Abstract
Clp ATPases are a unique group of ATP-dependent chaperones supporting targeted protein unfolding and degradation in concert with their respective proteases. ClpX is a representative member of these ATPases; it consists of two domains, a zinc-binding domain (ZBD) that forms dimers and a AAA+ ATP-binding domain that arranges into a hexamer. Analysis of the binding preferences of these two domains in ClpX revealed that both domains preferentially bind to hydrophobic residues but have different sequence preferences, with the AAA+ domain preferentially recognizing a wider range of specific sequences than ZBD. As part of this analysis, the binding site of the ClpX dimeric cofactor, SspB2, on ZBD in ClpX was determined by NMR and mutational analysis. The SspB C terminus was found to interact with a hydrophobic patch on the surface of ZBD. The affinity of SspB2 toward ZBD2 and the geometry of the SspB2–ZBD2 complex were investigated by using the newly developed quantitative optical biosensor method of dual polarization interferometry. The data suggest a model for the interaction between SspB2 and the ClpX hexamer.
Keywords: NMR, SspB, zinc-binding domain
Protein degradation is an essential component of biological regulation and protein quality control in all organisms. Cylindrical proteases, such as the proteasome, form large oligomers in which the proteolytic active sites are sequestered within an internal chamber. Access to the chamber is provided through narrow axial pores that exclude entry of large polypeptides and allow entry only of small peptides of ≈30 residues in length (1). These cylindrical proteases typically form complexes with ATPases associated with various cellular activities (AAA+) chaperones that denature substrates and then translocate them into the proteolytic chamber of the protease for degradation.
ClpXP of Escherichia coli forms such a complex (2). ClpX is the AAA+ ATPase, and it belongs to the Clp/Hsp100 family, whereas ClpP is a cylindrical serine protease consisting of two rings with 7-fold symmetry (3). ClpX has an N-terminal domain followed by a AAA+ domain (Fig. 6A, which is published as supporting information on the PNAS web site). The importance of the N-terminal domain of ClpX is evident from its absolute conservation across all sequenced genomes (4). Our group demonstrated that the N-terminal domain of ClpX is a C4-type zinc-binding domain (ZBD) that forms a very stable constitutive symmetric dimer in isolation and in full-length ClpX. ZBD binds one Zn(II) per monomer (4, 5). Hydrophobic residues that form the interface between two ZBD monomers are highly conserved throughout the sequenced genomes (5). The AAA+ domain forms a hexameric ring complex in a nucleotide-dependent manner from which the ZBD protrudes (Fig. 6B). ClpX unfolds proteins and then feeds them into ClpP for degradation.
Two proteins that were initially recognized as ClpX substrates are the phage proteins λO and MuA. More recently, ≈50 endogenous E. coli ClpX substrates were identified through a proteomics approach (6). ClpXP has also been implicated in the degradation of C-terminally SsrA-tagged proteins. GFP with an SsrA sequence added to its C terminus has typically been used as a model substrate to study such tagged proteins (7). The SspB2 cofactor enhances the degradation efficiency of C-terminally SsrA-tagged proteins by ClpXP (8). The SspB polypeptide can be divided into a substrate-binding domain that forms a dimer and a C-terminal unstructured domain that binds to the ZBD in ClpX (9). It has been proposed that SspB2 binds to SsrA-tagged proteins forming a complex that subsequently binds to the ClpX hexamer (10). Hence, the binding of SspB2, loaded with substrate, to ZBD in ClpX functions to hold the substrate in place as the AAA+ domain of ClpX pulls the substrate, starting from the recognition motif, through ClpX and into ClpP for degradation. It is established that both C-terminal tails in the SspB dimer are required to enhance the degradation of SsrA-tagged substrates by ClpXP (11). Although the ZBD in ClpX is required to bind the cofactor SspB2, the ZBD is not required to bind and degrade SsrA-tagged proteins (4).
Here we address the question of how the ZBD of ClpX recognizes substrates/cofactors. To this end, we initially used peptide array analysis to determine the general binding preferences of ZBD2 and compared those to the binding preferences of the AAA+ domain. Subsequently, we carried out NMR, mutational, and thermodynamic interaction studies to further characterize and map the binding site of SspB2 on ZBD2. The implications of our findings on SspB2–ClpX6 complex formation are discussed.
Results
ZBD and AAA+ Domains of ClpX Preferentially Bind to Hydrophobic Residues.
