) compared with AR+/y mice (B and C), such as spermatogonia (black arrows), spermatocyte (red arrows), round (blue arrows), and elongated spermatids (green arrows). (Scale bars: 160 μm, B and C; D and E, 40 μm.)
There are several possible reasons that PM-AR−/y mice have a decreased sperm count, yet their germ cell ratio remains unchanged. The defective nursing functions of Sertoli cells could slow down the maturation process of germ cells or influence the half-life of sperm. Reduced PM cell contractility in the PM-AR−/y mice could also reduce the sperm count in the epididymis. Together, these factors might contribute to the slowdown or delay in the spermatogenesis and decreased sperm output, thus contributing a quantitative rather than a qualitative defect.
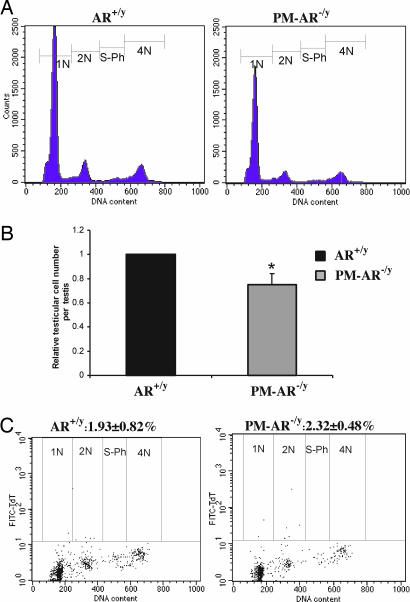
Decreased Germ Cell Number per Testis and Comparable Apoptotic Germ Cells in PM-AR−/y Mice Determined by Flow Cytometry
Total germ cell number was determined by flow cytometry (Fig. 4 A) and normalized to AR+/y average germ cell number (Fig. 4 B). We found that germ cell number per testis in PM-AR−/y mice decreased to 74.8% of AR+/y mice (n= 5; *, P < 0.05 vs. AR+/y). This finding is consistent with the decreased testis size. Fig. 4C is representative of the flow cytometry dot plot of germ cells isolated from the AR+/y and PM-AR−/y mice and processed for TUNEL. Four cell populations contained TUNEL-positive cells: 1N, 2N, S-ph, and 4N cells. The overall incidence of apoptotic cells in all testicular cells for three pairs of mice was 2.32 ± 0.48% in PM-AR−/y mice when compared with 1.93 ± 0.82% in AR+/y mice; however, the difference is not statistically significant (n= 3; P > 0.05).
Fig. 4.
Analyses of germ cell number per testis and apoptotic germ cells from AR+/y and PM-AR−/y by flow cytometry. (A) Representative DNA histograms of the testicular germ cells show PM-AR−/y mice have fewer germ cells per testis. (B) Quantitation by normalizing to AR+/y average germ cell number show that germ cell number per testis from PM-AR−/y mice decreased to 74.8% of AR+/y (n= 5; ∗, P < 0.05 vs. AR+/y). (C) Representative flow cytometry dot plot of TUNEL assay for germ cells isolated from the AR+/y and PM-AR−/y mice. Four cell populations contained TUNEL-positive cells: 1N, 2N, S-ph, and 4N cells. The overall incidence of apoptotic cells in all testicular cells for three pairs of mice was 2.32 ± 0.48% in PM-AR−/y mice compared with 1.93 ± 0.82% in AR+/y mice; however, the difference is not significant (n= 3; P > 0.05).
