Abstract
Although protein structures are primarily encoded by their sequences, they are also critically dependent on environmental factors such as solvents and interactions with other molecules. Here we investigate how the folding-energy landscape of a short peptide is altered by interactions with another peptide, by performing atomistic replica-exchange molecular dynamics simulations of polyalanines in various environments. We analyzed the free-energy landscapes of Ala7 and Ala8 in isolation, near an α-helix template, and near a β-strand template. The isolated Ala7 and Ala8 at 270 K were mainly in polyproline II helix conformations and in equilibrium between the α-helix and polyproline II helix, respectively, in harmony with the experiment. Interestingly, we found remarkably strong secondary-structure “templating”; namely, the α-helix template enhanced α-helix conformation and the β-strand template induced β-strand conformation in the simulated Ala8. The α-helix template lowered the nearby dielectric constant, which strengthened hydrogen bonds in the simulated Ala8, leading to α-helix stabilization. The β-strand template provided hydrogen bond positions to the simulated Ala8, sharply inducing β-strand structure. With or without templates, the energy landscape of Ala8 is always funnel-like and centered at the α-helix conformation, whereas entropic contribution disfavors the α-helix, leading to subtle competition. Secondary-structure templating may play a critical role in protein conformation dynamics in the cellular environment.
Keywords: energy landscape, generalized Born, polyproline II, protein folding, replica exchange
Protein structures are primarily encoded by their sequences, and many proteins can spontaneously fold into their native structures. This robust refolding can be realized through organization of the funnel-like energy landscape (1). However, many cases where the protein environment makes the situation substantially more complex need further consideration, such as domain swapping in multidomain proteins (2), amyloid formation (3), and nonspecific macromolecular crowding (4). In general, environmental factors such as protein–protein and protein–solvent interactions change the energy landscape of proteins, leading to critical differences in conformational states, which could be an important source of biological diversity.
Environmental effects on protein conformation are very versatile, even for one of the simplest polypeptides, polyalanine, and its derivatives. Ala13, flanked by two ornithine- and alanine-rich peptides containing interior lysine, glutamic acid, or glutamine, favors an α-helix structure in water (5–7), which is consistent with the high propensity of alanine to take on an α-helix structure in the statistics of the Protein Data Bank (8). However, similar alanine-based peptides, Ala-Gly-Ala-Ala-Ala-Ala-Gly-Ala (9) and Ac-Lys-Ala14-Lys-methyl amide (NMe) (10), form amyloid fibrils in water. In a more complicated way, shorter alanines (Alan with n ≈ 10) flanked by Lys are in disordered form (11), and Ala7 flanked by two ornithines forms not an α-helix but a polyproline II (PPII) helix at low temperatures (12). The PPII helix has been proposed to be a significantly populated structure in the denatured state and has been studied intensively (13–20). The central Ala in Ac-Gly-Gly-Ala-Gly-Gly-NH2 predominantly takes PPII helix structure (21), which is supported by other studies (13–16). Alanine-based polypeptides take on various forms and thus could be an excellent means of addressing the physical basis of environmental effects on the protein energy landscape.
Here, to address how the energy landscape of a polyalanine is altered by its environment, we simulated polyalanines near spatially fixed templates of α-helix and β-strand conformations and compared their energy landscapes with those isolated in water. Although the simulated system is relatively simple, statistically sufficient sampling is not at all easy because the polypeptide can take α-helix, β-strand, PPII helix, and disordered states, which are well separated from each other in conformational space. Thus, highly balanced conformational sampling is required to obtain converged results. To this end, we performed replica-exchange molecular dynamics (REMD) simulation (22) with the CHARMM force field (23) plus the generalized Born (GB) implicit solvent model (24). Quite comprehensive (cumulatively 18 μs) REMD simulations were carried out to obtain well converged results. We first confirmed that the CHARMM plus GB model could consistently reproduce the PPII helix structure as the dominant conformation of the alanine heptamer. The PPII helix structure is stabilized by hydrogen bonds between the peptide backbone and water molecules (12) and is thought to be difficult to approximate by using implicit solvent models. However, it turned out that the GB solvation model well represented the heterogeneous nature of the local dielectric field, leading to a stabilized PPII helix structure.
Using this simulation method, we found that the α-helix template stabilized the α-helix and the β-strand template sharply induced the β-strand in the simulated polyalanine. The template of the α-helix decreased the local dielectric constant near the template, which strengthened the hydrogen bond in the simulated polypeptide and thus stabilized the α-helix. The β-strand template provided hydrogen bond positions to the simulated polyalanine when the latter took β-strand conformation and thus stabilized the β-strand very sharply. Energy landscape analysis revealed that the energy landscapes were always funnel-like and centered at the α-helix conformation, whereas conformational entropy disfavored the α-helix in all cases simulated, leading to subtle competition.
Results and Discussion
Isolated Polyalanines.