To determine the roles of the ZBD and AAA+ (ClpXΔZBD) domains of ClpX in substrate and cofactor recognition, peptide array analysis was carried out to identify residues that are preferentially bound by these two domains of the chaperone (Fig. 7, which is published as supporting information on the PNAS web site). Purified ZBD2 or AAA+ domains were incubated with peptide arrays containing a total of 3,717 C-terminally attached peptides whose sequences were derived from 26 different proteins, some of which are known ClpX substrates (see Materials and Methods). The ZBD is dimeric under all conditions used (4, 5). Because substrates entering into ClpX hexamer will bind to the interior chamber of the AAA+ ring, and because the oligomerization of the AAA+ ring requires the presence of nucleotides (12), peptide array analysis of the AAA+ domain was carried out in the absence of nucleotides to expose the putative polypeptide-binding sites that will be involved in substrate translocation and possibly unfolding. The binding of AAA+ to the peptide arrays in the presence of nucleotides was also carried out but was found to be significantly reduced (Fig. 8, which is published as supporting information on the PNAS web site); consequently, the proportion of binders was statistically insignificant, reflecting the fact that the hexamerized AAA+ ring can recognize only specific sequence tags such as the SsrA tag. Hence no further analysis of the data for AAA+ in the presence of nucleotides could be carried out.
Fig. 7 shows an example of the binding preferences of ZBD2 and AAA+ (in the absence of nucleotides) to arrays generated by using peptides derived from λO and MuA as detected by Western blot analysis. In general, ZBD2 and AAA+ bound different peptide sequences. For example, ZBD2 bound strongly to peptides derived from residues Gln49-Met67 of λO, whereas AAA+ preferentially bound to peptides derived from residues Ile7-Ser41, Pro215-Leu241, and Lys283-Leu299 of λO (Fig. 7). Because ZBD2 binds λO and is required for λO degradation by ClpXP (4), these results suggest that the region Gln49-Met67 of λO is essential for the recognition of λO by ClpX (see below). ZBD2 and AAA+ also recognize different MuA peptide sequences. MuA is 663 aa long. Interestingly, the C terminus of MuA, which was proposed to be the ClpX-recognition motif (13), was only weakly bound by ZBD2, and only residues Glu645-Lys661 were bound by the AAA+ domain (Fig. 7).
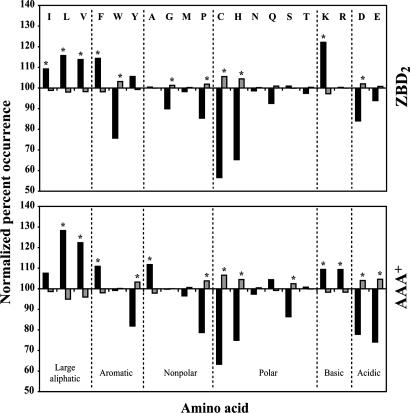
In analyzing the peptide array data, peptides were classified as binders of ZBD2 or AAA+ if their normalized percent intensity was ≥75% (see Materials and Methods); otherwise, they were classified as nonbinders. Separation of the peptides on the arrays into binder and nonbinder groups allowed for comparison of these groups with respect to amino acid occurrence. As can be seen in Fig. 1, both ZBD2 and AAA+ preferentially bound to sequences enriched in hydrophobic residues as well as in the positively charged lysine. Negatively charged residues were typically disfavored.
Fig. 1.
Analysis of ZBD- and AAA+-binding preferences using peptide arrays. Shown are the ZBD2 and AAA+ residue-binding preferences. The normalized percent occurrence for each amino acid was determined for both the binder (black bars) and nonbinder (gray bars) groups. Asterisk indicates that the difference between the occurrence of an amino acid in the binder group and on the array is significant at the 95% confidence level using the z test (22).
ZBD2 and AAA+ Domains of ClpX Preferentially Bind to Distinct Sequence Patterns.
To determine whether the ClpX domains have distinct sequence-binding preferences, and because a large data set has been obtained, binder and nonbinder peptide sequences were submitted to Teiresias (14) to search for short consensus sequences that may be responsible for ZBD2 and AAA+ binding. The Teiresias output included sequence patterns that varied in length but were generally 3–6 aa long. Patterns were analyzed as described in Materials and Methods to rank the consensus sequences that occurred more frequently in binders than in nonbinders (Table 1).
Table 1.