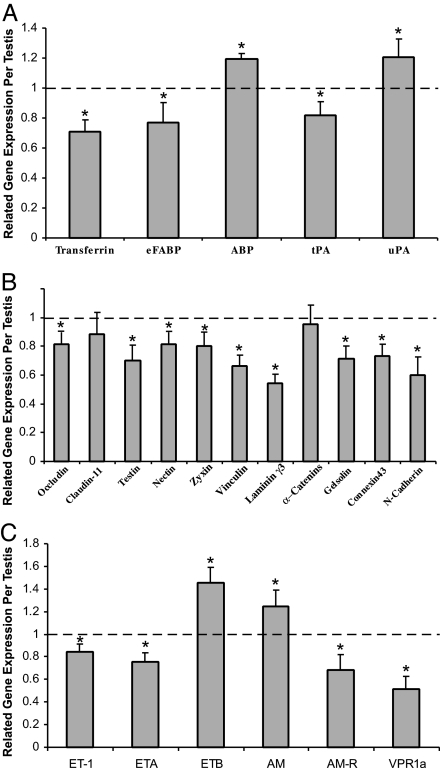
Altered Expression of Sertoli Cells' Functional Genes, Junction Genes, and PM Cells Contractility Related Genes in PM-AR−/y Testes by Real-Time PCR
To understand how the loss of AR in peritubular cells results in oligozoospermia, we selected mechanism-related genes for real-time PCR analyses. First, we found that Sertoli cells' functional genes that nourish germ cells and cooperate with germ cells in germ cell movement and spermiation, such as transferrin, epidermal fatty acid binding protein, and tissue-type plasminogen activator, significantly decreased as well as ABP and urokinase-type plasminogen activator significantly increased in PM-AR−/y testes (Fig. 5 A). Fig. 5B shows the junction genes including occludin, testin, nectin, zyxin, vinculin, lamininγ3, gelsolin, connection43, and N-cadherin all significantly decreased in PM-AR−/y testes; however, caludin-11 and α-catenins had no significant change. Among the PM cells contractility related genes, endothelin (ET) 1, ET receptor A, adrenomedullin (AM) receptor, and vasopressin receptor 1a were found significantly decreased, and ET receptor B and AM were significantly increased in PM-AR−/y testes (Fig. 5 C). These modulated genes can be further validated by either coculture system (20), isolated primary cells from AR+/y mice with AR-siRNA knockdown, or from PM-AR−/y mice with lentiviral AR restoration. Moreover, these cell culture systems might also allow us to further dissect the molecular mechanism of how AR modulates these genes at either the transcriptional and/or translational level. Results from future studies might then help us to better understand the roles of AR in spermatogenesis and male fertility.
Fig. 5.
Gene expression changes in testes of three 14-wk-old PM-AR−/y male mice compared with WT littermates. (A) Changes in expression of Sertoli cells' functional genes related to nourishing germ cells and cooperating with germ cells in germ cell movement and spermiation. (B) Tight junctions and anchoring junctions component genes. (C) PM cells contractility-related genes. RNA was extracted from testes and cDNA was prepared as described in Materials and Methods. Real-time PCR was used to measure cDNA levels relative to external control luciferase (∗, P < 0.05 vs. AR+/y). eFABP, epidermal fatty acid-binding protein; tPA, tissue-type plasminogen activator; uPA, urokinase-type plasminogen activator; ETA and ETB, ET receptor A and B, respectively; AM-R, AM receptor; VPR1a, vasopressin receptor 1a.
Discussion
PM-AR−/y Mice Have Reduced Sperm Count with Normal Fertility
PM cells have multiple roles in the testis. These include interacting with Sertoli cells, contributing to the contractile activity of testicular tubules, and thus taking part in the regulation of spermatogenesis and testicular function (14, 21).
In our knockout mice model, we found that loss of AR in PM cells resulted in 57% reduction in epididymal sperm count but normal fertility. This finding is not surprising as it was reported that an impact on fertility should have an estimated sperm count reduction to 10% or less (22). Furthermore, previous reports have demonstrated several animal models with reduced sperm count but normal fertility (22–26). ADP-ribosylation factor-like 4 (Arl4) is a GTP-binding protein that shuttles between the nucleus and intracellular organelles depending on GTP/GDP-binding status. Schürmann et al. (23) have demonstrated that Arl4 null mice have 30% reduction in testis weight and a 60% reduction in sperm count, but do not have a reduction in litter size or frequency. In addition, FSHβ has been reported to play a role in spermatogenesis and mice lacking FSHβ are also fertile despite having a reduced testis size and epididymal sperm count (25, 26). Furthermore, centromere protein B is a DNA-binding protein required for meiosis, and its deletion caused a reduction in testis weight by 30% and a 40% reduction in sperm count without a reduction in fertility (24). Finally, mice with disrupted protein inhibitor of activated STAT splice variant also show a similar phenotype with absolute testicular weight decrease of 23% and sperm count decrease of 19% (22). These phenotypes are similar to our PM-AR−/y mice model in which we observed a 25% reduction in testis weight and a 57% reduction in sperm count; however, normal fertility was observed compared with AR+/y littermates (Table 1).