In the simulation of isolated Ala7 and Ala8, we found a significant number of PPII helical conformations. For Ala7, the PPII helix was the dominant structure, with a content of 50% at 270 K, which gradually decreased with temperature, whereas α-helices and β-strands were hardly found (Table 1). We note that the ensemble contained many disordered structures that had neither α-helix, β-strand, nor PPII helix structure, the content of which was ≈50%. The PPII helix content was reduced to 32% for Ala8 at 270 K because α-helices became prominent at 31%, and thus Ala8 was in equilibrium between the PPII helix and the α-helix (Table 1). The large PPII helix content in Ala7 was consistent with previous experiments (12). The increase in α-helix content for longer polyalanines was also consistent with previous experiments (5, 11). In preliminary simulations, alanines (Alan with n ≤ 7) hardly formed α-helices, suggesting that they were too short to form α-helices and that they predominantly take the PPII helix form. Ala8 is the shortest polyalanine that forms the α-helix with modest stability, making Ala8 a fragile peptide chain, the conformation of which is critically altered by the environment.
Table 1.
Structural summary of simulation at 270 K
| Environment | α-Helix, % | β-Strand, % | PPII helix, % | |
|---|---|---|---|---|
| Ala8 | ||||
| Isolated | 30.8 (8.2) | 0.7 (0.2) | 32.1 (5.2) | |
| Near α-helix template | ||||
| 8-Å separation | 65.5 (7.2) | 0.7 (0.7) | 10.0 (2.8) | |
| 9-Å separation | 37.3 (11.3) | 14.5 (3.4) | 13.9 (5.7) | |
| 11-Å separation | 27.4 (10.2) | 2.7 (1.1) | 18.7 (4.0) | |
| Near β-strand template | ||||
| 8-Å separation, 2–8 ns | 16.3 (12.0) | 84.6 (14.0) | 6.7 (1.6) | |
| 8-Å separation, 10–16 ns* | 3.5 (1.4) | 97.1 (1.4) | 4.3 (2.6) | |
| 9-Å separation | 2.8 (1.2) | 92.8 (2.5) | 2.2 (0.7) | |
| 11-Å separation | 9.8 (8.0) | 78.4 (6.2) | 11.4 (0.7) | |
| Isolated, non-GB | ||||
| ε = 1 | 1.3 (0.8) | 46.4 (10.0) | 0.2 (0.1) | |
| ε = 2 | 47.4 (6.0) | 30.8 (8.1) | 1.3 (0.3) | |
| ε = 4 | 65.1 (0.2) | 2.5 (0.4) | 0.06 (0.06) | |
| ε = 10 | 36.2 (1.2) | 0.1 (0.01) | 0.03 (0.03) | |
| ε = 80 | 11.9 (0.6) | 0.1 (0.1) | 0.0 (0.0) | |
| Ala7 | ||||
| Isolated | 3.1 (1.4) | 2.9 (2.7) | 48.6 (3.3) | |
| Near α-helix template | 3.4 (3.4) | 0.0 (0.0) | 19.7 (2.3) | |
| Near β-strand template | 0.0 (0.0) | 96.0 (1.2) | 7.6 (0.3) | |
| Isolated, non-GB | ||||
| ε = 1 | 0.0 (0.0) | 48.8 (4.1) | 0.2 (0.03) | |
| ε = 2 | 25.5 (5.7) | 55.9 (6.4) | 0.4 (0.1) | |
| ε = 4 | 58.5 (1.9) | 2.03 (0.3) | 0.07 (0.03) | |
| ε = 10 | 34.3 (0.9) | 0.07 (0.03) | 0.0 (0.0) | |
| ε = 80 | 18.0 (0.7) | 0.07 (0.03) | 0.0 (0.0) | |
The standard deviations are in parentheses. We note that there exist many disordered conformations that do not have any secondary structures.
*The data are from samples during the 10- to -16-ns step.
The fact that our simulation with the GB model produced the PPII helix structure is somewhat surprising because water molecules are thought to play crucial roles in stabilizing the PPII helix structure (13, 15, 16, 25). Quantum mechanical calculations on Ac-Ala-methyl amide (NMe) in water showed that the lowest energy was in the PPII helix, which is stabilized by double-water bridges (13). [An opposing suggestion has been made (26).] Notably, the same molecule in vacuum showed no energy minimum around the PPII helix or α-helix. MD simulations of polyalanine in explicit water (17, 27) support the theory of a water bridge in the PPII helix. Consistently, the PPII helix in H2O is more stable than that in alcohols (28), DMSO, or 2H2O (14). These all support the idea that solvent–solute hydrogen bonds play major roles in stabilizing the PPII helix. Although water bridging is inherently molecular in nature, some aspects of water bridges seem to be captured by the heterogeneity of dielectric constants around the polypeptide in the GB model. Thus, our calculations could reproduce the PPII helix structure in Ala7. In fact, any uniform ε (thus non-GB model) simulation did not produce the PPII helix structure at all (Table 1); uniform ε simulation produced mainly β-hairpin with ε = 1, α-helix and β-hairpin coexistence with ε = 2, α-helix with ε = 4–10, and little secondary structure with ε = 80. The PPII helix is stabilized only via the spatially heterogeneous dielectric environment made by the peptide itself, which is efficiently taken into account in the GB model. We also note that, in the simulation with the GB model, typical conformations that contained the PPII helix contained disordered segments as well and thus were not “ideal” PPII helices, consistent with the view obtained from NMR- and small-angle x-ray scattering (SAXS)-based arguments (19, 29).