Peptide sequence patterns preferentially bound by ZBD2 and AAA+
| ZBD2 | AAA+ | ||
|---|---|---|---|
| Rank | Pattern | Rank | Pattern |
| 1 | [ST]xEx[ILMV] | 1 | [ST]Lxx[ILMV] |
| 2 | [ST]x[DE]xL | 2 | Rx[ILMV][ILMV] |
| 3 | V[AG]xx[ILMV] | 3 | [ILMV][ILMV][AG][AG] |
| 4 | S[ILMV]x[AG] | 4 | [ILMV]x[ILMV][ILMV][ILMV] |
| 5 | ALxx[ILMV] | 5 | [ILMV]xx[ST]L |
| 6 | [ILMV]I[QN] | 6 | [ILMV][ILMV][ILMV]x[AG] |
| 7 | [AG]I[ILMV] | 7 | [ILMV]xx[ILMV][ILMV][AG] |
| 8 | [FY]x[ILMV]xG | 8 | [ILMV]I[QN] |
| 9 | [AG][DE]xV | 9 | V[AG]xx[ILMV] |
| 10 | [KR][AG]xxL | 10 | T[ILMV]xxL |
| 11 | [ILMV]xL[ILMV]x[AG] | 11 | [ST][ILMV]L |
| 12 | [ILMV]x[ILMV][ILMV]xx[AG] | 12 | [ILMV]xAG |
| 13 | [ILMV][ILMV]x[ILMV]x[KR] | 13 | [ILMV]x[QN]L |
| 14 | [KR]x[ILMV][ILMV]x[ILMV] | 14 | [ILMV]xL[ILMV][ILMV] |
| 15 | [ST]xExL | 15 | VxL[ILMV] |
| 16 | [AG]x[ILMV]xI | ||
| 17 | LLxx[AG] | ||
| 18 | [ILMV]x[ILMV][ILMV]xA | ||
| 19 | [ILMV]xL[ILMV]x[AG] | ||
| 20 | [AG]I[ILMV] | ||
Residues in brackets indicate that any of the listed amino acids can be present at that position. x represents any amino acid. Patterns in bold are present in both ZBD2 and AAA+. Patterns are ranked according to FB (see Materials and Methods).
The utility of this approach is demonstrated by the identification of several consensus sequences previously implicated as possible ClpX-recognition sequences. For example, the strongest binding of ZBD2 to the λO array occurs in the region Q49FKVLLAILRKTYGWNKPM67 (Fig. 7A), and the patterns responsible for the binding may be [AG]I[ILMV] and [KR]x[ILMV][ILMV]x[ILMV] (ranked 7 and 14 in Table 1), corresponding to the λO region of A55IL57 and K51VLLAI56, respectively. As another example, it is known that SspB2 specifically binds ZBD in ClpX (4, 11). The binding is mediated by the last 10 residues in SspB (G156GRPALRVVK165) (9, 15), which contain the ZBD2 consensus sequence ALxx[ILMV] (ranked 5 in Table 1). Other such consensus sequences from this analysis can be found for the putative substrates identified by Flynn et al. (6).
Patterns common to both ZBD2 and AAA+ binders are shown in bold in Table 1. In general, it is not surprising that there are such common patterns indicating an equal role of both domains in binding these patterns. It should be noted that more patterns were found for AAA+ binders than for ZBD binders (Table 1). This might indicate that ZBD2 exercises more specificity in substrate recognition, whereas the AAA+ domain more promiscuously binds and releases unfolded proteins as they translocate through ClpX and into ClpP.
To verify some of the results of the peptide array analysis, the following experiments were carried out. Initially, the ClpXP-mediated degradation of λO was carried in the presence of 50 times excess of the peptides λO49–63, MuA653–663, or SspB154–165 (Fig. 9A, which is published as supporting information on the PNAS web site). According to the peptide array analysis, ZBD2 bound to peptides corresponding to λO49–63 but not to those corresponding to MuA653–663 (Fig. 7A). Consistent with these results, excess of λO49–63 peptide slowed down the ClpXP-mediated degradation of λO, whereas MuA653–663 did not affect the rate of degradation. In addition, the C terminus of SspB, SspB154–165 peptide, significantly slowed down λO degradation (Fig. 9A), suggesting that λO and SspB have similar or adjacent binding sites on ZBD in ClpX.
In another set of experiments, the peptides IYYITGESLKAVE (IYY) and DVGVLVISARKGE (DVG) were added at the N terminus of a sequence consisting of 6xHis tag followed by a tobacco etch virus recognition sequence and then GFP to form the constructs IYY-GFP and DVG-GFP, respectively. In the peptide array experiments, IYY and DVG peptides were preferentially bound by ZBD2 and AAA+, respectively. Consistent with those experiments, ELISA analysis confirmed that ZBD2 preferentially bound IYY-GFP, whereas AAA+ preferentially bound DVG-GFP (Fig. 9B). However, it should be noted that neither IYY-GFP nor DVG-GFP were unfolded or degraded by ClpXP (data not shown), indicating that, whereas some sequences are required for recognition by ClpX, other additional sequences might be required for unfolding and degradation.
SspB C Terminus Binds a Hydrophobic Patch on the Surface of ZBD2.