PM Cells' Physiological Roles as Contributor to Nursing Function of Sertoli Cells
The maintenance and control of testicular function and the process of spermatogenesis require cooperarative interactions among different cell types (27). Among the testicular cells, PM cells and Sertoli cells provide an example of a mesenchymal–epithelial cell interaction (1). Sertoli cells, often referred to as “nurse cells,” help with the formation of the seminiferous tubules and provide both cytoarchitectural and nutritional support for the developing germ cells. Sertoli cells provide an essential link between the interstitial and basal aspects of the testis and the developing germ cells within the tubule adluminal space (28). Therefore, regulation of Sertoli cell function directly affects germinal cell development. Results from earlier in vitro studies indicated that androgen/AR can act on PM cells to stimulate the production of a paracrine factor, termed P-Mod-S, which modulates a number of Sertoli cell functions, such as the secretion of transferrin, inhibin, and ABP and provides nutritional interactions essential for germ cell development (4, 5). Here, we found that loss of functional AR in PM cells might cause some functional defects of Sertoli cells and lead to oligozoospermia.
Quantitative PCR expression of PM-AR−/y showed decreases in transferrin and epidermal fatty acid-binding protein expression. Transferrin is secreted by differentiated Sertoli cells and is proposed to transport iron to the developing germ cells within the adluminal compartment of seminiferous tubule (29). The mutant hypotransferrinemic mouse model is found to be defective in spermatogenesis with low sperm counts (28, 30, 31). Furthermore, levels of seminal transferrin are proportional to sperm production in humans and might be an effective indicator of Sertoli cell function (32). Epidermal fatty acid-binding protein expression was found in Sertoli cells and proposed to be involved in uptake and transport of essential fatty acids for growth and function of the surrounding germ cells (33). A combined reduction in both transferrin and epidermal fatty acid-binding protein expressions might contribute to decreased Sertoli cell nourishing function in PM-AR−/y mice, resulting in decreased sperm count (Fig. 5A).
There are three types of junctions found in the testis, including inter-Sertoli cell tight junction, anchoring junction, and gap junction. They are important in germ cell movement in the seminiferous epithelium during spermatogenesis. Both caludin-11 and occludin are integral components of tight junctions between Sertoli cells at the site of the bloodtestis barrier. Testin, nectin, zyxin, vinculin, lamininγ3, α-catenins, gelsolin, and N-cadherin are components of anchoring junction (34). Our data show that some junction-related genes expression including occludin, testin, nectin, zyxin, vinculin, lamininγ3, gelsolin, connexin43, and N-cadherin are significantly decreased in PM-AR−/y testes compared with AR+/y testes, suggesting that loss of functional AR in PM cells might impair Sertoli cell junction and slow down the germ cell movement during spermatogenesis. These results might provide a partial explanation of the reported reduction in testis germ cell number and epididymal sperm count (Fig. 5B).
PM Cells' Physiological Roles Contributing to Contractility of Testicular Tubules.
An important role that PM cells play is contractions of the seminiferous tubule to induce peristalsis-like waves and impulses that aid the transport of spermatozoa through the tubular lumen and into the epididymis, hence its regulation might represent a key point in male fertility (6). Our in vivo studies show that PM-AR−/y mice might have some defects in contractility, thus resulting in decreased sperm output. Earlier studies demonstrated that ET-1 and AM are two balancing factors that control PM cell contraction (35–39). It is possible that androgen/AR may go through the modulation of ET-1 and AM (Fig. 5C) to influence the PM cell contractility that resulted in oligozoospermia.