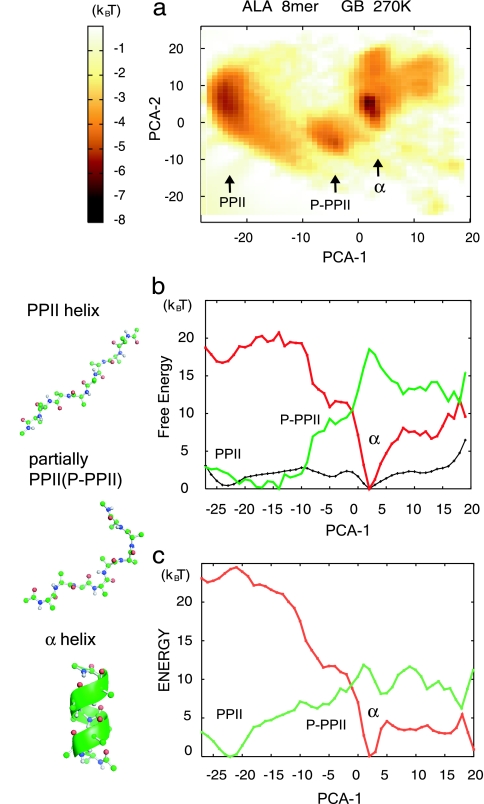
We next analyzed the free-energy landscape of the isolated Ala8. The peptide conformational space was large-dimension, but, for intuitive understanding, we needed a few axes by which major conformational states could be distinguished. For this purpose, it is standard to use the principal component (PC) axes, which tend to classify sampled conformations as much as possible (30–32). Fig. 1a depicts the computed free energy of Ala8 at 270 K in two dimensions spanned by the first and second PCs, PC1 (horizontal axis) and PC2 (vertical axis). Three free-energy basins were found that corresponded to the PPII helix at (−23, 6), the α-helix at (2, 5), and the partial PPII helix at (−5, −4), whereas the β-hairpin conformation basin, which should be at approximately (13, −20), was hardly found. We see that PC1 (horizontal axis) alone could distinguish the three free-energy minima. In fact, the contribution of PC1 was quite high at 83% and that of PC2 (vertical axis) was only 6%. PC1 was clearly correlated with the end-to-end distance (the correlation coefficient r = −0.944) and the radii of gyration (Rg) (r = −0.966), as was reported earlier (30). Thus, we show the 1D free-energy curve F(PC1) along PC1 in Fig. 1b as well as its energetic contribution E(PC1) and entropic contribution −TS(PC1) = E(PC1) − F(PC1). Note that we include the GB solvation free-energy term in the energetic contribution, although the GB energy includes entropic contribution from solvent. Although the PPII helix and the α-helix have almost the same free energies, the origins of their stabilities are different: the α-helix is stabilized by ≈17 kcal/mol (1 cal = 4.18 J) by the internal energy, leading to a funnel-like energy landscape centered around the α-helix (red in Fig. 1b). Conversely, the PPII helix is favored by nearly the same amount by the conformational entropy (green in Fig. 1b). The compensation between internal energy and entropy yields almost the same free energy at the α-helix and the PPII helix, which are separated by a small free-energy barrier of ≈3 kcal/mol. Similar results from MD simulation with explicit water were reported (33).
Fig. 1.
Free-energy landscape of Ala8 in isolation at 270 K calculated by using the GB solvation model. P-PPII, partially PPII helix. (a) 2D free-energy surface on the PC1 (PCA-1; horizontal) and PC2 (PCA-2; vertical) axes. The color chart on the right describes the free-energy value. (b) The (relative) free-energy curve F (shown in black), energetic contribution E (shown in red), and entropic contribution −TS (shown in green) are projected on PC1. (c) The energetic contribution in b was further decomposed into vdW energy (shown in red) and electrostatic energy (shown in green). Note that the latter includes the GB solvation energy. Protein structures were drawn by using PyMOL (DeLano Scientific, South San Francisco, CA).
We further decomposed the energy contribution into the van der Waals (vdW) energy and the whole electrostatic interaction including GB solvation energy: the results are plotted along PC1 in Fig. 1c. We see that the vdW interaction favored the α-helix (shown in red), whereas the electrostatic positive GB energy was lower in the PPII helix (shown in green). Amid the electrostatic contribution, the GB solvation free energy was particularly low at the PPII helix (data not shown), consistent with the reported observation that hydration plays a key role in the formation of the PPII helix.
Polyalanine Near an α-Helix Template.