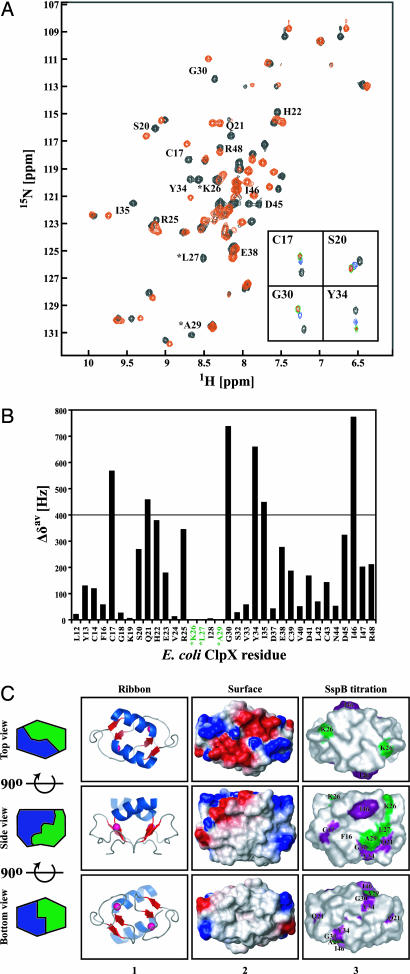
The peptide array analysis revealed that ZBD in ClpX might recognize a limited set of specific sequence patterns. To understand how ZBD2 recognizes such sequence patterns, we endeavored to map the binding site for SspB2 cofactor on ZBD2. To this end, a series of 1H, 15N heteronuclear sequential quantum correlation spectra were recorded of a uniformly 15N-labeled ZBD2 in the presence of increasing concentrations of SspB154–165 (Fig. 2A) or SspB2 (data not shown). SspB154–165 consists mostly of hydrophobic residues (P154RGGRPALRVVK165). The NH chemical shift assignments that we determined (5) were used. Upon addition of SspB154–165, the chemical shifts of several residues systematically moved as the peptide concentration was increased (Fig. 2A), and saturation was typically reached at peptide to ZBD2 concentration ratio between 2.5 and 5 (Fig. 2A Inset).
Fig. 2.
Binding of SspB154–165 to ZBD2 as monitored by NMR. (A) A heteronuclear sequential quantum correlation spectrum of 0.25 mM 15N-labeled ZBD2 in the absence (black) or presence of 5 mM of SspB154–165 (red). ∗, chemical shifts that disappear upon addition of peptide. Insets show the NH chemical shift changes of four residues in ZBD2 in the presence of 0 (black), 0.5 mM (blue), 2.5 mM (green), and 5 mM (red) of SspB154–165. (B) Shown are the chemical shift changes Δδav = [(Δδ1H)2 + (Δδ15N)2]1/2 in the presence of 5 mM SspB154–165. ∗, residues whose chemical shifts disappeared upon addition of 5 mM SspB154–165. (C) Ribbon and surface representation of ZBD dimer (Protein Data Bank ID code 1OVX) (5). Middle and Bottom are rotated 90o along the horizontal axis with respect to Top and Middle, respectively. In column 1, helices are colored blue, strands are red, and Zn(II) atoms are shown as pink spheres. In column 2, the electrostatic potential surface of ZBD2 is shown with negatively charged, positively charged, and hydrophobic surfaces in red, blue, and gray, respectively. In column 3, residues for which Δδav > 400 Hz and whose chemical shifts disappeared in the SspB154–165 titration experiments are colored purple and green, respectively. All structures were drawn by using PyMOL (http://pymol.sourceforge.net).
The NH groups of residues Cys17, Gln21, Gly30, Tyr34, Ile35, and Ile46 in ZBD2 (0.25 mM) shifted by >400 Hz (Δδav; see Materials and Methods) in the presence of 2.5 mM of SspB154–165 (Fig. 2B). Because the maximum shift observed was ≈800 Hz (Fig. 2B), chemical shifts more than half the maximum, i.e., >400 Hz, were arbitrarily considered to be significant. The chemical shifts of three ZBD residues (Lys26, Leu27, and Ala29) disappeared upon addition of SspB peptide (marked by an asterisk in Fig. 2 A and B). The same phenomenon was observed upon addition of SspB2 (data not shown). Thr1-Leu11, Ser15, Pro31, Cys36, and Glu49-Arg60 are not observed in the ZBD2 spectra.
ZBD2 forms a box-shaped structure with four main surfaces. The highly charged surface of ZBD2 is shown in Fig. 2C Upper (top view), two identical hydrophobic surfaces exist on both sides of the ZBD2 box (Fig. 2C, side view), and a fourth hydrophobic surface is present on the bottom (Fig. 2C, bottom view). Residues Cys17, Gln21, Leu27, Ala29, Gly30, Tyr34, and Ile46 are part of the ZBD2 side hydrophobic surfaces, whereas residue Lys26 contributes to the side hydrophobic surfaces through its long hydrophobic side chain (Cβ-ζ). Ile35 is part of the ZBD2 dimer interface and is not solvent-exposed. The results strongly indicate that the side hydrophobic surfaces of ZBD2 are the primary SspB2 binding sites in ClpX; hence, there are two potential SspB2-binding sites per ZBD dimer that can interact with the tails of SspB2.