Together, androgen/AR might modulate those spermatogenesis-related genes directly or indirectly. AR might indirectly go through PM cells to affect the gene expressions in Sertoli cells or directly modulate gene expression within the PM cells. By searching for potential androgen response element sequences to demonstrate direct AR modulation, several potential androgen response elements were found located in the 5′ promoter of AR regulated genes such as ET receptor A, ET receptor B, and AM receptor. Future studies using DNA microarray approaches should allow us to further dissect how AR functions in PM cells.
Materials and Methods
Generation of PM Cell-Specific AR−/y Mice
Protocols for use of animals were in accordance with the National Institutes of Health standards. Transgenic male mice (C57BL/6*SJL* 129S5/SvEvBrd) expressing Cre recombinase, under the control of the mouse Tagln promoter (17), were mated with floxed AR(C57BL/6) female mice (Fig. 1). The generation of floxed AR gene-targeted mice has been described (12, 15). The expression of transgelin promoter-driven Cre recombinase can efficiently and selectively delete the floxed AR gene in PM cells. PM-AR−/y mice express floxed AR and Cre alleles in tail genomic DNA. We genotyped 21-day-old pups from tail snips by PCR as described (12, 15).
LacZ Staining of Tissue Sections.
The transgenic Tagln Cre recombinase activity by 8 weeks of age was confirmed through breeding with the ROSA26-LacZ reporter line. The ROSA26-LacZ reporter line (Jackson Laboratories, Bar Harbor, ME) harbors a bacterial β-galactosidase reporter gene, the expression of which required Cre-mediated deletion of a floxed stop sequence separating the ROSA26 promoter and the β-galactosidase gene (18). Thus, the β-galactosidase gene is expressed only where Cre is expressed and active. Fresh dissected testes from double transgenic mice (Tagln Cre and ROSA26-LacZ) and ROSA26-LacZ transgenic control mice were frozen in Tissue-Tek, sectioned at 10 μm, and stained by using the β-galactosidase staining kit from Specialty Media (Billerica, MA) (17).
Flow Cytometry Analysis of Testicular Cells for DNA Content, Total Cell Number, and Apoptosis with TUNEL Assay.
A monocellular suspension of testicular cells was prepared as described (19, 40). Briefly, the tunica albuginea was removed, and the seminiferous tubules were minced in PBS to release the testicular cells. The minced tissue was gently aspirated for 2 min, and the cells were washed in PBS and spun down at 800 × g for 10 min. Cells were resuspended in PBS, filtered through 80-μm nylon mesh, fixed in cold 70% ethanol, and kept at 4°C until further analysis. For DNA content assay, 1 × 106 cells were washed twice with PBS and incubated in 500 μl of 0.2% pepsin for 10 min at 37°C. After centrifugation, the cells were stained with a solution containing 25 μg/ml propidium iodide, 40 μg/ml RNase, and 0.3% Tween-20 in PBS at room temperature for 20 min. The stained cells were analyzed with a FACScan flowcytometer (Becton Dickinson Immunocytometry, San Jose, CA). For calculating total cell number, equal volumes of cells from AR+/y and PM-AR−/y were removed after propidium iodide staining for analyses by a FACScan flow cytometer. The total cell number per testis was calculated by multiplying total volume by concentration (cells per ml). For TUNEL assay, we used an Apo-Direct kit (Calbiochem, San Diego, CA) and followed the manufacturer's suggested protocol.
Assessment of Serum Hormone Levels.
Male PM-AR−/y, AR−/y, and B6 AR+/y mice were killed at 14 wk. A midline sternotomy was performed and 1 ml of blood was drawn by cardiocentesis. After 15-min centrifugation at 3,000 × g, the serum was collected and stored at −20°C until analysis. Total testosterone, LH, and FSH levels were measured with ELISA kits (Assay Designs Inc., Ann Arbor, MI, and Amersham Biosciences, Bucks, U.K.).