Next, we simulated Ala8 near a spatially fixed template that was in the conformation of an α-helix made of Ala8. The distance between the centers of mass of the simulated and templated Ala8s was constrained to 8, 9, or 11 Å. With the α-helix template in which the distance separating centers of mass was constrained to 8 Å, marked enhancement of α-helix content was observed during comparison with the isolated Ala8 (Table 1). The enhancement was quickly reduced as the distance of separation increased: the effect was considerably weaker at a 9-Å separation and was not detectable at an 11-Å separation. Thus, the α-helix template effect seems to have quite a short range. Possibly, the discrete nature of water molecules may alter the distance dependence of the templating effect. Table 1 indicates that using the template with a 9-Å separation weakly induced β-sheet structure in the simulated Ala8. Visual inspection clarified, however, that although the induced β-sheet structure contained the single pair of hydrogen bonds characteristic to parallel β-sheets by the definition of the Definition of the Secondary Structure of Proteins (DSSP), the overall structure did not resemble β-sheet structure at all. When we used a more stringent definition of β-sheet, i.e., at least three consecutive hydrogen bonds, then the β-sheet content was <1%.
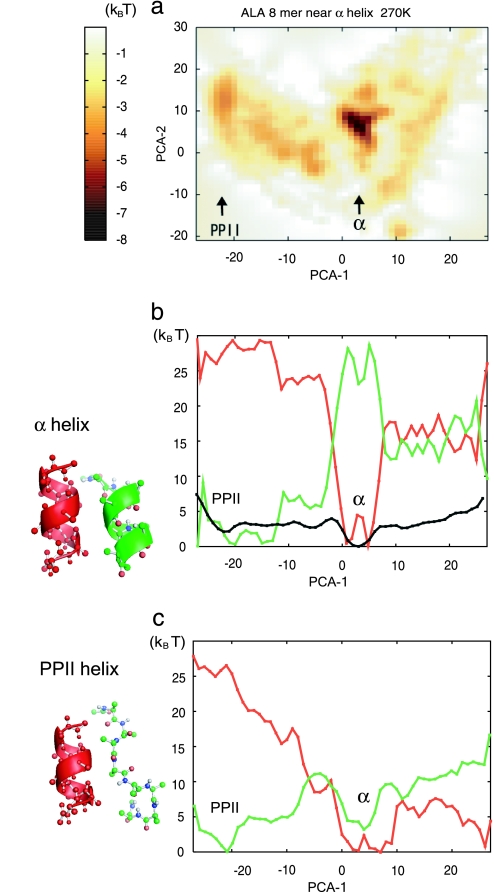
Fig. 2a depicts the free-energy landscape of the simulated Ala8 near the α-helix template with an 8-Å separation projected on PC1 and PC2. [Note that PC1 and PC2 used here are identical to those used for the isolated Ala8 (Fig. 1a), so we can directly compare the landscapes of the two systems.] Clearly, the landscape was significantly altered by the α-helix template. As it was in the isolated Ala8, PC1 here was a good coordinate by which the α-helix and the PPII helix were well separated. Thus, in Fig. 2b, we plotted the free-energy curve and its energetic and entropic components along PC1. The energy landscape was funneled around the α-helix structure, whereas entropic contribution was sharply anticorrelated with the energetic contribution as in the case of the isolated Ala8. However, in the case of the simulated Ala8, the energetic bias toward α-helix (≈25 kBT relative to PPII helix) was stronger than the entropic antibias; thus, the α-helix was ≈3 kBT more stable than the PPII helix in the free energy. To see why the energetic bias was strengthened by the α-helix template, we decomposed the energetic contribution into the electrostatic and the vdW interactions (Fig. 2c). In addition to the vdW energy (shown in red), the electrostatic contribution (shown in green) had the local minimum at the α-helix conformation, which was absent in the isolated Ala8 (see Fig. 1c). Thus, the template lowered the electrostatic energy of the simulated Ala8 at the α-helix conformation, which was mostly from the hydrogen bonds.
Fig. 2.
Free-energy landscape of Ala8 near the α-helix template with an 8-Å separation (shown in red) at 270 K calculated by the GB solvation model. Descriptions of a–c are identical to those given for Fig. 1.