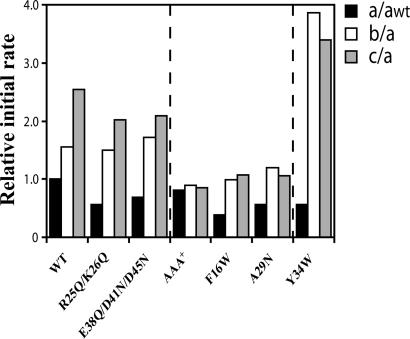
To further map the hydrophobic binding sites for SspB2 on ZBD2, systematic mutagenesis of the ZBD in ClpX was carried out. The degradation of GFP-SsrA was performed at 37°C in the presence of ClpP, ClpX mutants and different concentrations of SspB2 and was monitored by fluorescence (Fig. 3 and Fig. 10, which is published as supporting information on the PNAS web site). As expected, SspB2 does not enhance the ClpP-dependent degradation of GFP-SsrA if ClpX lacking ZBD (AAA+) is used, because ZBD is the binding site for SspB2 (4, 11). Several ClpX variants were generated with mutations in ZBD to identify mutants that behaved like AAA+ in the degradation assays. Only mutations of the hydrophobic surfaces of ZBD in ClpX (F16W and A29N) abolished the effect of SspB2, whereas replacing the positively or negatively charged residues on the charged surface of ZBD in ClpX with neutral residues (R25Q/K26Q and E39Q/D41N/D45N) did not prevent the SspB2-mediated enhancement of GFP-SsrA degradation (Figs. 3 and 10). Interestingly, under the conditions used for Fig. 3, the enhancing effect of SspB2 was stronger for ClpX(Y34W) as compared with ClpX WT (see also Fig. 10). Y34W is part of the hydrophobic surface of ZBD2 (Fig. 2C). All mutations shown in ZBD of ClpX do not significantly affect the secondary structure content, stability, or dimerization state of isolated ZBD2 compared with WT domain as judged by CD measurements, thermal denaturation, and size-exclusion chromatography (Fig. 11 A–C, which is published as supporting information on the PNAS web site). The mutation of other residues in ZBD that were identified by NMR as possible binding sites for the SspB C terminus (Fig. 2B) generally resulted in poorly behaved or misfolded ClpX.
Fig. 3.
Mutational analysis to determine the SspB2-binding site on ZBD in ClpX. Relative initial rates of the ClpP-dependent degradation of GFP-SsrA (3.9 μM monomer concentration) mediated by ClpX or different ClpX mutants (each at 1 μM monomer concentration) in the presence of ClpP (1.2 μM monomer concentration) are shown in the presence of 0 (a), 0.025 μM (b), and 0.165 μM (c) of SspB2. The initial rates a were normalized to awt (black bars), whereas the initial rates b (white bars) and c (gray bars) were normalized to the a of the respective mutant. Initial rates were determined from the first 200 seconds of the curves shown in Fig. 10.
The results from the mutational analysis (Fig. 3) correlate well with those from the NMR analysis (Fig. 2). The NH chemical shift for Ala29 disappeared upon addition of 2.5 mM SspB peptide or 125 μM SspB2 full length, and for Tyr34, Δδav was >400 Hz (Fig. 2B and data not shown). The NH chemical shift of Phe16 did not significantly change upon addition of SspB peptide or SspB2 protein, which is probably because this residue is part of the dimer interface (5); however, Phe16 is next to Cys17, which did show a Δδav >400 Hz in Fig. 2B. Hence, the NMR and mutagenesis analyses strongly suggest that residues Phe16, Ala29, and Tyr34 in ZBD of ClpX are part of or close to the SspB2-binding site, and that the binding mainly occurs through the C termini of the SspB dimer.
Measuring the Binding Affinity and Geometry of the Interaction of SspB2 to ZBD2.