Statistical Analyses.
Values were expressed as mean ± SD. We used Student's t test from software SPSS11.0 to compare values among the two groups. One-way ANOVA, which was followed by Tukey-Kramer multiple comparison test using the software SPSS11.0, was used to compare values among the three groups. P < 0.05 was considered significant.
Other Methods.
Immunohistochemical and histological analyses, fertility assessment, evaluation of epididymal sperm, and real-time quantitative RT-PCR were as described (12, 13).
Acknowledgments
This work was partly supported by National Institutes of Health Grants DK60948 and DK60912 and a George H. Whipple professorship endowment.
Abbreviations
- AR
androgen receptor
- T-AR−/y
total AR knockout
- PM-AR−/y
peritubular myoid cell-specific AR knockout
- PM
peritubular myoid
- S-AR−/y
Sertoli cell-specific AR knockout
- LH
leuteinizing hormone
- FSH
follicle-stimulating hormone
- AM
adrenomedullin
- ABP
androgen-binding protein
- Tagln
transgelin
smooth muscle protein 22-α
- ET
endothelin
- AR+/y
WT AR.
Footnotes
The authors declare no conflict of interest.
References
- 1.Anthony CT, Skinner MK. Biol Reprod. 1989;40:811–823. doi: 10.1095/biolreprod40.4.811. [DOI] [PubMed] [Google Scholar]
- 2.Vornberger W, Prins G, Musto NA, Suarez-Quian CA. Endocrinology. 1994;134:2307–2316. doi: 10.1210/endo.134.5.8156934. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Nie R, Prins GS, Saunders PT, Katzenellenbogen BS, Hess RA. J Androl. 2002;23:870–881. [PubMed] [Google Scholar]
- 4.Skinner MK, Fritz IB. Proc Natl Acad Sci USA. 1985;82:114–118. doi: 10.1073/pnas.82.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norton JN, Skinner MK. Endocrinology. 1989;124:2711–2719. doi: 10.1210/endo-124-6-2711. [DOI] [PubMed] [Google Scholar]
- 6.Romano F, Tripiciano A, Muciaccia B, De Cesaris P, Ziparo E, Palombi F, Filippini A. Contraception. 2005;72:294–297. doi: 10.1016/j.contraception.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Heinlein CA, Chang C. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 8.Chang CS, Kokontis J, Liao ST. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 9.Heinlein CA, Chang C. Endocr Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 10.Gray LE, Ostby J, Furr J, Wolf CJ, Lambright C, Parks L, Veeramachaneni DN, Wilson V, Price M, Hotchkiss A, et al. Hum Reprod Update. 2001;7:248–264. doi: 10.1093/humupd/7.3.248. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham GR, Huckins C. Endocrinology. 1979;105:177–186. doi: 10.1210/endo-105-1-177. [DOI] [PubMed] [Google Scholar]
- 12.Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, et al. Proc Natl Acad Sci USA. 2002;99:13498–13503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H, Yeh S. Proc Natl Acad Sci USA. 2004;101:6876–6881. doi: 10.1073/pnas.0307306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maekawa M, Kamimura K, Nagano T. Arch Histol Cytol. 1996;59:1–13. doi: 10.1679/aohc.59.1. [DOI] [PubMed] [Google Scholar]
- 15.Yeh S, Hu YC, Wang PH, Xie C, Xu Q, Tsai MY, Dong Z, Wang RS, Lee TH, Chang C. J Exp Med. 2003;198:1899–1908. doi: 10.1084/jem.20031233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins AS. Funct Integr Genomics. 2002;2:81–91. doi: 10.1007/s10142-002-0049-3. [DOI] [PubMed] [Google Scholar]