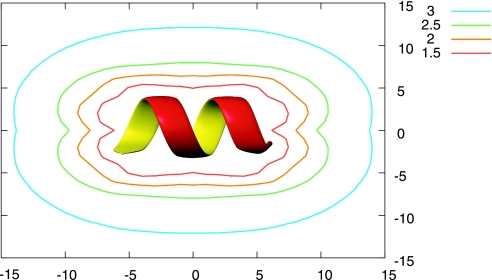
We then looked into the spatial dependence of the electrostatic environment around the α-helix template. Fig. 3 plots the dielectric constant field ε(x, y) around the α-helix template of Ala8. To estimate the dielectric constant within the GB formula, we introduced the second Ala8 and used its hydrogen bond between the carbonyl oxygen of Ala3 and the amide hydrogen of Ala7 as a probe. Therefore, we put an α-helix template at the origin and scanned the second Ala8 in the xy plane, keeping the helical axes of both helices parallel to the x axis. Clearly, the α-helix template lowered the dielectric constant around the template, strengthened the probe hydrogen bond of the α-helix structure, and thus resulted in the destabilization of the PPII helix. This corresponds to template-induced dehydration around the probe. We conclude that the α-helix template induced a local electrostatic environmental change that caused the stabilization of the second α-helix. This interpretation is parallel to many relevant observations and perspectives, which follow. Some fragments cleaved from the α-helical part of proteins do not form the α-helix (34). The α-helices and dimer formation of peptides with low α-helical propensity, such as GCN4-Glu9-Gly4 (35) require their collision and desolvation. The nucleation–condensation mechanism of protein folding suggests the partial ordering of long-range interactions in the transition state to stabilize the otherwise weak secondary structure (36, 37). The environmental effect of the α-helix template may resemble the solvent effect of alcohols. Alcohols enhance the α-helix (25, 38–41) and destabilize the PPII helix, which is correlated with the solvent polarity of alcohols (28). These results suggest that alcohols strengthen the electrostatic interactions, primarily hydrogen bonds, resulting in an increase in the α-helix (42) and a decrease in the PPII helix.
Fig. 3.
The contour map of the dielectric constant ε(x, y) on the xy plane. We calculated ε(x, y) using two Ala8 α-helices. A templated α-helix was put at the origin, and a second α-helix was scanned on the plane with both of the helical axes kept parallel to the x axis. The dielectric constant for the hydrogen bond between the carbonyl oxygen of Ala3 and the amide hydrogen of Ala7 in the second Ala8 helix was calculated by using the GB formula. PCA-1, PC1 axis; PCA-2, PC2 axis.
When the same simulation was performed for Ala7, the α-helix template did not induce the α-helix, probably because Ala7 is too short to form the α-helix even with the help of the template (Table 1). We also note that the PPII helix content was reduced with the α-helix template for both Ala8 and Ala7, suggesting template-induced desolvation.
Polyalanine Near a β-Strand Template.
We then simulated Ala8 near a template that was in the conformation of a β-strand made of Ala8, and the distance separating centers of mass was constrained to 8, 9, or 11 Å. The β-strand template remarkably enhanced the β-strand formation of the simulated Ala8 from 0.7% of the isolated peptide to 97% with the template with an 8-Å separation between centers of mass (Table 1). A similar enhancement was found for Ala7. Intuitively, this enhancement was caused by the β-strand template providing the template for making hydrogen bonds when the simulated Ala8 took on β-strand form. As the distance from the template increased, the β-strand content decreased slowly. Namely, with the template with the 11-Å separation, 78% of conformations contained the β-strand structure. Looking into these structures, we found that in these conformations the β-strand structure was formed at an edge of the chain by contacting the template, whereas the other end of the chain was far from the template: thus, the distance separating centers of mass was kept to the constrained values. Importantly, in these cases, the induction of β-strand structure occurred only in the neighborhood of the β-strand template. Thus, the templating effect of the β-strand template is also short in range. All β form antiparallel β-sheet with the template (data not shown), possibly because the β-strand template structure was taken from that in an ideal antiparallel β-sheet. Note that the peptide terminal was capped, so the terminal charge effect was not relevant.
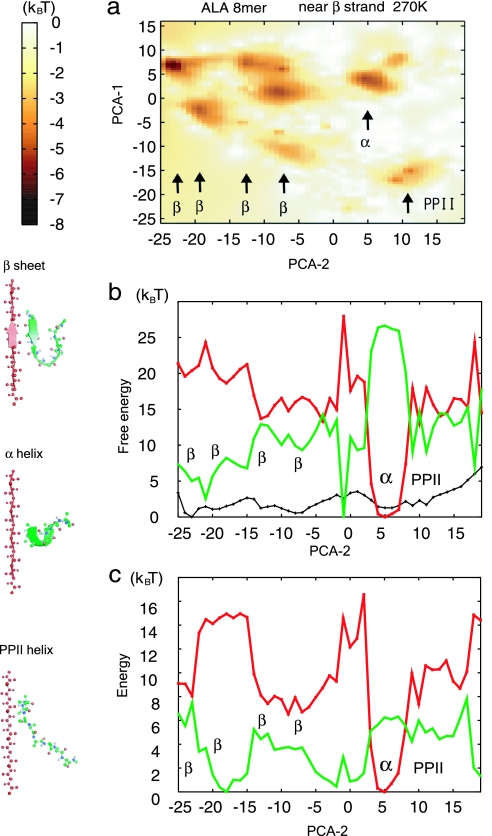
We then investigated the free-energy landscape of the Ala8 near the β-strand template with an 8-Å separation. Fig. 4a depicts the free energy on PC1 and PC2. We note that, although PC1 and PC2 used here were identical to those used before, the two axes were flipped for convenience, with PC1 on the vertical axis and PC2 on the horizontal axis. We found several free-energy minima on the left corresponding to various β-hairpin structures, whereas a single minimum for the α-helix and for the PPII helix was found on the right (Fig. 4a). We see that in this case PC2 is a better coordinate to separate the β-hairpin structure (PC2 between −25 and −5) from the α-helix (PC2 of ≈5) and the PPII helix (PC2 of ≈10), and thus we projected the free-energy surface as well as the energetic and entropic contributions on the PC2 in Fig. 4b. The free-energy curve along PC2 (shown in black) had the global minimum at −25 and a few more minima between −25 and −7, corresponding to β-hairpin structures. The α-helix and β-hairpin conformations were separated by the free-energy barrier of ≈4 kBT. Interestingly, even though the β-hairpin structure was dominant, the energy curve (shown in red in Fig. 4b) still shows a funnel-like shape centered at the α-helix, whereas entropic contribution favors the β-hairpin (shown in green in Fig. 4b). Compared with the isolated system, the system with the template had almost the same entropic contribution; thus, β-hairpin stabilization is attributed to the changes in interactions. We further analyzed the decomposed energies (Fig. 4c) and their intermolecular components and found that the dominant effect of the β-strand template was on the intermolecular electrostatics (hydrogen bonds) with some minor effects on vdW energy.