To further support the conclusions made above, isothermal titration calorimetry (ITC) experiments were carried out at 20°C to determine the binding constant of SspB154–165 peptide to the different ZBD2 mutants. ZBD2 WT bound SspB154–165 with a Kd of 34.13 ± 5.94 μM and n = 0.97 ± 0.07 (Fig. 12 A and B, which is published as supporting information on the PNAS web site), suggesting that only one site on the hydrophobic surfaces of ZBD2 can be saturated by the peptide. This could indicate that the binding of SspB154–165 to one surface prevents interactions with the second surface as a result of some slight but critical conformational changes. Other interpretations are also possible. The binding of SspB154–165 to ZBD2(Y34W) gave a Kd of 30.49 ± 4.09 μM and n = 3.08 ± 0.04 (Fig. 12 C and D), suggesting that the mutation of Y34 to Trp allowed for the binding of the peptide to more than one site on the hydrophobic surfaces of ZBD2(Y34W). Finally, as expected, ZBD2(F16W) and ZBD2(A29N) did not show any significant binding to SspB154–165 (Kd > 200 μM; data not shown). Hence, these results, combined with the NMR and mutational analyses, strongly suggest that residues F16 and A29 are required to preserve the SspB2-binding site on ZBD2.
A further understanding of the mode of interaction of SspB2 and SspB154–165 to ZBD2 was gained by using an AnaLight Bio200 (Farfield Scientific, Crewe, U.K.) instrument that uses a dual polarization interferometry detection method (16, 17). The instrument gives absolute measurements of thickness and density of crosslinked and bound biological molecules on films, with resolution levels of subpicogram per mm2 and subAngstrom, respectively. ZBD2 was crosslinked to the sensor chip by using Bis(Sulfosuccinimidyl)suberate (BS3). Because, BS3 reacts with primary amines, ZBD2 is probably preferentially crosslinked with its charged surface facing the sensor chip. The thickness of ZBD2 reached 1.93 ± 0.13 nm on the sensor chip (data not shown), close to the expected dimensions of ZBD2. After the establishment of a stable buffer baseline, protein or peptide was injected over the immobilized ZBD2. The thickness and density of the protein/peptide interacting with ZBD2 on the sensor chip were then measured; hence, the mass change on the surface of the sensor chip as a function of injected sample concentration was obtained (Fig. 13 A and B, which is published as supporting information on the PNAS web site).
The binding profile for SspB154–165 peptide was best fit to a single binding event with a Kd of 23.1 μM (Fig. 4A and C), close to the value obtained from ITC measurements described above. On the other hand, the changes in density and thickness as a function of SspB2 concentration (Fig. 13B), indicated there are (at least) two different binding events taking place between SspB2 and ZBD2. Fitting the mass change data to a model of two independent binding events gave a very good fit to the experimental points (Fig. 4B). The strong association reaction had a Kd of 0.85 μM (SspB2 to ZBD2) with a final thickness of 3.3 nm at saturation (Figs. 4B and 13C). The thickness obtained agrees with the expected dimension of a layer of SspB dimers, placed on their sides, on top of the ZBD dimers (Fig. 13C Upper) (5, 16, 18). The weak association reaction had a Kd of 11.0 μM (SspB2 to ZBD2) and a final thickness of 7.6 nm (Figs. 4B and 13C). The data suggest that the SspB dimer in this case is oriented vertically upon binding to ZBD2 (Fig. 13C). Also, at saturation, the stoichiometry of the interaction between SspB2 and ZBD2 was found to be 1:1. It was previously reported that the binding affinity of SspB154–165 to ClpX hexamer, SspB154–165 to ZBD dimer, and SspB dimer to ClpX hexamer has a Kd of 22.8, 20.0, and 1.3 μM, respectively (10, 11, 15), which are close to the values obtained in our analysis (Fig. 4C).
Fig. 4.
Determination of the binding affinity of SspB2 and SspB154–165 to ZBD2 using dual polarization interferometry. (A) Open circles represent the experimental binding data of SspB154–165 to ZBD2, whereas the solid line represents the theoretical fit to the data assuming a single binding event. (B) Open circles represent the experimental data for the binding of SspB2 to ZBD2, whereas the solid line represents the theoretical fit to the data assuming two independent binding events. Curves drawn with short- and long-dashed lines represent the theoretical binding curves assuming a single binding event with Kd values given in C corresponding to the high- and low-affinity interactions, respectively. (C) Binding parameters of SspB154–165 or SspB2 to ZBD2 obtained from the fits to the experimental data. Values in parentheses are standard deviations.
Using this technique, it is possible to estimate the area occupied by molecules on the surface of the chip. For the high-affinity interaction, SspB2 occupies an area corresponding to twice that occupied by a single ZBD2 (Fig. 13D). On the other hand, for the low affinity interaction, SspB2 occupies an area corresponding to a single ZBD2 (Fig. 13D). We propose that the strong binding interaction is due to the association of the SspB dimer to two ZBD dimers by using both C termini of SspB2, whereas the weak association interaction is due to the association of SspB2 to one ZBD2 through only one C terminus.