- 17.Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J, Kuhn M. Proc Natl Acad Sci USA. 2002;99:7142–7147. doi: 10.1073/pnas.102650499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 19.Jeyaraj DA, Grossman G, Petrusz P. Reprod Biol Endocrinol. 2003;1:48. doi: 10.1186/1477-7827-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhoeven G, Swinnen K, Cailleau J, Deboel L, Rombauts L, Heyns W. J Steroid Biochem Mol Biol. 1992;41:487–494. doi: 10.1016/0960-0760(92)90374-r. [DOI] [PubMed] [Google Scholar]
- 21.Verhoeven G, Hoeben E, De Gendt K. Andrologia. 2000;32:42–45. [PubMed] [Google Scholar]
- 22.Santti H, Mikkonen L, Anand A, Hirvonen-Santti S, Toppari J, Panhuysen M, Vauti F, Perera M, Corte G, Wurst W, et al. J Mol Endocrinol. 2005;34:645–654. doi: 10.1677/jme.1.01666. [DOI] [PubMed] [Google Scholar]
- 23.Schurmann A, Koling S, Jacobs S, Saftig P, Krauss S, Wennemuth G, Kluge R, Joost HG. Mol Cell Biol. 2002;22:2761–2768. doi: 10.1128/MCB.22.8.2761-2768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar TR, Wang Y, Lu N, Matzuk MM. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 26.Kumar TR. Reproduction. 2005;130:293–302. doi: 10.1530/rep.1.00660. [DOI] [PubMed] [Google Scholar]
- 27.Skinner MK. Ann NY Acad Sci. 1987;513:158–171. doi: 10.1111/j.1749-6632.1987.tb25006.x. [DOI] [PubMed] [Google Scholar]
- 28.Griswold MD. Biol Reprod. 1995;52:211–216. doi: 10.1095/biolreprod52.2.211. [DOI] [PubMed] [Google Scholar]
- 29.Skinner MK, Schlitz SM, Anthony CT. Endocrinology. 1989;124:3015–3024. doi: 10.1210/endo-124-6-3015. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein SE. J Lab Clin Med. 1987;110:690–705. [PubMed] [Google Scholar]
- 31.Jabado N, Canonne-Hergaux F, Gruenheid S, Picard V, Gros P. Blood. 2002;100:2617–2622. doi: 10.1182/blood-2002-04-1182. [DOI] [PubMed] [Google Scholar]
- 32.Sylvester SR, Griswold MD. J Androl. 1994;15:381–385. [PubMed] [Google Scholar]
- 33.Kingma PB, Bok D, Ong DE. Biochemistry. 1998;37:3250–3257. doi: 10.1021/bi972520l. [DOI] [PubMed] [Google Scholar]
- 34.Mruk DD, Cheng CY. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 35.Filippini A, Tripiciano A, Palombi F, Teti A, Paniccia R, Stefanini M, Ziparo E. Endocrinology. 1993;133:1789–1796. doi: 10.1210/endo.133.4.8404621. [DOI] [PubMed] [Google Scholar]
- 36.Tripiciano A, Filippini A, Giustiniani Q, Palombi F. Biol Reprod. 1996;55:25–31. doi: 10.1095/biolreprod55.1.25. [DOI] [PubMed] [Google Scholar]
- 37.Tripiciano A, Palombi F, Ziparo E, Filippini A. FASEB J. 1997;11:276–286. doi: 10.1096/fasebj.11.4.9068617. [DOI] [PubMed] [Google Scholar]
- 38.Rossi F, Guerrini L, Pasimeni G, Markouizou A, Fabbrini A, Santiemma V. Arch Androl. 2000;44:103–107. doi: 10.1080/014850100262263. [DOI] [PubMed] [Google Scholar]
- 39.Santiemma V, Rossi F, Guerrini L, Markouizou A, Pasimeni G, Palleschi S, Fabbrini A. Biol Reprod. 2001;64:619–624. doi: 10.1095/biolreprod64.2.619. [DOI] [PubMed] [Google Scholar]
- 40.Jeyaraj DA, Grossman G, Weaver C, Petrusz P. Biol Reprod. 2002;66:877–885. doi: 10.1095/biolreprod66.4.877. [DOI] [PubMed] [Google Scholar]