Fig. 4.
Free-energy landscape of Ala8 near the β-strand template with an 8-Å separation (shown in red) at 270 K was calculated by using the GB solvation model. (a) 2D free-energy surface on PC1 (PCA-1; vertical) and PC2 (PCA-2; horizontal) axes. Note that these axes are reversed from those in Figs. 1 and 2. (b) The (relative) free-energy curve F (shown in black), energetic contribution E (shown in red), and entropic contribution −TS (shown in green) are projected on PC2. (c) The energetic contribution in b was further decomposed into vdW energy (shown in red) and electrostatic energy (shown in green).
The β-strand template has a strong effect on enhancing the β tendency. The exposed β-sheet edge in proteins is prone to the aggregation and formation of amyloid fibrils. In fact, in natively β-sheet-rich proteins, the naked β-sheet edges are protected by several strategies, such as dimerization and covering by loops (43).
Discussion of Convergence of Sampling.
Although we used REMD techniques that significantly enhance conformational sampling, integration time per replica was 8 ns, which would be 2 orders of magnitude shorter than the physical time scale of relaxation of polyalanine dynamics. Here, we examine the convergence of the simulations based on the following criteria (22, 44). (i) All replica pairs have high and uniform exchange ratios. In the current REMD runs, the average acceptance ratio of replica exchange was sufficiently high (40–50%) and uniform. (ii) All replicas move around the whole temperature range. In our case, replicas traversed all of the temperatures, from 270 to 500 K, many times. (iii) No trapping in local minima on the energy landscape (see below).
As a more stringent test of convergence, we started all simulations with three completely different initial conformations, α-helix, extended, and PPII helix, observing discrepancy among them. In general, as simulation went on, the ensemble bias was monotonically, but slowly, reduced. Part of the statistical error in Table 1 may come from the residual bias. Because the templating effect was much stronger than the error bar, the result would not be affected too much by this bias.
During these investigations, we noticed that sampling for Ala8 with the β-strand template with an 8-Å separation was among the most difficult cases. The trajectories that started from the α-helix tended to be trapped on the α-helix for quite a long time. The number of replicas that contained the α-helix, initially 36, decreased nearly exponentially. The conformational transitions from α-helix to disordered and to β-strand preferentially occurred at relatively high temperatures, whereas relaxation was slower at lower temperatures. To test convergence, we extended the simulations up to 16 ns per replica. Results from the first 2–8 ns and those from the 10- to 16-ns samples are given in Table 1. We found that the α-helix content in the 2- to 8-ns samples was somewhat biased, an effect that lasted until ≈10 ns. However, the induction of β-strand structure had a stronger effect than the bias and thus was robust.
Materials and Methods
We performed the REMD simulations of Ala7 and Ala8 with an implicit solvent model in three different environments, the isolated polyalanine, the polyalanine near an α-helix template, and the polyalanine near a β-strand template. The peptide terminal charge was blocked, and thus Ala7 means Ac-Ala7-methyl amide (NMe) precisely. The template consisted of the same molecule as that simulated, the Alan template with n = 7 or 8. Template conformations were harmonically fixed either in ideal α-helix or antiparallel β-strand. In simulations with the templates, the center of mass of simulated Alan was harmonically constrained to a position separated by 8, 9, or 11 Å from the template's center of mass and to a position perpendicular to the helical or strand axis. We note that, in Protein Data Bank statistics, the peaks of distances between two helical axes are 9–11 Å (45).
The CHARMM param19 extended atom force field was used for polypeptide interactions (23). We used the GB implicit solvent model of Dominy and Brooks (24) to approximate hydration free energy. This energy has been used and validated especially for small peptides (46). We used the dielectric constant value of 80 for water and 1 for the interior of the protein at all temperatures simulated. We note that, although absolute secondary-structure contents inevitably depend on the force field used (26), the templating effect, i.e., the difference in conformational ensemble between the isolated and templated systems, would be more robust in nature. For comparison, we also performed REMD simulations of isolated Ala7 and Ala8, instead of the GB solvation model, using the uniform dielectric constants ε = 1, 2, 4, 10, and 80.