Discussion
Rigorous analysis of the binding of ZBD and AAA+ domains of ClpX to a large peptide library allowed us to identify novel binding preferences for these two ClpX domains. Although ZBD and AAA+ domains favor binding to hydrophobic residues, these domains preferentially recognize different amino acid sequences. This suggests that the ZBD and AAA+ domains bind to different classes of polypeptides and, hence, “filter” the interaction between ClpX and its putative substrates.
The binding of SspB2 to ZBD2 occurs mainly through the interaction of the C termini of SspB2 to hydrophobic patches present on the surface of ZBD2, as determined by NMR and mutagenesis analyses (Figs. 2 and 3). The interaction is significantly enhanced by the binding of two C-terminal tails of SspB2 to ZBD2 (Fig. 4C). Interestingly, residue Leu161 at the C terminus of SspB is highly conserved and is essential for the enhancement of the degradation of SsrA-tagged proteins by ClpXP (15). Therefore, it would be reasonable to propose that Leu161 of SspB is one of the C-terminal residues that directly interact with the hydrophobic patches on ZBD2 surface.
In principle, three SspB dimers can bind to one ClpX hexamer. However, it has been experimentally determined that only one SspB2 binds to a ClpX hexamer at a given time (19). We had proposed earlier that the ZBD domains in ClpX hexamer might come together to form a trimer of dimers at one stage during the chaperone functional cycle (5). Fig. 5 shows the four possible modes by which two tails of SspB2 might interact with the ZBD trimer of dimers. In Fig. 5, it is assumed that the ClpX AAA+ ring is below the ZBD2s. Based on our binding analysis (Figs. 4, 12, and 13) and on published literature (10, 11, 15), the binding mode of Fig. 5D in which the two tails of SspB2 interact with the same ZBD2 is unlikely to occur, especially that an SspB tail can bind to only one site on ZBD2 according to ITC measurements (Fig. 12). In Fig. 5 A and C, the two tails of SspB2 do not interact “symmetrically” with the ZBD surface and would have to be differently kinked; this is not the case for the binding mode of Fig. 5B. Furthermore, in Fig. 5C, both tails are underneath the ZBD oligomer, between the AAA+ ring and the ZBD2s. We consider the modes of binding in Fig. 5 A and C to be possibly disfavored. Hence, the binding mode of Fig. 5B might be most likely. This mode of binding would also be in agreement with our recently published data suggesting a nucleotide-dependent block movement of the ZBD2 toward the AAA+ ring in ClpX (20).
Fig. 5.
Binding mode of SspB2 to ZBD trimer of dimers. (A–D) Shown are the different possible modes of binding between two C-terminal tails of SspB2 and the ZBD trimer of dimers shown in surface representation. ZBD residues implicated in interacting with SspB C terminus by NMR and mutagenesis are in a darker color. The AAA+ ring of ClpX is assumed to be below the ZBD trimer of dimers. Arrowheads represent the N termini of the tails of SspB2 and point toward the rest of SspB2 body, whereas the slashes represent the C termini of the tails. Tails in dashes and in solid colors are below and above the plane of the ZBD oligomer, respectively. The three ZBD dimers might be in a more open and less planar configuration than shown.
In the model of Fig. 5B, one tail binds to the top of one ZBD2, whereas the other tail binds to the bottom of the second ZBD2. The third ZBD2 can be prevented from interacting with other cofactors or substrates by the folded domain of the bound SspB2. The possible movements of the ZBD2 can then drive the bound SspB2 closer to the AAA+ ring to deliver the SsrA-tagged substrate, whereas the tails of SspB2 reposition the other two ZBD2s away from the entry pore. This model implies that the enhancing activity of SspB2 is due in part to its ability to direct the movement of ZBD2s and to regulate the delivery of tagged substrates in addition to increasing the local concentration of those substrates near ClpX.
Materials and Methods
Protein Purification and Peptide Synthesis.
Proteins were expressed, purified, and manipulated as described (4). Peptides were purchased from Dalton Chemical Laboratories (Toronto, ON, Canada). CD measurements of 15 μM ZBD2 WT and mutants were carried out in buffer A (25 mM Tris·HCl, pH 8/150 mM NaCl/1 mM DTT) by using Jasco (Easton, MD) J-810. Degradation assays were typically carried out in buffer B (25 mM Hepes, pH 7.5/5 mM MgCl2/5 mM KCl/0.03% Tween-20/10% glycerol), as described (4).
Peptide Array Experiments.
Peptide arrays were prepared by using an AutoSpot ASP 222 spot synthesizer (intavis AG) according to a standard spot synthesis protocol. Each peptide was 13 aa long, with a frame shift of 2 aa along the protein sequence, for a total of 3,717 peptides tested. Three independent peptide array incubation experiments were analyzed by using a procedure similar to that of Rüdiger et al. (21). Further details are given in Supporting Text, which is published as supporting information on the PNAS web site.