To enhance sampling, we used the REMD method developed by Sugita and Okamoto (22) for all simulations. We used 24 replicas for Ala7 simulations and 36 replicas for Ala8 simulations. In both cases, the temperatures of replicas were distributed exponentially in the range of T = 270–500 K, in which we found that the peptide can sample both ordered and disordered states. Exchange attempts were made after every 1 ps. After each exchange attempt, velocities were reassigned from the Maxwell–Boltzmann distribution (47). The replica temperatures were maintained by the Nosé–Hoover method (48), with the coupling constant (Q) = 10. The SHAKE algorithm (49) was applied to bonds connected to hydrogen atoms. We used a 1-fs time step for the stability of the simulations. Each REMD simulation was initiated by the following protocol: From initial configurations (see below), we made a 100-step steepest-descent minimization and a 500-step conjugate-gradient minimization, followed by a 50-ps MD simulation with the temperature increasing from 0 K to the temperature specified for each replica. Then, a 200-ps MD simulation was performed at a temperature specified for equilibrating velocities; the final structures of this simulation became the initial structures of the REMD simulation.
The REMD simulation time was 8 ns for each replica for all simulations but that of Ala8 with a β-template with an 8-Å separation, on which, to test convergence, we conducted longer simulations (16 ns per replica). For each case, the last 6 ns of data was used for analysis. Peptide configurations were saved every 1 ps, before the temperature exchange attempt. In all cases, we used three different initial configurations, extended, α-helical, and PPII-helical, for testing the sampling convergence. From each initial configuration, two independent REMD runs were carried out (i.e., total of six runs) for an isolated Ala8 and Ala8 with an α-helix template. One REMD run for each of the three initial configurations was performed for all other cases. For a single REMD run, the integration time summed over all replicas was 192 ns for Ala7 and 288–576 ns for Ala8, and the cumulative simulated time in the 72 REMD runs was ≈18 μs.
All simulations were carried out with the CHARMM simulation package (c28b1) (50). The REMD simulation was implemented as perl script for the CHARMM package.
We analyzed the α-helix and PPII helix contents of polypeptide configurations by the backbone dihedral angles (φ, ψ). Following the Lifson–Roig model (51), we stated that the ith residue is in an α-helix when dihedral angles i − 1, i, and i + 1 are within the range (φ, ψ) = (−57 ± 30°, −47 ± 30°). The same rule of assignment was used for the PPII helix with the range (φ, ψ) = (−78 ± 30°, 145 ± 30°). The β-strand contents were calculated by using the DSSP program (52). For confirmation, the α-helix content calculated when the DSSP was used was essentially the same as that calculated when the Lifson–Roig model was used (data not shown).
To analyze the free-energy landscape, we used PC analysis (30–32). First, we picked up 72,000 configurations (in total) from 2.0- to 8.0-ns snapshots of all replicas for simulations of (i) the isolated Ala8 (mostly α-helix and PPII helix), (ii) Ala8 near the β-strand template (β-strand rich), (iii) the isolated Ala8 in the uniform dielectric constant ε = 1 (β-hairpin rich), and (iv) the isolated Ala8 in the uniform dielectric constant ε = 4 (α-helix rich). These conformations were superimposed on a reference conformation, an α-helix conformation. Then, the variance–covariance matrix of the ensemble was “diagonalized” to get eigenvectors (PC axes) and eigenvalues [the standard deviations of the conformational distribution along the ith PC axis (λi)]. λi/Σiλi is regarded as the relative contribution of the distribution along the ith PC.