NMR Spectroscopy.
Uniformly 15N-labeled ZBD2 was prepared by growing the E. coli strain BL21 gold in minimal media containing 15NH4Cl. The NMR sample concentration was typically 0.25 mM of ZBD2 in buffer C (20 mM sodium phosphate, pH 7.6/150 mM NaCl/10% D2O). NMR spectra were recorded at 20°C on a 500 MHz Varian Spectrometer. 1H, 15N heteronuclear single quantum correlation experiments were carried out in the absence or presence of SspB2 full length or SspB154–165 peptide. Changes in the chemical shifts of ZBD2 NH groups (Δδav) were derived from [(Δδ1H)2 + (Δδ15N)2]1/2, where Δδ is the chemical shift change expressed in Hz.
Measuring Binding Affinities and Geometries.
ITC experiments were performed at 20°C. Twenty-nine 10-μl injections of 1.1 or 2 mM SspB154–165 were added to 1.4 ml of 70 μM ZBD2 or ZBD2 mutants. Peptide and proteins were resuspended in buffer D (50 mM potassium phosphate, pH 8/75 mM NaCl/1 mM DTT). The thermograms were fit to a one-site model by using Origin 7 software (OriginLab, Northampton, MA). ITC experiments were performed by using a Microcal (Amherst, MA) VP-ITC and were repeated three times. Other binding experiments were performed by using an AnaLight Bio200 dual waveguide interferometer instrument from Farfield (16). Experiments were performed in buffer D at a flow rate of 0.05 ml/min. ZBD2 (0.5 mg/ml) was crosslinked to both channels of the sensor chip by incubating with [Bis(sulfosuccinimidyl) suberate] (BS3) (Pierce, Rockford, IL). Free BS3 was blocked by using 10 mg/ml glucosamine. After the establishment of a stable buffer baseline, SspB154–165 or SspB2 was injected into one channel, whereas the second channel was used as a reference.
Supplementary Material
Acknowledgments
We thank Dr. Monika Niggermann for synthesizing some of the peptide arrays. We also thank Dr. Jimin Wang (Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT) for helpful discussions. G.T. holds a Natural Sciences and Engineering Research Council (NSERC) PGSD2 Fellowship. J.Y. was supported by an NSERC Undergraduate Student Research Award. R.S. acknowledges financial support from a European Molecular Biology Organization postdoctoral fellowship. W.A.H. is Canadian Institutes of Health Research New Investigator. This research was supported in part by a grant from the Canadian Institutes of Health Research (to W.A.H.).
Abbreviations
- AAA+
ATPases associated with various cellular activities
- ZBD
zinc-binding domain
- ITC
isothermal titration calorimetry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Thompson MW, Maurizi MR. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 2.Schirmer EC, Glover JR, Singer MA, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 3.Wang J, Hartling JA, Flanagan JM. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 4.Wojtyra UA, Thibault G, Tuite A, Houry WA. J Biol Chem. 2003;278:48981–48990. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson LW, Wojtyra U, Houry WA. J Biol Chem. 2003;278:48991–48996. doi: 10.1074/jbc.M307826200. [DOI] [PubMed] [Google Scholar]
- 6.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 7.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 8.Levchenko I, Seidel M, Sauer RT, Baker TA. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 9.Dougan DA, Weber-Ban E, Bukau B. Mol Cell. 2003;12:373–380. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Bolon DN, Grant RA, Baker TA, Sauer RT. Mol Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Mol Cell. 2004;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- 12.Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 13.Levchenko I, Yamauchi M, Baker TA. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 14.Rigoutsos I, Floratos A. Bioinformatics. 1998;14:55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Mol Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- 16.Cross GH, Reeves AA, Brand S, Popplewell JF, Peel LL, Swann MJ, Freeman NJ. Biosens Bioelectron. 2003;19:383–390. doi: 10.1016/s0956-5663(03)00203-3. [DOI] [PubMed] [Google Scholar]
- 17.Swann MJ, Peel LL, Carrington S, Freeman NJ. Anal Biochem. 2004;329:190–198. doi: 10.1016/j.ab.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Song HK, Eck MJ. Mol Cell. 2003;12:75–86. doi: 10.1016/s1097-2765(03)00271-5. [DOI] [PubMed] [Google Scholar]
- 19.Wah DA, Levchenko I, Baker TA, Sauer RT. Chem Biol. 2002;9:1237–1245. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- 20.Thibault G, Tsitrin Y, Davidson T, Gribun A, Houry WA. EMBO J. 2006;25:3367–3376. doi: 10.1038/sj.emboj.7601223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore DS, McCabe GP. Introduction to the Practice of Statistics. New York: Freeman; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.