Acknowledgments
We thank Jeffrey Saven and Takatsugu Hirokawa for helpful comments. This work was supported in part by the Water and Biomolecules Program of the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Abbreviations
- PPII
polyproline II
- MD
molecular dynamics
- REMD
replica-exchange MD
- GB
generalized Born
- PC
principal component
- vdW
van der Waals.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Cho SS, Levy Y, Cheung MS, Levine H, Wolynes PG, Onuchic JN. Proc Natl Acad Sci USA. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Minton AP. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 5.Spek EJ, Olson CA, Shi ZS, Kallenbach NR. J Am Chem Soc. 1999;121:5571–5572. [Google Scholar]
- 6.Marqusee S, Robbins VH, Baldwin RL. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholtz JM, York EJ, Stewart JM, Baldwin RL. J Am Chem Soc. 1991;113:5102–5104. [Google Scholar]
- 8.Chou PY, Fasman GD. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 9.Gasset M, Baldwin MA, Lloyd DH, Gabriel JM, Holtzman DM, Cohen F, Fletterick R, Prusiner SB. Proc Natl Acad Sci USA. 1992;89:10940–10944. doi: 10.1073/pnas.89.22.10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forood B, Perez-Paya E, Houghten RA, Blondelle SE. Biochem Biophys Res Commun. 1995;211:7–13. doi: 10.1006/bbrc.1995.1770. [DOI] [PubMed] [Google Scholar]
- 11.Ingwall RT, Scheraga HA, Lotan N, Berger A, Katchalski E. Biopolymers. 1968;6:331–368. doi: 10.1002/bip.1968.360060308. [DOI] [PubMed] [Google Scholar]
- 12.Shi Z, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Proc Natl Acad Sci USA. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han WG, Jalkanen KJ, Elstner M, Suhai S. J Phys Chem B. 1998;102:2587–2602. [Google Scholar]
- 14.Eker F, Cao XL, Nafie L, Huang Q, Schweitzer-Stenner R. J Phys Chem B. 2003;107:358–365. [Google Scholar]
- 15.Poon CD, Samulski ET, Weise CF, Weisshaar JC. J Am Chem Soc. 2000;122:5642–5643. [Google Scholar]
- 16.Woutersen S, Hamm P. J Chem Phys. 2001;114:2727–2737. [Google Scholar]
- 17.Garcia AE. Polymer. 2004;45:669–676. [Google Scholar]
- 18.Kentsis A, Mezei M, Gindin T, Osman R. Proteins. 2004;55:493–501. doi: 10.1002/prot.20051. [DOI] [PubMed] [Google Scholar]
- 19.Zagrovic B, Lipfert J, Sorin EJ, Millett IS, van Gunsteren WF, Doniach S, Pande VS. Proc Natl Acad Sci USA. 2005;102:11698–11703. doi: 10.1073/pnas.0409693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pappu RV, Rose GD. Protein Sci. 2002;11:2437–2455. doi: 10.1110/ps.0217402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Chen K, Santini PA, Shi ZS, Kallenbach NR. J Am Chem Soc. 2003;125:8092–8093. doi: 10.1021/ja035551e. [DOI] [PubMed] [Google Scholar]
- 22.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 23.Neria E, Fischer S, Karplus M. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 24.Dominy BN, Brooks CL. J Phys Chem B. 1999;103:3765–3773. [Google Scholar]
- 25.Weise CF, Weisshaar JC. J Phys Chem B. 2003;107:3265–3277. [Google Scholar]
- 26.Hu H, Elstner M, Hermans J. Proteins. 2003;50:451–463. doi: 10.1002/prot.10279. [DOI] [PubMed] [Google Scholar]
- 27.Sreerama N, Woody RW. Proteins. 1999;36:400–406. [PubMed] [Google Scholar]
- 28.Liu ZG, Chen K, Ng A, Shi ZS, Woody RW, Kallenbach NR. J Am Chem Soc. 2004;126:15141–15150. doi: 10.1021/ja047594g. [DOI] [PubMed] [Google Scholar]
- 29.Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA. Proc Natl Acad Sci USA. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda K, Galzitskaya OV, Nakamura H, Higo J. J Comput Chem. 2003;24:310–318. doi: 10.1002/jcc.10160. [DOI] [PubMed] [Google Scholar]
- 31.Kitao A, Hayward S, Go N. Proteins. 1998;33:496–517. doi: 10.1002/(sici)1097-0134(19981201)33:4<496::aid-prot4>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 32.Garcia AE. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 33.Mu YG, Stock G. J Phys Chem B. 2002;106:5294–5301. [Google Scholar]
- 34.Bai YW, Karimi A, Dyson HJ, Wright PE. Protein Sci. 1997;6:1449–1457. doi: 10.1002/pro.5560060709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisner WK, Sosnick TR. Proc Natl Acad Sci USA. 2004;101:13478–13482. doi: 10.1073/pnas.0404057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daggett V, Fersht AR. Trends Biochem Sci. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Uzawa T, Ishimori K, Morishima I, Takahashi S, Konno T, Akiyama S, Fujisawa T. Proc Natl Acad Sci USA. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiraki K, Nishikawa K, Goto Y. J Mol Biol. 1995;245:180–194. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- 39.Uversky VN, Narizhneva NV, Kirschstein SO, Winter S, Lober G. Folding Des. 1997;2:163–172. doi: 10.1016/s1359-0278(97)00023-0. [DOI] [PubMed] [Google Scholar]
- 40.Luo PZ, Baldwin RL. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 41.Hong DP, Hoshino M, Kuboi R, Goto Y. J Am Chem Soc. 1999;121:8427–8433. [Google Scholar]
- 42.Vila JA, Ripoll DR, Scheraga HA. Proc Natl Acad Sci USA. 2000;97:13075–13079. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson JS, Richardson DC. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hukushima K, Nemoto K. J Phys Soc Jpn. 1996;65:1604–1608. [Google Scholar]
- 45.Chothia C, Levitt M, Richardson D. J Mol Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 46.Bursulaya BD, Brooks CL. J Phys Chem B. 2000;104:12378–12383. [Google Scholar]
- 47.Pitera JW, Swope W. Proc Natl Acad Sci USA. 2003;100:7587–7592. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoover WG. Phys Rev A At Mol Opt Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 49.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Chem. 1977;23:327–341. [Google Scholar]
- 50.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 51.Lifson S, Roig A. J Chem Phys. 1961;34:1963–1974. [Google Scholar]
- 52.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]